Abstract
The PICALM (CALM) gene, whose product is involved in clathrin-mediated endocytosis, has been identified in two recurring chromosomal translocations, involving either MLL or MLLT10 (AF10). We developed a mouse model of CALM-AF10+ leukemia to examine the hypothesis that disruption of endocytosis contributes to leukemogenesis. Exclusion of the C-terminal portion of CALM from the fusion protein, which is required for optimal binding to clathrin, resulted in the development of a myeloproliferative disease, while inclusion of this domain led to the development of acute myeloid leukemia and changes in gene expression of several cancer-related genes, notably Pim1 and Crebbp. Nonetheless, the development of leukemia could not be attributed directly to interference with endocytosis or consequential changes in proliferation and signaling. In leukemia cells, full-length CALM-AF10 localized to the nucleus with no consistent effect on growth factor endocyctosis, and suppressed H3K79 methylation regardless of the presence of clathrin. Using FRET analysis, we show that CALM-AF10 has a propensity to homo-oligomerize, raising the possibility that the function of endocytic proteins involved in chimeric fusions may be to provide dimerization properties, a recognized mechanism for unleashing oncogenic properties of chimeric transcription factors, rather than disrupting the internalization of growth factor receptors.
Keywords: AML, MPD, endocytosis, Dot1l, H3K79 methylation, oligomerization
Introduction
Endocytosis has long been regarded as a mechanism of signaling attenuation through the degradation of signaling receptors. However, emerging evidence suggests that endocytic proteins are involved in additional cellular processes, including mitosis, and the regulation of signaling, cell cycle, apoptosis and cell fate determination (Lanzetti and Di Fiore, 2008). Given their involvement in a variety of physiological processes, an alteration in expression of endocytic pathway proteins is predicted to play a role in cancer. In support of this hypothesis, changes in expression levels of the endocytosis regulators, HIP1, RAB25 and NUMB, have been linked to human cancer (Bradley et al., 2007; Cheng et al., 2004; Colaluca et al., 2008), and chromosomal translocations involving genes encoding endocytic proteins, such as the clathrin heavy chain (CTLC), clathrin assembly lymphoid myeloid leukemia (PICALM/CALM), EPS15, HIP1, and CBL genes, have been identified in human hematological malignant diseases (Bernard et al., 1994; Chikatsu et al., 2003; Dreyling et al., 1998; Fu et al., 2003; Wechsler et al., 2003). It is not known, however, whether the fusion proteins resulting from these translocations disrupt endocytic processes, and whether this contributes to malignant transformation. Here, we address this issue for acute leukemias with such translocations.
The t(10;11) (p13;q14) is a recurring translocation observed in both acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML), and has been shown to involve the fusion of the (MLLT10/AF10) gene on chromosome 10, with the (PICALM/CALM) gene on chromosome 11 (Carlson et al., 2000; Dreyling et al., 1996; Dreyling et al., 1998). AF10 functions as a putative transcription factor binding DNA through an AT hook motif, and interacting with the SWI/SNF chromatin remodeling complex (Debernardi et al., 2002; Linder et al., 2000). The CALM protein binds clathrin and regulates clathrin-mediated endocytosis through its ability to regulate the orderly progression of clathrin-coat formation (Meyerholz et al., 2005; Tebar et al., 1999). Recent data suggests that CALM-AF10 interacts with the histone H3 lysine 79 (H3K79)-specific methyltransferase DOT1L, through the OM-LZ domain in AF10. This interaction leads to local H3K79 hypermethylation on the Hox loci and upregulation of Hoxa cluster genes (Caudell et al., 2007; Deshpande et al., 2006; Okada et al., 2006). Expression of CALM-AF10 can also lead to a global reduction in H3K79 methylation, by sequestering DOT1L from chromatin, and this correlates with increased chromosome instability (Lin et al., 2009). These particular transformation activities can be attributed to the AF10 component of the fusion protein, whereas the contribution of the fused CALM moiety to the transformation process remains unclear.
CALM has been shown to undergo nucleocytoplasmic shuttling (Vecchi et al., 2001), suggesting that its fusion to AF10 might contribute to transcriptional deregulation; however, whether CALM itself possesses transcriptional activity remains unclear (Archangelo et al., 2006; Vecchi et al., 2001). Additionally, the fusion of CALM to AF10 could lead to a disruption of the endocytic process and enhancement of growth factor receptor signaling. Lastly, CALM’s contribution to the oncogenic potential of AF10 could be attributed to its scaffolding properties. Another endocytic protein, EPS15, has been shown to contribute to oncogenic activation of MLL-EPS15 by promoting dimerization of MLL (So et al., 2003). By analogy, CALM may promote self-association of the CALM-AF10 fusion. In fact, oligomerization of chimeric fusion transcripts, including MLL, RUNX1/AML1 and RARA fusions, has emerged as a powerful mechanism for activating their oncogenic properties (So and Cleary, 2004).
To decipher CALM’s role in CALM-AF10+ leukemias, we set out to determine whether defective endocytosis contributes to the development of leukemia. Notably, the clathrin-binding domain of CALM-AF10 influenced the severity of disease. Although the presence of this domain was required for CALM-AF10 to inhibit transferrin endocytosis in a transfected human cell line, internalization of growth factor receptors and their signaling was not affected in leukemia cells lines or in primary leukemia cells isolated from mice transplanted with cells transduced with CALM-AF10. Moreover, the presence of CALM-AF10 in transduced mouse cells led to a global reduction in H3K79 methylation, independent of clathrin expression. However, using FRET analysis we showed that CALM2091AF10 has a propensity to homo-oligomerize, suggesting that the oncogenic function of the CALM domain may be to confer oligomerization properties that influence transcription, rather than through its effect on endocytosis. Our results suggest the possibility that oligomerization via the CALM domain could increase the binding affinity of AF10-interacting proteins, such as DOT1l, thereby modifying epigenetic patterns, altering transcriptional properties and promoting the development of AML.
