Abstract
Elucidating the factors influencing genetic differentiation is an important task in biology, and the relative contribution from natural selection and genetic drift has long been debated. In this study, we used a regression-based approach to simultaneously estimate the quantitative contributions of environmental adaptation and isolation by distance on genetic variation in Boechera stricta, a wild relative of Arabidopsis. Patterns of discrete and continuous genetic differentiation coexist within this species. For the discrete differentiation between two major genetic groups, environment has larger contribution than geography, and we also identified a significant environment-by-geography interaction effect. Elsewhere in the species range, we found a latitudinal cline of genetic variation reflecting only isolation by distance. To further confirm the effect of environmental selection on genetic divergence, we identified the specific environmental variables predicting local genotypes in allopatric and sympatric regions. Water availability was identified as the possible cause of differential local adaptation in both geographic regions, confirming the role of environmental adaptation in driving and maintaining genetic differentiation between the two major genetic groups. In addition, the environment-by-geography interaction is further confirmed by the finding that water availability is represented by different environmental factors in the allopatric and sympatric regions. In conclusion, this study found that geographical and environmental factors together created stronger and more discrete genetic differentiation than isolation by distance alone, which only produced a gradual, clinal pattern of genetic variation. These findings emphasize the importance of environmental selection in shaping patterns of species-wide genetic variation in the natural environment.
Keywords: Genetic variation, Environment, Geography, Niche modeling, Environmental adaptation
Introduction
Elucidating the processes underlying the origin and maintenance of genetic variation in natural populations is a fundamental task in biology. The detailed characterization of genetic variation may reveal the demographic history and population structure of a species (Bryc et al. 2010; Novembre et al. 2008; Novembre & Stephens 2008; Platt et al. 2010; Sharbel et al. 2000; van Heerwaarden et al. 2011). This information also enables further analyses, such as association mapping for complex traits (Atwell et al. 2010; Huang et al. 2010; Yu et al. 2006) and the identification of genes that co-vary with specific environmental factors (Coop et al. 2010; Eckert et al. 2010; Hancock et al. 2010; Manel et al. 2010), both aiming at understanding the genetic basis of local adaptation and the mechanisms underlying evolutionary changes. However, despite the fundamental importance of studying natural genetic variation and the availability of diverse methods of describing patterns of genetic variation, (Engelhardt & Stephens 2010; Gao et al. 2007; Jombart et al. 2009; Pritchard et al. 2000), still few studies have tried to investigate the relative contributions of factors affecting genetic differentiation across a species range.
It is widely acknowledged that genetic differentiation is strongly influenced by two processes: isolation by distance and differential local adaptation (Nosil et al. 2008; Nosil et al. 2005; Slatkin 1987; Wright 1931; Wright 1943). Under isolation by distance, the major factor limiting interbreeding is the physical distance, and populations diverge via genetic drift or clinal selective factors correlated with geographical distance. Because neighboring populations often have only minor differences in local environments (for example, day-length across latitude) and therefore minor reductions of immigrant or hybrid fitness, substantial gene flow could occur among adjacent populations. As a consequence, the amount of gene flow is mainly restricted by geographical distance, and genome-wide divergence, as revealed by neutral genetic markers, is expected to be clinally correlated with geographical distance. In contrast, when migration occurs between nearby populations adapted to distinct environments, fitness of immigrants or hybrids may be reduced by natural selection (Nosil et al. 2005), and the resulting reduction of genetic exchange may facilitate or maintain genetic divergence (Thibert-Plante & Hendry 2010). Under this process, an abrupt change in local environment (for example, elevation change over a few kilometers) may cause substantial reduction of immigrant fitness, resulting in discrete, rather than continuous pattern of genetic differentiation. Therefore, the degree of genetic differentiation inferred from neutral loci is expected to correlate more with differences in local environment than with geographical distance. Although examples, theories, and reviews exist for the two processes (Engelhardt & Stephens 2010; Nosil et al. 2008; Nosil et al. 2005; Orr & Smith 1998; Rundle & Nosil 2005; Schluter 2001; Schluter & Conte 2009; Templeton 2008; Thibert-Plante & Hendry 2010; Wang & Summers 2010), few studies have jointly considered the relative importance of isolation by distance and local adaptation on genetic variation at a species-wide scale (but see Cushman et al. 2006; Freedman et al. 2010; Pease et al. 2009). By combining population structure estimation and niche modeling, here we statistically separate and quantify the effects of isolation by distance and local adaptation on genetic divergence patterns in the wild mustard species Boechera stricta.
For divergent selection to facilitate or maintain population differentiation, the environmental differences between lineages should be higher than within species or populations (Coyne & Orr 2004). Therefore, niche modeling has been used to identify possible environmental factors contributing to population differentiation (Hübner et al. 2009; Kozak et al. 2008; Kozak & Wiens 2006; Nakazato et al. 2008). However, many environmental factors are highly correlated with each other and with geographical distance. To avoid spurious correlations, it is necessary to control for neutral processes when estimating the relationship between environment and genetic structure (Dyer et al. 2010; McCormack et al. 2010). Using geographical distance as a covariate, we investigate the contribution of environmental factors to independent axes of genetic differentiation in Boechera stricta. With isolation by distance as the null model (Novembre et al. 2008; Novembre & Stephens 2008; Platt et al. 2010; Sharbel et al. 2000), we attribute an axis of genetic differentiation to isolation by distance when only geographical distance has significant effect on this axis, or when we are unable to separate the effects of geography and environment due to their strong correlation. On the other hand, after controlling for geography, significant effects of environmental factors are expected when local adaptation drives or maintains genetic divergence.
Previous research has identified three major genetic groups within Boechera stricta (Song et al. 2009). A contact zone between the two most diverged groups (East and West) is found in the Rocky Mountains in Idaho, USA. During the last glacial maximum, this contact zone was mostly unsuitable habitat for this species or was covered by montane glaciers (Brunelle & Whitlock 2003; Hostetler & Clark 1997), suggesting that the current overlap is a zone of secondary contact after historical allopatry. Despite the existence of this contact zone, less than 3% of sampled genotypes were admixed (Song et al. 2009); nevertheless, fertile and healthy hybrids can be produced in the laboratory. Both observations suggest the existence of an extrinsic reproductive isolating mechanism other than isolation by distance or intrinsic hybrid inviability. If natural selection imposed by environmental factors contributes to divergence and prevents current hybridization between the two genetic groups, we may be able to identify environmental factors as significant predictors of genotypic differentiation in both allopatric and sympatric regions. Additionally, the significant predictors should reflect the same underlying causal factors in both regions. In contrast, if reproductive isolation is caused by factors not related with environmental selection, while several environmental factors may be identified in the allopatric regions due to correlations among geography, genetic structure, and environments, no relationship between environmental factors and genetic divergence should exist in the contact zone.