Results
CALM-AF10 inhibits clathrin-mediated endocytosis in 293T cells
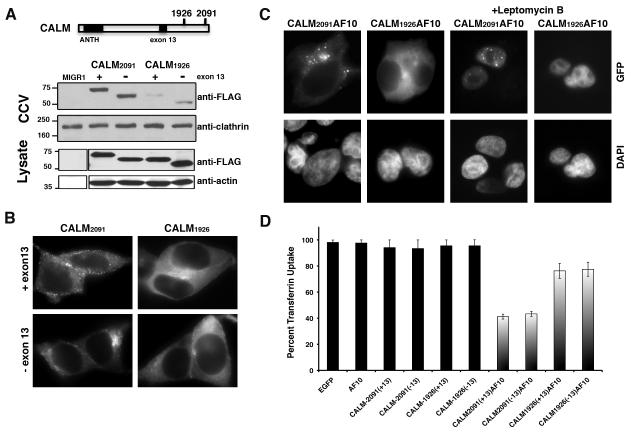
In patients with the t(10;11)(p13;q14), eight alternative CALM-AF10 fusion transcripts have been identified, since there are two breakpoints in CALM and four breakpoints in AF10 (Bohlander et al., 2000; Carlson et al., 2000). All fusion proteins retain the OM-LZ domain of AF10. Relevant to this study, there are two breakpoints in CALM at positions 1926 and 2091(GenBank accession number U45976), referred herein as CALM1926AF10 and CALM2091AF10 (Fig. 1A). The CALM2091AF10 transcript encodes a protein that retains almost the entire coding region of CALM (only the last four amino acids of CALM are replaced by AF10), whereas CALM1926AF10 is missing the C-terminal 59 amino acids of CALM. CALM associates with clathrin heavy chain and co-immunoprecipitation studies using fragments of CALM demonstrated that the C-terminal domain of CALM was required for optimal binding to clathrin heavy chain (Tebar et al., 1999). To examine the interaction of truncated CALM with clathrin, we cloned CALM ending at breakpoint 1926 or breakpoint 2091, with or without exon 13, which encodes a DPF motif that binds the endocytic adaptor AP2. In transfected 293T cells, we demonstrated that CALM2091, but not CALM1926, was enriched in clathrin-coated vesicles (Fig. 1A), and only CALM2091 displayed a punctate pattern of staining, typical of clathrin-coated pits (Fig. 1B). Exclusion of exon 13 didn’t change the pattern, since there are additional AP2 binding sites (Meyerholz et al., 2005)). Next, we engineered constructs expressing chimeric fusion proteins of CALM1926AF10 or CALM2091AF10 with the green fluorescent protein (GFP). An examination of the fusion proteins revealed that CALM2091AF10-GFP localized to the cytoplasm with aggregates of variable size and number, whereas CALM1926AF10-GFP displayed a much more diffuse pattern throughout the cytoplasm (Fig. 1C). When cells were treated with leptomycin B, a chemical that blocks nuclear export, both CALM-AF10 fusion proteins localized to the nucleus suggesting that CALM-AF10 shuttles to and from the nucleus in 293T cells. Interestingly, CALM2091AF10-GFP retained a punctate appearance in the nucleus, whereas CALM1926AF10-GFP was diffusely localized within the nucleus. Clathrin (a major binding partner of CALM) did not co-localize with CALM2091AF10 clusters within the nucleus (Supplementary Fig. S1), suggesting that CALM2091AF10 clustering occurs independently of clathrin association. Together, these results suggest that CALM2091AF10 retains a region of CALM that is critical for clathrin binding, but it has a tendency to form microscopic aggregates regardless of cellular localization or its association with clathrin. The presence of these aggregates can be attributed to the presence of the clathrin-binding region of CALM and not the GFP tag, as this behavior is not displayed by CALM1926AF10-GFP.
Fig. 1. CALM-AF10 inhibits clathrin-mediated endocytosis in 293T cells.
(A) Schematic diagram of the CALM protein indicating nucleotide positions where fusion to AF10 occurs. The large black box refers to the ANTH domain (PIP2-binding) and the small box refers to the location of exon 13, which is alternatively spliced and contains a DPF peptide motif involved in, but not essential for, AP2 binding. Clathrin-coated vesicle fractions (CCV) or total cell lysates, isolated from NIH3T3 cells infected with retroviral vectors expressing FLAG-tagged CALM constructs ending at breakpoint 1926 or 2091, with or without exon 13, were resolved by electrophoresis and blotted with FLAG, clathrin and actin-specific antibodies. (B, C) 293T cells were transfected with chimeric transcripts of CALM2091, CALM1926, CALM2091AF10 or CALM1926AF10 fused to GFP. Expressed proteins were detected by GFP fluorescence and nuclei were visualized by DAPI staining. To block nuclear export, transfected cells were treated with 20 nM leptomycin B, where indicated. Original magnification × 787.5 for all panels. (D) Forty-eight hours after transfection with GFP-fused constructs, a total of 150 GFP+ 293T cells were scored for uptake of fluorescent transferrin, from three independent experiments (50 cells per experiment). There was a significant difference in transferrin uptake between control (EGFP-N1) and CALM1926AF10 (P<0.02) and control and CALM2091AF10 (P<0.0001), with and without exon 13.
To determine whether CALM-AF10 disrupts internalization, we first examined transferrin endocytosis, as a marker for clathrin-mediated internalization. The uptake of fluorescently-labeled transferrin was examined in GFP-positive 293T cells expressing the GFP-tagged chimeric fusion proteins (Fig. 1D). Approximately 92-96% of cells expressing either the CALM or AF10 individual moiety internalized transferrin. However, only ~40% of cells expressing CALM2091-AF10 and ~80% of cells expressing CALM1926-AF10 endocytosed transferrin. This suggests that expression of CALM2091-AF10 impairs the ability of 293T cells to undergo transferrin endocytosis, consistent with the fact that CALM2091-AF10 retains the clathrin-binding region of CALM.
The absence of the clathrin-binding region of CALM alters the phenotype of CALM-AF10+ myeloid neoplasms
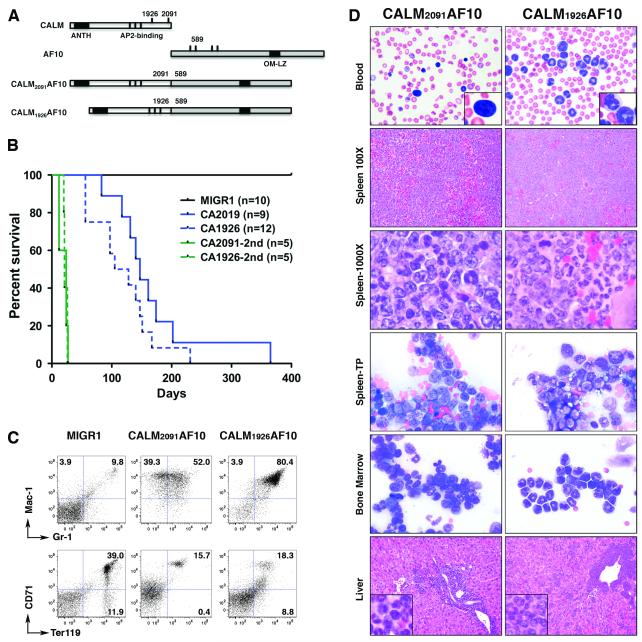
To examine the role that the CALM moiety, and how its ability to bind to clathrin, might modulate disease phenotype, fetal liver progenitors were infected with retroviral vectors expressing CALM-AF10 and transplanted into lethally-irradiated BALB/c recipient mice. In contrast to controls (MIGR1), mice receiving progenitors expressing either CALM2091AF10 or CALM1926AF10 succumbed within 8 months to disease with 100% penetrance (median latency, 147 and 117 days, respectively) (Fig. 2B). Moribund CALM2091AF10+ mice were generally anemic (7/9= 78%), whereas, fewer CALM1926AF10+ mice had anemia (2/12=17%) (Table 1). CALM1926AF10+ mice displayed slightly higher leukocyte counts (median 164 × 106 ml−1) than CALM2091AF10+ mice (median: 127 × 106 ml−1). Analysis of blood smears revealed that the increased leukocyte counts were due to granulocytosis. In all mice succumbing to disease, spleens were markedly enlarged (3-9 fold); however, enlarged lymph nodes and thymus were observed more frequently in CALM1926AF10+ mice. In both, a large percentage (41-96%) of the cells in the bone marrow and spleen were GFP+, a surrogate marker for CALM-AF10. CALM-AF10+ cells were Mac-1(CD11b)hi/Gr-1med-hi, indicative of granulocytes or their progenitors (Fig. 2C). Histological analysis of bone marrow and spleen revealed an enrichment of myeloid blasts adjacent to moderately differentiated granulocytic cells in CALM2091AF10+ mice, whereas a massive enrichment of mature granulocytes without enrichment of myeloid blasts was observed in CALM1926AF10+ mice (Fig. 2D). In both, non-hematopoietic tissues, such as the liver, were infiltrated with myeloid cells, and the disease was transplantable to secondary recipients with a slightly lesser degree of myeloid maturation as compared to primary disease (Fig. 2A, Supplementary Table S1), as is commonly observed after secondary transplant (personal communication, Scott Kogan). Spectral karyotype analysis of CALM2091AF10+ (n=6) and CALM1926AF10+ (n=4) splenocytes revealed a normal karyotype in all cases (Table 2).
Fig. 2. The absence of the clathrin-binding region of CALM alters the phenotype of CALM-AF10+ myeloid neoplasms.
(A) Schematic representation of CALM, AF10 and CALM-AF10 fusion proteins. The two breakpoints in CALM, and one of the four breakpoints in AF10 are indicated by black lines and the base pair position. The two CALM-AF10 fusion proteins used in this study are shown. (B) Survival curves of mice injected with progenitor cells transduced with CALM2091AF10, CALM1926AF10 or MIGR1 (control vector). CA= CALM-AF10; 2ND = secondary recipients of primary MPD or AML cells. (C) Fluorescence-activated cell sorter analysis of cells isolated from spleen of CALM-AF10 mice. Analysis of GFP-gated cells reveals an enrichment of myeloid lineage-specific markers (CD11b+Gr-1med-hi) and decrease in erythroid cells (CD71+Ter119+) in CALM2091AF10 and CALM1926AF10 mice compared to control (MIGR1) mice. (D) Peripheral blood, bone marrow aspirates and spleen touch preparations (TP) were stained with Wright-Giemsa. Paraffin-embedded liver and spleen sections were stained with hematoxylin and eosin. Original magnification ×100 for spleen and liver; ×500 for blood, spleen-TP and bone marrow; ×1000 for spleen and insets.
Table 1.