In this study, we address the following questions: (i) What is the relative contribution of isolation by distance and environmental adaptation on independent genetic axes showing distinct patterns of differentiation? (ii) When environmental adaptation is inferred, can we further confirm this by identifying the same causal environmental variable in both allopatric and sympatric regions?
Material and Methods
Study species
Boechera stricta (Brassicaceae) is a wild perennial mustard species and a close relative of Arabidopsis thaliana (Mitchell-Olds 2001; Oyama et al. 2008). This species is native to western North America, occupying wide geographical, altitudinal, and environmental ranges (Song et al. 2006). Although polyploidy or apomixis occur in this genus (Schranz et al. 2005), B. stricta genotypes are predominantly diploid and sexual, with approximately 95% selfing rate (Song et al. 2006). With 46 genotypes, previous research has identified three genetic groups within this species (Song et al. 2009). To obtain more detailed information on genetic variation across the distribution range and to examine the multi-dimensional niche space of these genetic groups, we used 239 genotypes sampled from relatively un-disturbed environments in western North America.
Genotyping
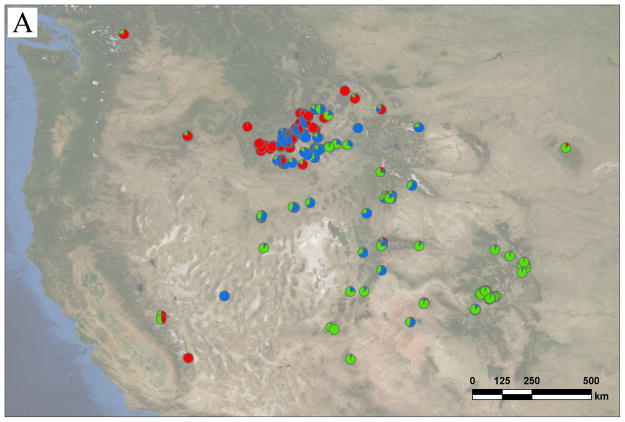
Seeds of Boechera stricta were collected from about 250 locations across western North America and grown in the Duke Greenhouse. One individual was randomly chosen as representative of each collection site, a sampling scheme also used in previous studies (Manel et al. 2003; Platt et al. 2010). Because genetic variation within local populations is low (Song et al. 2006), this sampling scheme maximizes genetic diversity for a given sample size. Trichome morphology was examined for species confirmation (Rollins 1993), and the ploidy was estimated by flow cytometry (Partec, Munster, Germany) or the number of alleles in microsatellite loci, leaving 239 diploid individuals, each from different locations (Figure 1A). Seventeen microsatellite markers used in a previous study (Table S1, Song et al. 2006) were genotyped, and the PCR primers were modified for fluorescently-labeled M13-tailing (Boutin-Ganache et al. 2001). PCR products were processed with Applied Biosystems 3730, and alleles were called with GeneMarker (SoftGenetics, State College, PA, USA).
Figure 1. Collection sites and STRUCTURE results for Boechera stricta.
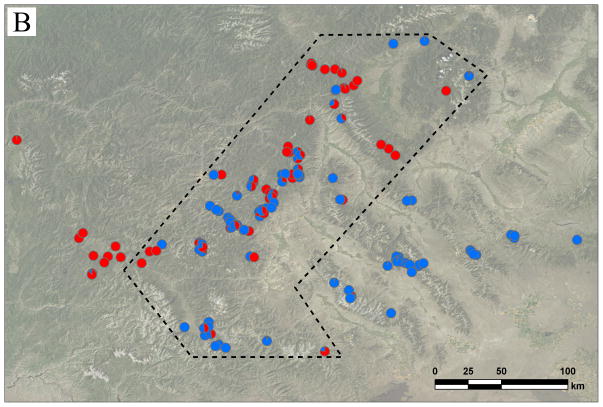
Each pie chart represents one individual randomly chosen from one location. Different colors in each pie chart represent STRUCTURE posterior probabilities that the individual belongs to each genetic group. A) The distribution of three genetic groups across western North America. Red = West; blue = North; green = South. Notice the narrow contact zone between West and East (comprised of North + South), and the clinal distribution between North and South genetic groups. B) The distribution of West and East genetic groups around the contact zone. Red = West; blue = East. Region encompassed by the dashed line is regarded as ‘sympatric zone’.
Genetic analysis
Two major methods have been employed to identify population structure (Engelhardt & Stephens 2010). Admixture-based models, such as STRUCTURE (Pritchard et al. 2000), estimate the proportion of each sample’s genome derived from an ancestral genetic group. The other method, principal component analysis (PCA), uses multivariate statistics to depict the genetic structure and is free from many population genetics assumptions underlying STRUCTURE (Gao et al. 2007; Jombart et al. 2009). Although the two methods differ in model assumptions and methodologies, a recent study (Engelhardt & Stephens 2010) showed that both approaches are special cases of matrix factorization with different constraints, and while admixture-based models are more suitable for discrete and partially admixed populations (such as secondary contact after historical allopatry), PCA is more useful with continuous patterns of differentiation (such as isolation by distance). Here, we employed advantages of both methods to investigate population structure within Boechera stricta. We have not employed methods that incorporate geographic information while assigning genetic structure (for example, Guillot et al. 2005) because our goal is to investigate the population structure based on genetic information per se, with the contributions from geography and environment to be estimated subsequently.
With STRUCTURE, three replicates were run for each k value (k = 2 and 3), following previous results (Song et al. 2006). We tried other k values (k from 4 to 10) but do not explicitly report the results here because we focused on the major genetic differentiation pattern in this study and other k values did not produce clear patterns (data not shown). Within each run, a total of two million iterations were conducted with the first one million as burn-in. In our definition, a genotype was regarded as belonging to a pure group if the Bayesian posterior probability was higher than 0.8. In addition, principal coordinate analysis (PCOA) was conducted with GenAlEx (Peakall & Smouse 2006). GenAlEx first calculated a pairwise genetic distance matrix based on the allele states. The PCOA axes and scores were then obtained by performing multidimensional scaling on this matrix. In theory, PCOA is equivalent to principal component analysis (PCA) if the initial distance matrix is calculated as Euclidean distance. Therefore, the PCOA result generated by GenAlEx can be viewed as the PCA of allele states within Boechera stricta.
We used customized Perl scripts to compare the range of FST values between genetic groups identified by STRUCTURE. Instead of bootstrapping among loci (Goudet 2001), our script performed bootstrap resampling of individuals within each genetic group. This approach gave us the advantage of retaining information from all loci while accounting for the spatial and temporal unevenness in field seed collection. One thousand bootstrapped data sets were generated by randomly resampling individuals from each group. Each data set was transformed into the input data format of FSTAT (Goudet 2001), and FST was calculated as the proportion of between-group to total genetic variation by package HIERFSTAT (Goudet 2005) in R (http://www.r-project.org/).