Primary myeloid neoplasms in mice transplanted with CALM-AF10 expressing fetal liver cells
| Mouse | Break- point |
Survival (days) |
WBC (109/L) |
RBC (1012/L) |
HGB (g/dL) |
Spleen Size (g) |
% GFP+ (BM/SPL) |
%Mac-1+Gr-1+ (BM/SPL)* |
% Blasts | Disease |
|---|---|---|---|---|---|---|---|---|---|---|
| 2524 | 2091 | 83 | 54 | 4.1 | 7 | 0.9 | 96/82 | 80/32 | 52 | AML |
| 2772 | 2091 | 117 | >200 | 8.1 | 15.1 | 0.2 | 96/44 | 81/72 | 34a | AML |
| 2526 | 2091 | 131 | 102 | 5.3 | 9.2 | nd | 95/41 | nd/88 | 62 | AML |
| 2617 | 2091 | 140 | 169 | 7 | 12.1 | 0.5 | 95/90 | 88/89 | 4 | MPD |
| 2618 | 2091 | 147 | 84 | 5.8 | 8.1 | 0.2 | 88/51 | 67/67 | 74 | AML |
| 2619† | 2091 | 161 | 15 | 4.6 | 10 | nd | nd | nd | nd | † |
| 2616b | 2091 | 174 | High‡ | Low ‡ | nd | 0.7 | 51/64 | 9/8 | 20b | AML |
| 3164 | 2091 | 202 | 151 | 3 | 6.9 | 0.4 | 96/80 | 18/24 | 90 | AML |
| 2903 | 2091 | 365 | >200 | 5.4 | 9.9 | 0.7 | 91/80 | 52/47 | 50 | AML |
| Median | 2091 | 147 | 127 | 5.4 | 9.6 | 0.5 | 95/72 | 67/57 | 51 | |
| 2576 | 1926 | 56 | >200 | 7.7 | 14.3 | 0.3 | 76/46 | 75/70 | 7 | MPD |
| 2577 | 1926 | 56 | 90 | 6.5 | 10.6 | 0.3 | 73/49 | 74/73 | 17 | MPD |
| 2578 | 1926 | 56 | >200 | 7.5 | 14 | 0.5 | 80/45 | 82/78 | 8 | MPD |
| 2580 | 1926 | 97 | 78 | 5.2 | 9.4 | 0.7 | 90/64 | 88/76 | 4 | MPD |
| 2874 | 1926 | 97 | >200 | 6.2 | 11.4 | 0.4 | 95/81 | 85/74 | 1 | MPD |
| 2662 | 1926 | 105 | 195 | 8.3 | 13 | 0.7 | 80/79 | 94/92 | 4 | MPD |
| 2579 | 1926 | 128 | 85 | 6.6 | 11.4 | nd | nd | nd | 1a,c | MPD |
| 2963 | 1926 | 140 | 179 | 6.5 | 10.7 | 0.4 | 92/50 | 9/26 | 92 | AML |
| 3193 | 1926 | 147 | 90.1 | 6.8 | 11.5 | 0.7 | 87/64 | 54/37 | 1a,c | MPD |
| 2904 | 1926 | 151 | 195 | 6.6 | 11.4 | 0.6 | 93/66 | 58/42 | 3 | MPD |
| 2709 | 1926 | 167 | 149 | 4.1 | 7.4 | 0.7 | 86/47 | 47/43 | 13a,b | MPDb |
| 2960 | 1926 | 231 | 94 | 8 | 13 | 0.3 | 60/90 | 77/76 | 2 | MPD |
| Median | 1926 | 117 | 164 | 6.6 | 11.4 | 0.5 | 86/64 | 75/73 | 4 | |
| MIGR1 | N/A | ~ 730 | 2-11 | 6-10 | 11-15 | ≤ 0.1 | 65-95 | 35-45 / 3-6 | nd | None |
WBC, white blood cells; RBC, red blood cells; HGB, hemoglobin; BM, bone marrow; SPL, spleen; nd, not determined.
Analysis was performed on GFP+ cells.
Counts obtained 5 days prior to death; tissues isolated from dead mouse suggest there are immature granulocytic elements in spleen and bone marrow.
Based on blood smears, since counts not obtained Anemic mice are indicated in bold type.
blast count is from blood; in all other samples a differential blast count was obtained from scoring 200 bone marrow cells
sheets of blasts in spleen
sheets of segmented neutrophils in spleen and/or liver, lymph node
Table 2.
Spectral karyotype analysis of CALM-AF10 myeloid neoplasms
| Case No. | Mouse | Breakpoint | Recipient | Tissue | Karyotype [no. cells] |
|---|---|---|---|---|---|
| 34270 | 2524 | 2091 | Primary | Spleen | 40,XY[19]/42,XY,+15,+17[1] |
| 34495 | 2526 | 2091 | Primary | Spleen | 40,XY[11] |
| 34582 | 2573 | 2091 | Secondary | Spleen | 40,XY[8]/80,XXYY[2] |
| 34583 | 2574 | 2091 | Secondary | Spleen | 40,XY[10] |
| 34752 | 2565 | 2091 | Secondary | Spleen | 40,XY[11] |
| 34756 | 2563 | 2091 | Secondary | Spleen | 40,XY[10] |
| 35094 | 2577 | 1926 | Primary | Spleen | 40,XX[10] |
| 35095 | 2578 | 1926 | Primary | Spleen | 40,XY[5]/40,XX[6] |
| 35093 | 2576 | 1926 | Primary | Spleen | 40,XY[10] |
Thus, the CALM2091AF10+ mice develop AML, consistent with previous studies that also examined mouse models with a CALM-AF10 fusion with the 2091 breakpoint (Caudell et al., 2007; Deshpande et al., 2006). However, the CALM1926AF10+ mice do not fulfill the Bethesda proposal for AML based on several features (Kogan et al., 2002): (1) < 20% nonlymphoid blasts were observed in the blood, marrow and spleen-median percent blasts was 4% for CALM1926AF10+ mice versus 51% for CALM2091AF10+ mice (P=0.009); (2) only some mice showed cytopenias: 2/7 (17%) CALM1926AF10+ mice had low RBC counts (<6 ×1012/L), vs. 7/9 (78%) for CALM2091AF10+; and (3) complete maturation of myeloid forms to segmented neutrophils were observed in the bone marrow and spleen. The CALM1926AF10+ mice do, however, meet several criteria for myeloid leukemia, including increased numbers of nonlymphoid hematopoietic cells in blood, bone marrow spleen and other tissues, and disease was transplantable to secondary recipients (Fig. 2), suggesting that these mice can be classified on the border between myeloproliferative disease (MPD) and MPD-like leukemia (personal communication, S. Kogan). Analysis of CALM-AF10 expression in spleen from diseased mice revealed variable expression, regardless of the specific breakpoint, confirming that differences in phenotype were not merely due to different CALM-AF10 expression levels (Supplementary Fig. S2). Moreover, Deshpande et al. (2006) first reported that CALM-AF10+ AMLs have a unique Mac1+ B220+ phenotype. Similar to Caudell et al. (2007), we observed that ~50% of the AMLs were Mac1+B220+, whereas almost all MPDs had this population of cells, suggesting that both fusion proteins can give rise to this unique population of cells (Supplemental Table S2).
Microarray analysis identified 28 over-expressed and 87 under-expressed genes in CALM2091AF10+ AMLs compared to CALM1926AF10+ MPDs (Supplementary methods, Supplementary Table S3). Although the changes were small (median fold change of 2.3), Ingenuity Pathway analysis revealed a strong association of differentially expressed genes with cellular growth, hematopoiesis and cancer. We confirmed that Pim1 and Klf9 (BtebI) were increased in AML samples, and Crebbp (CBP) and Itgam (Mac-1) were downregulated in AMLs relative to MPDs (Fig. 3A). The Itgam and Klf9 genes are markers for myeloid differentiation (Lian et al., 2002). The serine (Ser)/threonine (Thr) kinase Pim1 gene is a proto-oncogene that enhances the development of lymphoma and leukemia (Selten et al., 1985; van Lohuizen et al., 1989). The Crebbp gene encodes a transcriptional coactivator that has been shown to have tumor suppressor activity (Kung et al., 2000). The oncogenic potential of CALM-AF10 has been linked to expression of Hoxa cluster genes (Caudell et al., 2007; Dik et al., 2005; Okada et al., 2006). Hoxa5, Hoxa7, and Hoxa9 were increased relative to control MIGR1 cells, regardless of which CALM-AF10 fusion was expressed (Fig. 3B). Thus, CALM2091AF10 resulted in the development of AML, whereas CALM1926AF10 induced an invasive MPD-like leukemia, with concomitant changes in gene expression of cancer-related genes, and myeloid differentiation markers.
Fig. 3. Differential gene expression in CALM2091AF10 AMLs and CALM1926AF10 MPDs.
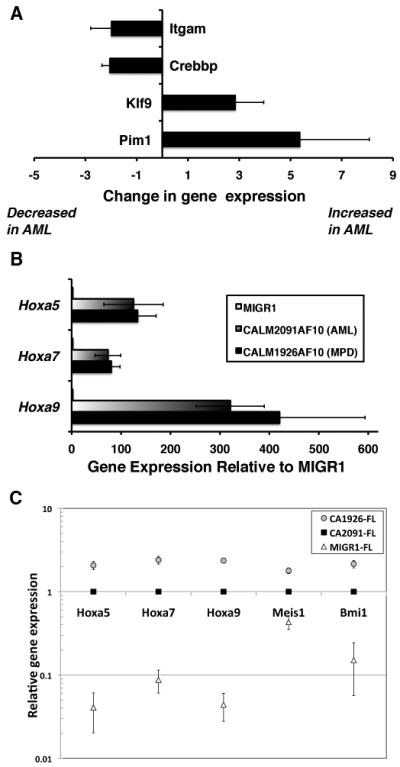
(A) RNA expression levels were quantified by real-time RT-PCR. Fold change in RNA expression in spleen cells from mice with AML relative to MPD is shown. A positive value indicates the gene is up-regulated in AML, and a negative value indicates it is down-regulated in AML compared to MPD samples. (B) Hoxa cluster gene expression relative to MIGR1 control (normalized to 1). Whereas Hoxa genes were significantly up-regulated compared to controls, there was no difference between AML and MPD samples. Results are expressed as the mean ± standard error from three independent experiments. (C) Fetal liver (FL) progenitors, infected with retroviral vectors expressing either CALM2091AF10 or CALM1926AF10 were grown in vitro for 10-12 days; changes in gene expression relative to the MIGR1 control were examined by real-time PCR (four independent experiments). To assess differences between CALM-AF10 constructs, relative gene expression was set as ‘1’ for CALM2091AF10+ cells. Hoxa5, Hoxa7, Hoxa9, Meis1, and Bmi1 gene expression was consistently increased ~2 fold in CALM1926AF10+ cells vs. CALM2091AF10+ cells (P<0.01).
To exclude the possibility that changes in gene expression were merely due to a comparison of blasts versus mature neutrophils, we examined changes in direct target genes, and noted some differences in gene expression as compared to leukemia cells (Supplementary Fig. S3). For example, CALM2091AF10 expression did not result in high Pim1 expression in vitro. As anticipated, Hoxa cluster genes, Meis1, and Bmi1 were elevated (5-50 fold) in CALM-AF10+ cells. Interestingly, expression of these genes was consistently and significantly ~2 fold higher (P<0.01) after CALM1926AF10 expression compared to CALM2091AF10 (Fig. 3C). As these genes are associated with stem cell proliferation and leukemogenesis, the elevated expression in CALM AF10+ 1926 cells may account for why the mice with MPD succumb more rapidly than the mice with AML (median survival 117d vs. 147d).
Growth factor receptor internalization and proliferation is not altered in CALM-AF10+ leukemia cells
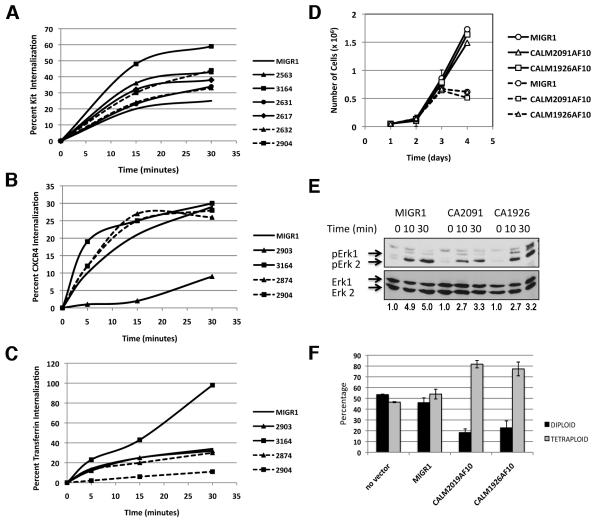
CALM2091AF10+ is an effective inhibitor of transferrin endocytosis in vitro (Fig. 1D) and mediator of acute leukemia in mice (Fig. 2), raising the possibility that defective endocytosis may contribute to leukemogenesis. The KIT, CXCR4 and TF receptors internalize via a clathrin-mediated pathway (Broudy et al., 1999; Hopkins, 1985; Signoret et al., 1997). Internalization of these receptors in freshly isolated bone marrow cells was highly variable with no significant difference from controls, suggesting that expression of CALM2091-AF10 or CALM1926-AF10 does not consistently affect internalization in AMLs or MPDs (Fig. 4A-C). As signaling and endocytosis are tightly coupled, we examined whether expression of CALM-AF10 had an effect on the proliferation of BaF3 cells, a well-established IL-3 dependent hematopoietic cell line model commonly used to test oncogene effects on growth and proliferation. Expression of CALM-AF10 did not affect the growth of BaF3 cells in response to low and high levels of IL-3 (Fig. 4D) and did not enhance Erk signaling in response to IL-3 (Fig. 4E). A greater percentage of tetraploid metaphases were observed in CALM-AF10+ BaF3 cells (Fig. 4F), suggesting that CALM-AF10 is physiologically active in these cells, since its expression is thought to contribute to genomic instability (Lin et al., 2009).