Environmental variables
Environmental variables with a resolution of 1 km2 were downloaded from publicly available databases. Elevation and nineteen biologically-relevant climatic variables (Bioclim variables) were downloaded from World Clim (Hijmans et al. 2005), and five topographical variables (aspect, slope, flow direction, flow accumulations, and compound topographical index) were downloaded from the HYDRO1k database of U.S. Geological Survey (USGS). Based on latitude and longitude, data layers were overlaid in ArcGIS 9 (ESRI, Redlands, CA, USA), and environmental factors from Boechera stricta collection sites were extracted with Hawth’s Tools (http://www.spatialecology.com/htools/tooldesc.php). In addition, we manually measured ‘distance to the nearest stream’ with the resolution of one meter in Google Earth. Some environmental factors were excluded due to high correlation (r > 0.9 in some pairs of variables), finally leaving six climatic and four topographical variables (Table S2). The six climatic variables were chosen as the representatives of four major clusters in the hierarchical clustering analysis of climatic variables (data not shown), and these variables represent the mean and variation of temperature and precipitation and their interaction effect. All environmental variables were log-transformed and standardized prior to statistical analyses due to their skewed distribution. Latitude and longitude were also transformed in the following regression-based but not distance-matrix-based analyses.
Niche modeling
The genetic analyses identified three major genetic groups, forming two contrasting patterns of genetic differentiation within B. stricta - the discrete East-West and the continuous North-South divergence. To dissect the effect of natural selection (environment, isolation by adaptation) and genetic drift (geography, isolation by distance) on the two distinct patterns of genetic differentiation, we first performed Mantel tests to assess the correlations among genetic, environmental, and geographic distance matrices. Pairwise genetic distance among genotypes was calculated by GenAlEx (described above), and the environmental distance matrix was obtained by calculating the Euclidean distance between pairs of collection sites from the ten environmental variables. The great-circle geographic distance, the nearest distance between two points on the Earth surface, was obtained by package ‘fields’ (http://CRAN.R-project.org/package=fields) of R using un-transformed latitude and longitude values. We did not employ more complex geographical distance measurements, such as least-cost path (Storfer et al. 2007), because the dispersal distance of B. stricta is only a few meters (Mitchell-Olds, personal observation), a much smaller scale than the resolution of the environmental data layers used in this study. To account for the correlation among these three distance matrices, partial Mantel tests were further conducted to estimate the contribution of environmental distance to genetic distance while accounting for the effect of geographic distance. Both Mantel and partial Mantel tests were performed with package ‘vegan’ (http://CRAN.R-project.org/package=vegan) of R, and significance was determined by 1000 permutations.
However, while partial Mantel tests can examine the significance of correlations among matrices, recent reports (Balkenhol et al. 2009; Legendre & Fortin 2010) show that such distance-based methods have less statistical power and do not correctly estimate the amount of total variation explained by predictor variables. To quantitatively estimate the relative influence of genetic drift and environmental adaptation on genetic differentiation, we combined the genetic principal component analysis (PCA) and geographical and environmental discriminant function analysis (DFA) into a multiple regression framework:
where GEN, GEO, and ENV are the genetic, geographic, and environmental ‘scores’ of each genotype. Each genotype has its unique positions in the multivariate genetic, geographic, and environmental spaces, and the corresponding scores are projections on axes that best distinguish genetic groups in each multivariate space. Notice that we employed DFA rather than PCA for geographical and environmental factors because PCA axes only capture most variation among all samples, but not necessarily the geographical or environmental differences between genetic groups. These scores provide a metric to quantify how geographical and environmental factors predict genetic variation between Boechera genetic groups. Thus, the GEN score is simply the projection on the genetic PCA axes. For GEO and ENV, discriminant function analyses (DFA) were first performed between the inferred genetic groups being compared, and the geographic and environmental score of every individual (including hybrids) was calculated from the coefficients of each variable identified by DFA. DFA was performed with the ‘MASS’ package (http://CRAN.R-project.org/package=MASS) in R, and multiple regression was performed with JMP 8 (SAS, Cary, NC). Proportion of genetic variation explained by GEO or ENV, after accounting for the effect of each other, was calculated from Type III sum of squares. The entire analysis was conducted separately for the East-West and North-South comparisons. We chose genetic PCA values rather than STRUCTURE posterior probabilities as responses because PCA axes are independent by definition. This allowed us to model the contribution from environment and geography to one genetic differentiation pattern (e.g., East-West, PC1) with minimal interference from the other pattern (e.g., North-South, PC2). In contrast, the posterior probabilities given by STRUCTURE are constrained so that all values sum to 1. Nevertheless, using STRUCTURE posterior probability as response variable yields qualitatively similar results (data not shown).
To further identify whether the two categories of environmental factors (climatic and topographical, Table S2) have different contributions to the spatial distribution of ‘pure genotypes’ in sympatric and allopatric regions, a similar regression analysis was performed by separating the ENV factor into CLIM (six climatic variables) and TOPO (four topographical variables):
In these regression analyses, we were able to quantitatively estimate the contribution of each predictor variable to the genetic structure of B. stricta by using the genetic PCA scores as response variables. However, PCA scores reflect the genetic variation both within and between genetic groups. Therefore, we used multiple logistic regression to identify specific environmental variables contributing mainly to the between-group differentiation, with ‘pure genetic group’ (a binary categorical variable) as response and twelve factors (latitude, longitude, and ten environmental factors) as predictor variables. Because putting all predictors in a full model simultaneously would cause over-fitting of the model, we first used automatic forward selection of predictors in JMP 8 and then manually removed non-significant variables. We set the alpha value for each iteration of the forward selection process as 0.01, a somewhat stringent significance criterion, to prevent type I error generated during multiple steps of model comparison and to limit the number of predictor variables in the final model.
In analyses involving the comparison between East and West genetic groups in the sympatric or allopatric regions, three collections from central Montana (MacDonald Pass Trailhead, Elkhorn, and Brackett Creek) were removed because, due to limited sampling, we were not certain about the existence of a contact zone there.