Fig. 4. Internalization and proliferation is not altered in CALM-AF10+ hematopoietic or leukemia cells.
Internalization of KIT (A), CXCR4 (B), or TF (C) receptor, gated on GFP+ bone marrow cells isolated from CALM2091AF10+ AML mice (solid lines) or CALM1926AF10+ MPD mice (dashed lines), compared to control MIGR1 mice. (D) Growth curve of trypan blue-excluded viable BaF3 cells expressing CALM-AF10 or control MIGR1 vector grown in low (0.01 ng/ml, dashed line) or high (1ng/ml, solid line) levels of IL-3,. (E) BaF3 cells expressing CALM-AF10 or control MIGR1 vector were serum starved, treated with IL-3 for times indicated, and lysates were immunoblotted with a phospho-Erk 1/2 antibody. No increase in Erk phosphorylation, normalized to Erk 2 levels, was observed in CALM-AF10 expressing cells. (F) To estimate the frequency of tetraploid metaphase cells, BaF3 cells were treated with Colcemid™, and metaphase cells were prepared with standard cytogenetic techniques. A higher percent of tetraploid metaphase cells were found in BaF3 cells expressing CALM-AF10 compared to MIGR1 controls. An average of three independent experiments is shown. In each experiment, 30-50 metaphases were scored for each line.
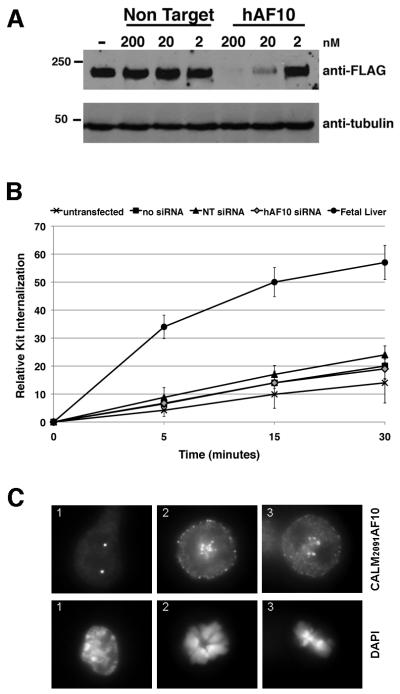
To understand why expression of CALM-AF10 did not have major effects on endocytosis or proliferation of hematopoietic cells, we generated three IL-3 independent cell lines from the malignant cells of CALM2091-AF10+ and CALM1926-AF10+ mice (Supplemental Table S4 and Fig. S4). All cell lines efficiently internalized the TF receptor. The KIT+ line (CA2091-CL1) displayed impaired KIT internalization; however, we could not prove that CALM-AF10 was directly responsible for impaired KIT internalization, since a 95% decrease in CALM-AF10 expression by siRNA was unable to restore KIT internalization to wild type levels in CA2091-CL1 cells (Fig. 5A,B). To explore why CALM-AF10 did not have a major effect on endocytosis in leukemia cells, as we observed with transfected 293T cells, the CA2091-CL1 cell line was stained with an anti-FLAG or anti-CALM antibody to detect expression of CALM2091AF10 (Fig. 5C). Similar to 293T cells, large punctate clusters variable in size and number were observed in interphase cells; however, they localized predominantly to the nucleus, where they did not co-localize with clathrin (Supplementary Fig. S5A, B). Although clathrin-mediated endocytosis ceases during early mitosis, it has been shown that clathrin localizes to mitotic spindles to aid congression of chromosomes (Royle et al., 2005). Interestingly in CA2019CL1 metaphase cells, CALM2091AF10 co-localizes with clathrin (Fig. 5C, Supplementary Fig S5B). The nuclear localization of CALM-AF10 in the CA2091-CL1 leukemia cell line and its apparent lack of effect on internalization or signaling downstream of growth factor receptors suggests that CALM-AF10 expression likely does not influence endocytic trafficking during leukemogenesis, but rather fusion of the CALM moiety to AF10 plays another crucial role.
Figure 5. CALM2091AF10 localizes to the nucleus and does not influence Kit internalization in a myeloid line derived from CALM2091AF10+ AML cells.
(A) Protein expression of CALM2091AF10, detected using an anti-FLAG antibody, 48 hours after nucleofection with siRNA specific for human AF10 (hAF10) or non-targeting (NT) control siRNA in mouse CA2091-CL1 cells, an IL-3 dependent line derived from CALM-AF10+ AML mouse cells. (B) CA2091-CL1 cells were transfected with siRNAs: NT-non-targeting; hAF10-specific for CALM-AF10. Cells were treated with 200ng/ml SCF for times indicated, and KIT receptor remaining on the cell surface was measured. Fetal liver cells are shown as a positive control for KIT internalization. Knock-down of CALM-AF10 expression (hAF10 siRNA) does not increase KIT internalization. (C) Localization of CALM2091AF10, expressed in CA2091-CL1 cells, during interphase (1) and metaphase (2,3). Original magnification, 787.5X.