Results
Genetic structure in Boechera stricta
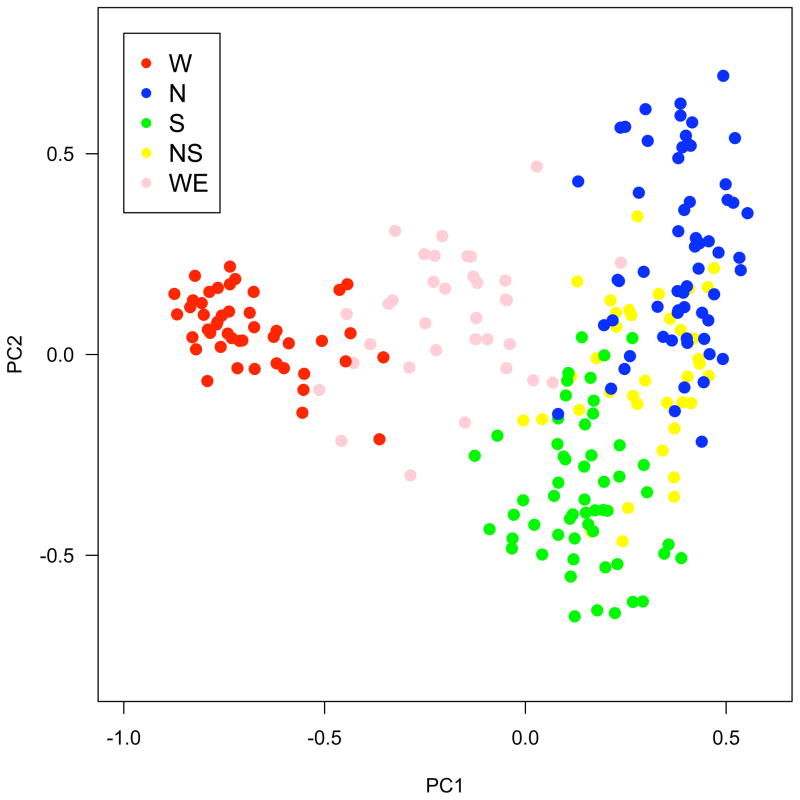
Our larger sample confirms previous results (Song et al. 2009), in that STRUCTURE identified three major groups (North, South, and West) when k = 3 (Figure 1A). When setting k = 2, North and South merged into one group while West remained distinct. This result was consistent with PCA (Figure 2). While the PC axis explaining the largest fraction (40.43%) of genetic variation distinguished West versus the two other groups, the axis accounting for 17.23% of the variation separated North from South groups. Both results were consistent with previous findings that West was most diverged from the two other genetic groups. Therefore, North and South lineages will be referenced collectively as the ‘East’ genetic group at some points in the following discussion. This pattern was also supported by the FST distribution from bootstrap resampling of ‘pure genotypes’ (mean FST between East and West = 0.30, with 95% CI from 0.28 to 0.32; North and South = 0.18, with 95% CI from 0.16 to 0.21).
Figure 2. Genetic principal component analysis (PCA) of 239 Boechera stricta accessions.
PC1 explains 40.4% and PC2 explains 17.2% of total genetic variation. Accessions were colored based on STRUCTURE results with k = 3, and a genotype belongs to a ‘pure genetic group’ (W = West, N = North, S = South) only when the corresponding posterior probability is higher than 0.8. ‘NS’ and ‘WE’ denote North-South hybrids and West-East (East = North + South) hybrids, respectively. Notice the distinct distribution patterns between West-East along PC1 (discrete) and North-South along PC2 (continuous).
Noticeably, North and South groups are distributed continuously along the second principal component axis (PC2, Figure 2). In contrast, although most of the West genotypes were sampled in the Idaho contact zone, they were genetically distinct from the North group in PC1, suggesting mechanisms other than geographic isolation may contribute to their genetic differentiation. Therefore, our niche modeling focused on two distinct comparisons: a species-wide comparison of East vs. West, and a North vs. South comparison within the more continuously distributed East group.
Contribution of environment versus geography to population structure
Mantel tests showed that for both East-West and North-South divergence, all three distance matrices (genetic, environmental, and geographic) were highly correlated (all P ≤ 0.002). In partial Mantel tests, environmental distance remained a significant predictor of genetic distance after accounting for geographic distance only in the East-West (P = 0.001) but not in the North-South comparison (P = 0.185).
The genetic PC1 values (from all samples) correspond to the genetic scores of East-West divergence. Within the East group, PC2 scores correspond to North-South divergence (Figure 2). In both cases, quantitative results from multiple regression revealed similar pattern as the partial Mantel tests (Table 1). While the full models explained comparable amounts of total genetic variation in both contrasts between groups (42.77% for East-West and 50.84% for North-South), environmental factors gave significant prediction only for East vs. West divergence (21.60%, P < 0.001) but not between North and South (0.87%, P = 0.107), while controlling for geographic effect. In the North-South comparison (Table 1), any predictor only explained a small portion of genetic variation after accounting for the contribution of other predictors. This reflects the strong correlation between geography and environment in the North-South comparison (Pearson’s correlation coefficient r = 0.95, P < 0.001). In contrast, this correlation was less pronounced (r = 0.66, P < 0.001) in the East-West divergence pattern.
Table 1.
Proportion of genetic variation explained by environmental (ENV) and geographical (GEO) effects or their interaction in the East-West (species-wide, genetic PC1) and the North-South (within-East, genetic PC2) genetic divergence patterns.
| Predictors | East-West Proportion explained (%) | P value | North-South Proportion explained (%) | P value |
|---|---|---|---|---|
| Full model | 42.77 | < 0.001 | 50.84 | <0.001 |
| -ENV | 21.60 | < 0.001 | 0.87 | 0.107 |
| -GEO | 0.06 | 0.608 | 1.15 | 0.065 |
| -ENV*GEO | 4.80 | < 0.001 | 0.74 | 0.139 |
| Error | 57.23 | 49.16 |
These results suggest that isolation by distance played a fundamental role in the divergence between North and South genetic groups. On the other hand, when controlling for geographical factors, the importance of environmental selection was highly significant in East-West divergence. Next, we focused on identifying the specific environmental factors contributing to the ecological differentiation between East and West lineages.
Identifying sources of environmental selection
By separating ten environmental variables into six climatic and four topographical variables (Table S2), similar regression analyses identified the relative contribution of the two categories of environmental variables to the genetic divergence between East and West genetic groups in sympatric and allopatric regions (Figure 1B, Table 2). In the allopatric region, climatic factors explained 8.17% (P < 0.001) of total genetic variation, about three times the contribution of topographical factors (2.66%, P = 0.001). These results were reversed in the sympatric region, where topographical factors predicted 5.68% (P = 0.002) of East-West genetic divergence, but climatic factors alone had little effect (0.67 %, P = 0.278).
Table 2.
Proportion of East-West (genetic PC1) genetic variation explained by climatic (CLIM), topographical (TOPO), geographical (GEO), or the interaction effects in the allopatric or sympatric regions.
| Predictors | Allopatric Proportion explained (%) | P value | Sympatric Proportion explained (%) | P value |
|---|---|---|---|---|
| Full model | 71.69 | <0.001 | 41.39 | <0.001 |
| -CLIM | 8.17 | <0.001 | 0.67 | 0.278 |
| -TOPO | 2.66 | 0.001 | 5.68 | 0.002 |
| -GEO | 3.18 | <0.001 | 3.32 | 0.017 |
| -CLIM*TOPO | 5.63 | <0.001 | 0.44 | 0.381 |
| -CLIM*GEO | 0.35 | 0.231 | <0.01 | 0.995 |
| -TOPO*GEO | 0.15 | 0.430 | 0.01 | 0.878 |
| -CLIM*TOPO*GEO | 3.25 | <0.001 | 1.27 | 0.136 |
| Error | 28.31 | 58.61 |
Logistic regression confirmed the importance of climate in allopatry and topography in sympatry for the genetic divergence between East and West lineages (Table 3). In the allopatric region, while most environmental variables differed significantly between East and West genotypes in simple logistic regression (data not shown), only ‘winter precipitation’ (a climatic variable, P < 0.001) and longitude (P < 0.001) were significant in multiple logistic regression. For sympatric genotypes, ‘distance to the nearest stream’ (a topographical variable, P < 0.001) and latitude (P < 0.001) were significant in multiple logistic regression. Noticeably, this pattern was also reflected by the significant interaction effect between environment and geography in the previous multiple regression (Table 1).
Table 3.
P values based on likelihood ratio tests in multiple logistic regressions on East-West genotypes (a binary response variable) in the allopatric or sympatric regions.
| Predictors1 | Allopatric | Sympatric |
|---|---|---|
| Winter precipitation | <0.001 | |
| Distance to the nearest stream | <0.001 | |
| Latitude | <0.001 | |
| Longitude | <0.001 |
Only significant predictors in multiple logistic regression are reported. Refer to Table S2 for a full list of all variables used.
Discussion
Recent years have witnessed the rise of landscape genetics, a research area combining molecular population genetics and landscape ecology (Manel et al. 2003; Storfer et al. 2007; Storfer et al. 2010). As summarized by Storfer et al. (2007), the study of landscape genetics includes several major research categories, using a broad range of approaches to examine geographical patterns of genetic variation. Nevertheless, most studies focus on the effects of geographical and environmental factors on current gene flow among local populations. Phylogeography, on the other hand, differs from landscape genetics in the broader spatial and longer temporal scale considered (Manel et al. 2003; Storfer et al. 2007). However, despite its larger spatio-temporal scale, phylogeographic analyses to date have concentrated primarily on the effect of historical neutral processes on the pattern of genetic variation, and the role of environmental adaptation is not often considered (Hickerson et al. 2010). Here we combine the consideration of environmental factors from landscape genetics and the broad spatio-temporal scale of phylogeography in order to separate the effects of neutral processes and environmental adaptation on the species-wide pattern of genetic variation. We regard the pattern of genetic variation within Boechera stricta as created via the long-term accumulation of reproductive isolation among the three major genetic groups, rather than the result of recent gene flow between local populations. Hence, this research has larger spatio-temporal scale than most landscape genetics studies. While most studies investigating within-species genetic variation are mainly exploratory rather than hypothesis driven (Storfer et al. 2010), our approach specifically tests whether different patterns of genetic differentiation (distinct or continuous) are driven by heterogeneous contributions from geography and environment.
In this study, we investigated the population structure of Boechera stricta and then performed sequential tests to examine the role of environmental factors in shaping the pattern of species-wide genetic variation. First, we investigated the relative contributions of isolation by distance and environmental adaptation to two contrasting patterns of genetic divergence: East-West (discrete) and North-South (continuous). After the importance of environmental adaptation was demonstrated in the East-West divergence, we then examined the allopatric versus sympatric portions of the species range in order to infer the contributing environmental factors.
Contribution of environment versus geography to population structure
Many studies have investigated the evolutionary processes that drive population differentiation (Hübner et al. 2009; McCormack et al. 2010; Nakazato et al. 2008; Pease et al. 2009). While most examples focus on either isolation by distance or environmental adaptation, our study is one of the first to jointly estimate the relative influence of these two forces on multivariate genetic differentiation at a species-wide level, and to identify distinct patterns at different levels of population structure (also see Cushman et al. 2006; Freedman et al. 2010; Pease et al. 2009). Here, we used neutral molecular markers to represent the pattern of genomic background divergence and used this estimated divergence as a surrogate for the historical accumulation of reproductive isolation. Therefore, our goal in this study is to use the degree of reproductive isolation as response variable and estimate the effect from environmental adaptation, using isolation by distance as background control. This is in contrast to many other studies, which controlled for population structure when searching for phenotype-environment correlation (Keller et al. 2009; Keller & Taylor 2008), gene-environment association (Coop et al. 2010; Eckert et al. 2010; Freedman et al. 2010; Hancock et al. 2010), or gene-phenotype association both in the whole-genome (Yu et al. 2006) and the single gene level (Korves et al. 2007; Samis et al. 2008). Specifically, using a multiple regression framework, we tested the contribution from isolation by distance and environmental adaptation at the two hierarchical levels of genetic differentiation and found heterogeneous effects from the two contributing factors across the species range. While isolation by distance alone is sufficient to explain the moderate and continuous North-South divergence, environmental variables show larger contribution than geographical factors in the discrete divergence between East and West. Thus, when environmental adaptation is involved, it may create or maintain higher genetic divergence than isolation by distance alone.
In this study, we incorporated genetic principal component analyses (PCA) and discriminant function analyses (DFA) of multivariate geographical and environmental data sets into a multiple regression framework. This regression-based approach enables the quantitative estimation of genetic variation explained by environmental and geographic factors and their interaction effects, which could not be correctly estimated by partial Mantel test and its derivatives (Balkenhol et al. 2009; Legendre & Fortin 2010; Manel et al. 2003). Similar regression-based approaches have examined the contributions of environment and geography to genetic variation (e.g., Sork et al. 2010), and the dimensions of environmental variables were usually reduced via PCA rather than DFA, and multiple PCA axes were often used. Instead, we examined factors contributing to each of the two hierarchical levels of population structure, and therefore, we chose DFA in order to identify the axis best distinguishing the environmental differences between genetic groups in the hierarchical level being investigated. In addition, our study may be the first to demonstrate the interaction effect between geography and environment in shaping natural genetic variation: In Boechera stricta, the significant GEO*ENV interaction effect in Table 1 is further confirmed by the finding that different environmental variables contribute to the East vs. West divergence in sympatric and allopatric regions (Table 2 and 3).