CALM-AF10’s interaction with clathrin does not influence global H3K79 hypomethylation
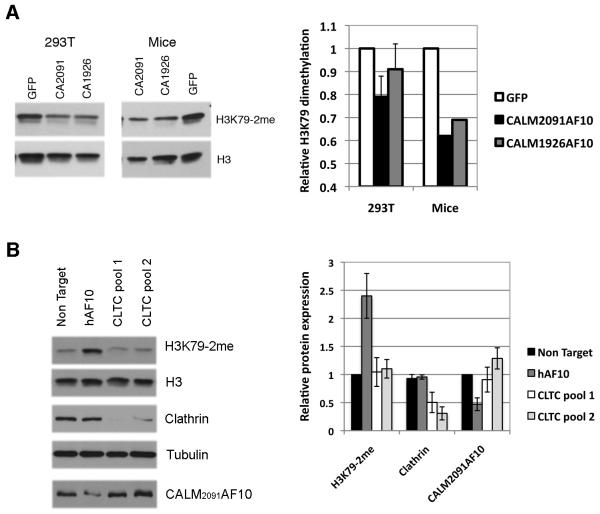
AF10 interacts with the H3K79 methyltransferase, DOT1L, and it has been hypothesized that CALM-AF10 prevents AF10 from targeting DOT1L to chromatin, leading to global H3K79 hypomethylation (Lin et al., 2009). Whether the interaction of other proteins with the CALM portion of CALM-AF10 influences DOT1L recruitment is unknown. In agreement with previous results, we observed a decrease in dimethylated H3K79 (H3K79-2me) after CALM2091AF10 expression compared to GFP control, in both 293T cells and cells from leukemic mice (Fig. 6A). Although subtle, expression of CALM1926AF10 did not appear to be as effective as CALM2091AF10 at lowering H3K79 methylation. These subtle shifts in H3K79 methylation may have a consequential impact on gene expression. Hoxa cluster genes, whose transcription is influenced by local H3K79 methylation, were expressed at 2-fold higher levels in CALM1926AF10+ cells vs. CALM2091AF10+ cells (Fig. 3C).
Fig. 6. CALM-AF10 interaction with clathrin does not influence global H3K79 methylation.
(A) Nuclear extracts, prepared from 293T cells transfected with CALM-AF10 constructs or leukemia cells from individual transplanted CALM-AF10+ mice, were immunoblotted for H3K79-2me and histone H3. Quantification of H3K79-2me Western blot immunostaining, normalized to H3, confirms a decrease in H3K79 methylation upon CALM2091AF10 expression compared to GFP control in 293T and mouse cells. Global hypomethylation after CALM1926AF10 expression is less pronounced. (B) Nuclear extracts were prepared from the CALM2091AF10+ CA2091-CL1 cell line, transfected with non-targeting (NT), CALM-AF10 (hAF10) or clathrin specific siRNA pools (CLTC-pool1, pool2). A representative blot indicates H3K79 methylation is increased after knock-down of CALM-AF10, but not clathrin (~92% knockdown for CLTC-pool1). Quantification of the average H3K79-2me, clathrin and CALM2091AF10 protein levels (Right panel, 3 independent experiments).
These data also raised the possibility that clathrin’s interaction with CALM2091AF10 may influence recruitment of DOT1L to its targets and, thereby, alter H3K79-2me levels. The CA2091-CL1 cell line was transfected with siRNA pools directed against CALM-AF10 or clathrin heavy chain and global H3K79 methylation was examined by Western blot analysis of nuclear extracts. Whereas knock-down of CALM2091AF10 restored H3K79 methylation as anticipated, knock-down of clathrin had no effect (Fig. 6B). These results suggest that the clathrin/CALM-AF10 interaction does not influence DOT1L recruitment and methylation of its targets, although it remains possible that residual clathrin expression may be sufficient to sequester CALM-AF10 with DOT1L away from chromatin-associated AF10.
CALM2091AF10 forms homo-oligomers
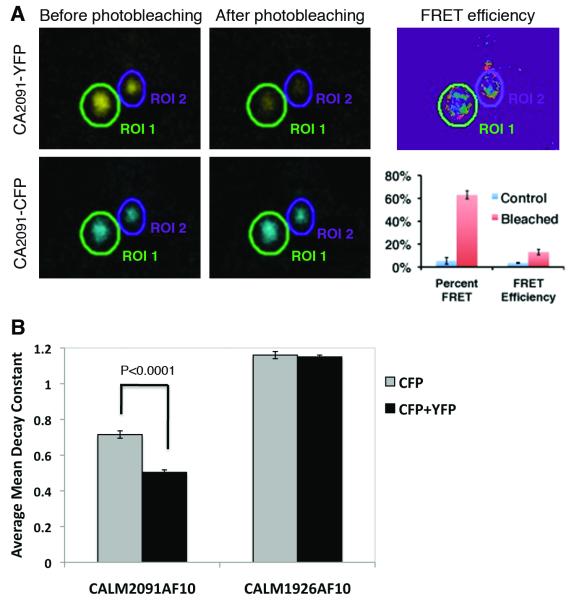
The localization of CALM2091AF10, in large punctate clusters, was reminiscent of some MLL fusions where oligomerization of the fusion proteins has been shown to play a critical role in promoting leukemogenesis. To test whether the clusters of CALM2091AF10 were interacting through homo-oligomerization, we undertook FRET analysis using two distinct detection methods: acceptor photobleaching and donor photobleaching. In acceptor photobleaching, energy transfer from CFP (donor) to YFP (acceptor) is reduced upon acceptor bleaching, resulting in increased donor fluorescence (Kenworthy, 2001). FRET efficiencies were determined for a region of interest (ROI), drawn around each aggregate of CALM2091AF10 (Fig. 7A). As a control, FRET was calculated in unbleached samples where energy transfer from CFP to YFP should not occur, and accordingly only FRET efficiencies above 3.5% were considered positive. For the experimental sample, 63% of the ROIs displayed FRET, with an average FRET efficiency of 13%, suggesting that CALM2091AF10 forms homo-oligomers.
Fig. 7. Self-association properties of CALM2091AF10 visualized by acceptor and donor photobleaching FRET.
(A) 293T cells were transfected with chimeric transcripts of CALM2091AF10 fused to YFP and CFP (CA2091-YFP + CA2091-CFP). A representative image of YFP and CFP fluorescence before and after photobleaching and corresponding FRET efficiency is shown. The percent of CALM2091AF10 aggregates displaying FRET and the average FRET efficiency for bleached regions (n=147) and control (non-bleached) regions (n= 97) ± the standard error of the mean, for three individual experiments are shown. (B) 293T cells were transfected with CA2091-CFP + CA2091-YFP, CA1926-CFP + CA1926-YFP, CA2091-CFP or CA1926-CFP. Cells were bleached using the donor excitation wavelength of 458 nm, a time-lapse of images was captured, and a fluorescence decay constant was measured. Mean decay constants in cells expressing both CA2091-CFP and CA2091-YFP (n=110) have a slower decay constant than CA2091-CFP alone (n=88) (P<0.0001), suggesting the occurrence of FRET.