The possibility that environmental factors contribute to the North-South divergence pattern in B. stricta cannot be ruled out, however. Indeed, several studies have found phenotypic divergence and local adaptation among populations along latitudinal gradients (Arthur et al. 2008; Colautti et al. 2009; Hopkins et al. 2008; Leinonen et al. 2009; Mitchell-Olds et al. 2007; Montague et al. 2008; Stinchcombe et al. 2004). As shown by several examples (Hübner et al. 2009; Platt et al. 2010), when both environmental variables and axes of genetic differentiation are highly correlated with geography, it is difficult to statistically identify the causal factors. This is analogous to the well-known issue of population structure in genome-wide association studies (Bergelson & Roux 2010; Marchini et al. 2004). Like association studies, which control false positives by incorporating population structure into the model (Yu et al. 2006), here we employ a similar approach by using isolation by distance as our null model (Novembre et al. 2008; Novembre & Stephens 2008; Platt et al. 2010; Sharbel et al. 2000) and then examine the effect of environmental variables on genetic differentiation while controlling for geographical factors. The importance of performing such controls is illustrated by a recent study (McCormack et al. 2010), in which, contrary to previous results not accounting for geographical effects, no niche divergence was detected between taxa after such controls were implemented. Similarly, another study (Zellmer & Knowles 2009) used landscape data from three different time periods to model concurrent genetic differentiation among frog populations, and after controlling the effect from each other, they found only contemporary landscape features, rather than historical ones, significantly predict genetic differentiation. Our approach is conservative, since we infer the existence of environmental adaptation only when environment factors explain significant genetic variation in addition to what is already accounted for by geography. If the effects of geography and environment cannot be separated due to their strong correlation, we conservatively attribute genetic differentiation patterns to isolation by distance. Thus, in some circumstances a strong correlation between environment and geography may obscure causal influences of natural selection due to environmental factors.
Nevertheless, even if the North-South divergence in B. stricta is under natural selection from undetected clinal environmental factors, such selection may not cause obvious immigrant or hybrid inviability between adjacent local populations. Under such clinal pattern, although obvious local adaptation may be detected between distant populations (Etterson 2004; Leinonen et al. 2009), there may be little environmental difference among nearby populations. For example, if day length mediates local adaptation between North and South genetic groups, the limited variation in day length between neighboring populations will cause little reduction in gene flow. This clinal pattern is in sharp contrast to the East-West divergence, where two genetically distant populations reside in environmentally distinct locations separated only by a few kilometers. Indeed, given the predominant role of isolation by distance in the North-South divergence of Boechera stricta and in Arabidopsis thaliana, a close relative having similar breeding system (Platt et al. 2010; Sharbel et al. 2000), our finding that environmental selection played a large role in the discrete East-West divergence pattern further illustrates the importance of environmental selection in facilitating or maintaining genetic divergence.
Identifying sources of environmental selection
After the importance of local environment was demonstrated in the East-West divergence, we examined possible environmental factors underlying this divergence pattern to further confirm the role of environmental variables and the GEO*ENV interaction effect in shaping genetic variation in B. stricta. If natural selection by environmental differences were driving phenotypic differentiation during historical allopatry and maintaining reproductive isolation after secondary contact, local genotypes should be consistently associated with predictable environmental conditions. We found similar underlying mechanisms influencing genetic differentiation in allopatric and sympatric regions (Table 2 and 3). In the allopatric region, West genotypes occur in habitats with higher winter snowfall, which provides greater water availability in summer. In the sympatric area, West genotypes occur in riparian sites near streams, where they may experience higher and more consistent levels of soil moisture. In contrast, East genotypes occur on high elevation mountain slopes where ephemeral moisture is supplied by rainfall and snowmelt in spring and early summer. Therefore, during historical allopatry, climatic differences likely drove the phenotypic divergence between the two genetic groups. Upon secondary contact, this trait divergence causes the two genetic groups to occur in distinct habitats based on topography, because climatic variation in the contact zone is low relative to the species range across western North America. In addition, the importance of controlling for geographical factors is again emphasized. While most variables are significant predictors of local East-West genotypes in simple logistic regression (data not shown), the putatively most important factors would be identified only when the effect of geography (latitude or longitude) is controlled in multiple logistic regression (Table 3).
The possibility cannot be totally ruled out, however, that other correlated factors (such as local fauna or other plant competitors) contribute to local adaptation of East and West genotypes, rather than direct effects of water availability. Nevertheless, the importance of soil moisture is supported by preliminary greenhouse and field observations (Lee and Mitchell-Olds, unpublished data). Phenotypic differentiation is significant in a common greenhouse environment, where East genotypes show higher tolerance of drought. Also, observations in the field suggest that in their native moist riparian sites, West genotypes have greater fruit production than East genotypes, possibly due to the longer flowering duration and larger vegetative size. In contrast, slower flowering of West genotypes makes them more susceptible to the late summer drought typical of Eastern habitat on montane slopes. In addition to reciprocal immigrant inferiority (Nosil et al. 2005), their difference in flowering time may also reduce the chance of hybridization, causing assortative mating. Although the genome-wide neutral genetic divergence between East and West may have arisen by genetic drift during historical allopatry, natural selection can be the force currently maintaining such differentiation in the sympatric zone, given the lack of intrinsic hybrid incompatibility.
Recently, methods have been developed to predict species distribution based on inferred environments at collection sites (Phillips et al. 2006; Thomassen et al. 2010). However, our results show that even if the same underlying factor (water availability) determines the distribution of East and West lineages in B. stricta, distinct environmental variables (‘winter precipitation’ or ‘distance to nearest stream’) may represent this underlying factor in different geographical regions. Therefore, in this study we do not attempt to predict the distribution of these genetic groups. In addition, the lack of a ‘distance to the nearest stream’ data layer with the resolution in meters may compromise the accuracy and statistical power of such modeling methods. We suggest that future studies involving environmental niche modeling should incorporate understanding of the biology and ecology of the target species before applying a universal model to continental-scale distributions.
Concluding remarks
This study jointly estimates the relative contribution of isolation by distance versus environmental adaptation to genetic divergence across a species range. In B. stricta, the East-West axis of genetic differentiation, incorporating the joint influences of isolation by distance and environmental adaptation, explains more species-wide genetic variation than the North-South genetic axis, where only the effect of isolation by distance is significant. In addition, our inference of environmental adaptation contributing to East-West divergence also is supported by preliminary observations from laboratory and field. In summary, this research emphasizes the role of ecological factors in the creation and maintenance of genetic differentiation.
Supplementary Material
Acknowledgments
We thank V. Sork and two anonymous reviewers for valuable comments on the manuscript. We are also grateful to J. Anderson and C. Olson-Manning for helpful discussion and B.-H. Song for helping set up the genotyping system. This research was supported by grants from NIH (R01 GM086496) and NSF (EF-0723447). C.-R. Lee was supported by the Hung Taiwan-Duke University Fellowship.
Biography
Cheng-Ruei Lee is a graduate student who is interested in the genetic basis of natural-selection-mediated evolutionary changes. Professor Thomas Mitchell-Olds studies the functional basis of evolutionary forces influencing ecologically important genetic variation. This study is the first step of a large-scale project, which will further utilize greenhouse experiments, field reciprocal transplants, QTL mapping, genotypic selection analysis, and population genomics to investigate the genetic basis of ecological speciation between East and West genetic groups in Boechera stricta.
Footnotes
Data Accessibility
Sample locations, microsatellite data, and environmental variables deposited at DRYAD: doi:10.5061/dryad.6rs51
References
- Arthur AL, Weeks AR, SgrÒ CM. Investigating latitudinal clines for life history and stress resistance traits in Drosophila simulans from eastern Australia. Journal of Evolutionary Biology. 2008;21:1470–1479. doi: 10.1111/j.1420-9101.2008.01617.x. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjalmsson BJ, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkenhol N, Waits LP, Dezzani RJ. Statistical approaches in landscape genetics: an evaluation of methods for linking landscape and genetic data. Ecography. 2009;32:818–830. [Google Scholar]
- Bergelson J, Roux F. Towards identifying genes underlying ecologically relevant traits in Arabidopsis thaliana. Nat Rev Genet. 2010;11:867–879. doi: 10.1038/nrg2896. [DOI] [PubMed] [Google Scholar]
- Boutin-Ganache I, Raposo M, Raymond M, Deschepper C. M13-Tailed Primers Improve the Readability and Usability of Microsatellite Analyses Performed with Two Different Allele-Sizing Methods. BioTechniques. 2001;31:24–27. [PubMed] [Google Scholar]
- Brunelle A, Whitlock C. Postglacial fire, vegetation, and climate history in the Clearwater Range, Northern Idaho, USA. Quaternary Research. 2003;60:307–318. [Google Scholar]
- Bryc K, Auton A, Nelson MR, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proceedings of the National Academy of Sciences. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colautti RI, Maron JL, Barrett SCH. Common garden comparisons of native and introduced plant populations: latitudinal clines can obscure evolutionary inferences. Evolutionary Applications. 2009;2:187–199. doi: 10.1111/j.1752-4571.2008.00053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using Environmental Correlations to Identify Loci Underlying Local Adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- Cushman SA, McKelvey KS, Hayden J, Schwartz MK. Gene flow in complex landscapes: Testing multiple hypotheses with causal modeling. American Naturalist. 2006;168:486–499. doi: 10.1086/506976. [DOI] [PubMed] [Google Scholar]
- Dyer RJ, Nason JD, Garrick RC. Landscape modelling of gene flow: improved power using conditional genetic distance derived from the topology of population networks. Molecular Ecology. 2010;19:3746–3759. doi: 10.1111/j.1365-294X.2010.04748.x. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Bower AD, GonzÁLez-MartÍNez SC, et al. Back to nature: ecological genomics of loblolly pine (Pinus taeda, Pinaceae) Molecular Ecology. 2010;19:3789–3805. doi: 10.1111/j.1365-294X.2010.04698.x. [DOI] [PubMed] [Google Scholar]
- Engelhardt BE, Stephens M. Analysis of Population Structure: A Unifying Framework and Novel Methods Based on Sparse Factor Analysis. PLoS Genet. 2010;6:e1001117. doi: 10.1371/journal.pgen.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etterson JR. Evolutionary potential of Chamaecrista fasciculata in relation to climate change. II. Genetic architecture of three populations reciprocally planted along an environmental gradient in the great plains. Evolution. 2004;58:1459–1471. doi: 10.1111/j.0014-3820.2004.tb01727.x. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Thomassen HA, Buermann W, Smith TB. Genomic signals of diversification along ecological gradients in a tropical lizard. Molecular Ecology. 2010;19:3773–3788. doi: 10.1111/j.1365-294X.2010.04684.x. [DOI] [PubMed] [Google Scholar]
- Gao H, Williamson S, Bustamante CD. A Markov Chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. FSTAT: A program to estimate and test gene diversities and fixation indices (version 2.9.3) 2001 www2.unil.ch/popgen/softwares/fstat.htm.
- Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes. 2005;5:184–186. [Google Scholar]
- Guillot G, Mortier F, Estoup A. GENELAND: a computer package for landscape genetics. Molecular Ecology Notes. 2005;5:712–715. [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proceedings of the National Academy of Sciences. 2010;107:8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson MJ, Carstens BC, Cavender-Bares J, et al. Phylogeography’s past, present, and future: 10 years after. Molecular Phylogenetics and Evolution. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol 2005 [Google Scholar]
- Hopkins R, Schmitt J, Stinchcombe JR. A latitudinal cline and response to vernalization in leaf angle and morphology in Arabidopsis thaliana (Brassicaceae) New Phytologist. 2008;179:155–164. doi: 10.1111/j.1469-8137.2008.02447.x. [DOI] [PubMed] [Google Scholar]
- Hostetler SW, Clark PU. Climatic controls of Western U.S. Glaciers at the last glacial maximum. Quaternary Science Reviews. 1997;16:505–511. [Google Scholar]
- Huang X, Wei X, Sang T, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42:961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- Hübner S, Höffken M, Oren E, et al. Strong correlation of wild barley (Hordeum spontaneum) population structure with temperature and precipitation variation. Molecular Ecology. 2009;18:1523–1536. doi: 10.1111/j.1365-294X.2009.04106.x. [DOI] [PubMed] [Google Scholar]
- Jombart T, Pontier D, Dufour AB. Genetic markers in the playground of multivariate analysis. Heredity. 2009;102:330–341. doi: 10.1038/hdy.2008.130. [DOI] [PubMed] [Google Scholar]
- Keller SR, Sowell DR, Neiman M, Wolfe LM, Taylor DR. Adaptation and colonization history affect the evolution of clines in two introduced species. New Phytologist. 2009;183:678–690. doi: 10.1111/j.1469-8137.2009.02892.x. [DOI] [PubMed] [Google Scholar]
- Keller SR, Taylor DR. History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecology Letters. 2008;11:852–866. doi: 10.1111/j.1461-0248.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- Korves TM, Schmid KJ, Caicedo AL, et al. Fitness effects associated with the major flowering time gene FRIGIDA in Arabidopsis thaliana in the field. Am Nat. 2007;169:E141–E157. doi: 10.1086/513111. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Graham CH, Wiens JJ. Integrating GIS-based environmental data into evolutionary biology. Trends in Ecology & Evolution. 2008;23:141–148. doi: 10.1016/j.tree.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Kozak KH, Wiens JJ. Does niche conservatism promote speciation? A case study in North American salamanders. Evolution. 2006;60:2604–2621. [PubMed] [Google Scholar]
- Legendre P, Fortin M-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Molecular Ecology Resources. 2010;10:831–844. doi: 10.1111/j.1755-0998.2010.02866.x. [DOI] [PubMed] [Google Scholar]
- Leinonen PH, Sandring S, Quilot B, et al. Local adaptation in European populations of Arabidopsis lyrata (Brassicaceae) American Journal of Botany. 2009;96:1129–1137. doi: 10.3732/ajb.0800080. [DOI] [PubMed] [Google Scholar]
- Manel S, Poncet BN, Legendre P, Gugerli F, Holderegger R. Common factors drive adaptive genetic variation at different spatial scales in Arabis alpina. Molecular Ecology. 2010;19:3824–3835. doi: 10.1111/j.1365-294X.2010.04716.x. [DOI] [PubMed] [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends In Ecology & Evolution. 2003;18:189–197. [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- McCormack JE, Zellmer AJ, Knowles LL. Does niche divergence accompany allopatric divergence in Aphelocoma Jays as predicted under ecological speciation?: insights from tests with niche models. Evolution. 2010;64:1231–1244. doi: 10.1111/j.1558-5646.2009.00900.x. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T. Arabidopsis thaliana and its wild relatives: a model system for ecology and evolution. Trends Ecol Evol. 2001;16:693–700. [Google Scholar]
- Mitchell-Olds T, Willis JH, Goldstein DB. Which evolutionary processes influence natural genetic variation for phenotypic traits? Nat Rev Genet. 2007;8:845–856. doi: 10.1038/nrg2207. [DOI] [PubMed] [Google Scholar]
- Montague JL, Barrett SCH, Eckert CG. Re-establishment of clinal variation in flowering time among introduced populations of purple loosestrife (Lythrum salicaria, Lythraceae) Journal of Evolutionary Biology. 2008;21:234–245. doi: 10.1111/j.1420-9101.2007.01456.x. [DOI] [PubMed] [Google Scholar]
- Nakazato T, Bogonovich M, Moyle LC. Environmental factors predict adaptive phenotypic differentiation within and between two wild Andean tomatoes. Evolution. 2008;62:774–792. doi: 10.1111/j.1558-5646.2008.00332.x. [DOI] [PubMed] [Google Scholar]
- Nosil P, Egan SP, Funk DJ. Heterogeneous genomic differentiation between walking-stick ecotypes: “isolation by adaptation” and multiple roles for divergent selection. Evolution. 2008;62:316–336. doi: 10.1111/j.1558-5646.2007.00299.x. [DOI] [PubMed] [Google Scholar]
- Nosil P, Vines TH, Funk DJ. Perspective: Reproductive Isolation Caused by Natural Selection against Immigrants from Divergent Habitats. Evolution. 2005;59:705–719. [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MR, Smith TB. Ecology and speciation. Trends in Ecology & Evolution. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- Oyama R, Clauss M, Formanová N, et al. The shrunken genome of Arabidopsis thaliana. Plant Systematics and Evolution. 2008;273:257–271. [Google Scholar]
- Peakall ROD, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease K, Freedman A, Pollinger J, et al. Landscape genetics of California mule deer (Odocoileus hemionus): the roles of ecological and historical factors in generating differentiation. Molecular Ecology. 2009;18:1848–1862. doi: 10.1111/j.1365-294X.2009.04112.x. [DOI] [PubMed] [Google Scholar]
- Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. [Google Scholar]
- Platt A, Horton M, Huang YS, et al. The Scale of Population Structure in Arabidopsis thaliana. PLoS Genet. 2010;6:e1000843. doi: 10.1371/journal.pgen.1000843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RC. The Cruciferae of Continental North America. Stanford University Press; Stanford, CA: 1993. [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecology Letters. 2005;8:336–352. [Google Scholar]
- Samis KE, Heath KD, Stinchcombe JR. Discordant Longitudinal Clines in Flowering Time and Phytochrome C in Arabidopsis thaliana. Evolution. 2008;62:2971–2983. doi: 10.1111/j.1558-5646.2008.00484.x. [DOI] [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends in Ecology & Evolution. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proceedings of the National Academy of Sciences. 2009;106:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Dobes C, Koch MA, Mitchell-Olds T. Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae) American Journal of Botany. 2005;92:1797–1810. doi: 10.3732/ajb.92.11.1797. [DOI] [PubMed] [Google Scholar]
- Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: biogeography and post-glacial colonization of Europe. Molecular Ecology. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- Slatkin M. Gene flow and the geographic structure of natural populations. Science. 1987;236:787–792. doi: 10.1126/science.3576198. [DOI] [PubMed] [Google Scholar]
- Song B-H, Windsor AJ, Schmid KJ, et al. Multilocus Patterns of Nucleotide Diversity, Population Structure and Linkage Disequilibrium in Boechera stricta, a Wild Relative of Arabidopsis. Genetics. 2009;181:1021–1033. doi: 10.1534/genetics.108.095364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BH, Clauss MJ, Pepper A, Mitchell-Olds T. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Molecular Ecology. 2006;15:357–369. doi: 10.1111/j.1365-294X.2005.02817.x. [DOI] [PubMed] [Google Scholar]
- Sork VL, Davis FW, Westfall R, et al. Gene movement and genetic association with regional climate gradients in California valley oak (Quercus lobata Née) in the face of climate change. Molecular Ecology. 2010;19:3806–3823. doi: 10.1111/j.1365-294X.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, et al. A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proceedings of the National Academy of Sciences. 2004:0306401101. doi: 10.1073/pnas.0306401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storfer A, Murphy MA, Evans JS, et al. Putting the ‘landscape’ in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- Storfer A, Murphy MA, Spear SF, Holderegger R, Waits LP. Landscape genetics: where are we now? Molecular Ecology. 2010;19:3496–3514. doi: 10.1111/j.1365-294X.2010.04691.x. [DOI] [PubMed] [Google Scholar]
- Templeton AR. The reality and importance of founder speciation in evolution. Bioessays. 2008;30:470–479. doi: 10.1002/bies.20745. [DOI] [PubMed] [Google Scholar]
- Thibert-Plante X, Hendry AP. When can ecological speciation be detected with neutral loci? Molecular Ecology. 2010;19:2301–2314. doi: 10.1111/j.1365-294X.2010.04641.x. [DOI] [PubMed] [Google Scholar]
- Thomassen HA, Cheviron ZA, Freedman AH, et al. Spatial modelling and landscape-level approaches for visualizing intra-specific variation. Molecular Ecology. 2010;19:3532–3548. doi: 10.1111/j.1365-294X.2010.04737.x. [DOI] [PubMed] [Google Scholar]
- van Heerwaarden J, Doebley J, Briggs WH, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proceedings of the National Academy of Sciences. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IJ, Summers K. Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Molecular Ecology. 2010;19:447–458. doi: 10.1111/j.1365-294X.2009.04465.x. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zellmer AJ, Knowles LL. Disentangling the effects of historic vs. contemporary landscape structure on population genetic divergence. Molecular Ecology. 2009;18:3593–3602. doi: 10.1111/j.1365-294X.2009.04305.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.