Donor photobleaching is based on the principle that processes such as FRET, which shorten the excited-state lifetime of a fluorophore, also protect it from photobleaching, making decay kinetics an efficient measure of FRET efficiency (Kenworthy, 2001). Fluorescence decay from cells expressing only CALM-AF10-CFP or CALM-AF10-CFP plus CALM-AF10-YFP was measured, and decay constants were determined by fitting to single exponential curves (Fig. 7B). Measurements from numerous aggregates from multiple cells show that the mean decay constant for cells expressing CALM2091AF10-CFP alone (0.72±0.014, n=88) was significantly greater (p=1.6×10−16) than that for cells co-expressing CALM2091AF10-CFP and CALM2091AF10-YFP (0.50±0.021, n=110). These data are consistent with the occurrence of FRET between CFP and YFP fluorophores, and again suggest that homo-oligomerization of CALM2091AF10 occurs within large aggregates. No difference in decay constant was observed when a similar experiment was performed with CALM1926AF10, suggesting that this fusion protein does not homo-oligomerize as readily as CALM2091AF10.
Discussion
Several endocytic proteins have been identified as fusion partners of transcription factors or tyrosine kinases in hematopoietic malignancies, and it has been proposed that disruption of the endocytic process may contribute to disease progression (Lanzetti and Di Fiore, 2008). To explore this hypothesis further, we examined the functional contribution of the CALM moiety in CALM-AF10-mediated transformation. CALM2091AF10+ and CALM1926AF10+ mice developed disease with a similar latency (P=0.1), yet CALM2091AF10 affects the severity of the disease, suggesting a potential role for clathrin. However, we did not identify deficits in endocytosis, proliferation or signaling in CALM-AF10+ leukemia or hematopoietic cell line models. In contrast, inclusion of the clathrin-binding domain (CALM2091AF10) was required to inhibit transferrin endocytosis in 293T cells efficiently. As many studies of endocytic function rely heavily on the use of epithelial cell lines, our studies emphasize caution in interpreting these results in a larger biological context. For example, CALM-AF10 localized predominantly in the cytoplasm of 293T cells (where it may have transiently interacted with clathrin-coated vesicles), but was found predominantly in the nucleus of the CA2091CL1 leukemia cell line, providing a potential explanation for the different effects on endocytosis in the two cell types.
Experimental results suggest that DOT1L and H3K79me2 are regulated during progression of the cell cycle, in which H3K79me2 levels increase during M phase (Feng et al., 2002). During interphase, clathrin plays a key role in membrane trafficking; however, when the cell enters mitosis, membrane trafficking ceases and a portion of clathrin is targeted to the mitotic spindle where it stabilizes kinetochore fibers (Royle et al., 2005). In the CALM2091AF10+ leukemia cell line, CALM-AF10 and clathrin generally segregated to either the nucleus or cytoplasm, respectively, during interphase; however, they co-localized during metaphase (Fig. 5, Supplementary Fig. S5). Therefore, we examined whether the DOT1L-sequestering effect of CALM-AF10 was influenced by interaction of the CALM moiety with clathrin. However, knock-down of clathrin had no effect on global H3K79me2 levels in a CALM AF10+ 2091 cell line, leaving open the question of how CALM-AF10 may act as a dominant negative competitor of AF10 to regulate global H3K79 hypomethylation. The genomic instability observed in CALM-AF10+ cells is speculated to be associated with global H3K79 hypomethylation (Lin et al., 2009). Given that disruption of clathrin has also been associated with genomic instability (Lemmon et al., 1990; Royle et al., 2005), and that CALM-AF10 colocalizes with clathrin during metaphase in the leukemia cell line tested, our data raises the intriguing possibility that clathrin/CALM-AF10 interaction may also contribute to increased genomic instability.
It has been proposed that oligomerization of chimeric transcription factors may act as a universal oncogenic amplifier that enhances the transcriptional properties by augmenting and broadening the binding affinity to interacting proteins and target DNA sequences. The nature of the endocytic machinery, characterized by their self-assembly properties and ability to form higher-order complexes, may lend itself to promoting dimerization or oligomerization of transcription factors. Moreover, previous work suggests that the homo-oligomerization of AF10 may itself enhance AF10-mediated transcription (Forissier et al., 2007). In our mouse model, we observed a slightly more aggressive disease with a form of CALM-AF10 (CALM2091AF10) that has a tendency to form macroscopic aggregates, regardless of cellular location, and could be shown to oligomerize by FRET analysis. The homo-oligomerization of CALM-AF10 could be envisaged to compete with endogenous AF10 for the recruitment of DOT1L, a known player in CALM-AF10-mediated transformation. Additionally, AF10 binds to GAS41 (Harborth et al., 2000), a protein previously found to interact with the SWI/SNF complex, which in turn binds to the nuclear mitotic apparatus protein, NuMA (Bhattacharya et al., 2002), which can be phosphorylated by Pim1. Alteration of the binding affinity of these interactions through oligomerization, could influence chromatin organization and transcriptional regulation, thereby, contributing to malignant transformation. Artificial oligomerization of AF10, as has been done for MLL (So et al., 2003), could be used to address this hypothesis and rule out other oncogenic contributions by CALM.
In patients with CALM-AF10+ T-ALL there appears to be a correlation between the AF10 fusion and leukemic phenotype (Asnafi et al., 2003). To date, there is no known association between CALM or AF10 fusion breakpoint and leukemic phenotype in AML patients; however, varying degrees of maturation and leukemia phenotype (AML M0-M5) have been noted (Carlson et al., 2000). Also, CALM-AF10 fusions are found in <1% of AML cases, and only a small number of cases have been well-characterized. Nonetheless, mouse models have proven to be a powerful biological tool in deciphering the underlying mechanisms of disease. In this study, we exploited the alternate CALM fusions to decipher the functional contribution of the CALM protein in CALM-AF10-mediated transformation. Our data suggest that increased proliferation through perturbed endocytosis is not a major contributing factor to oncogenic transformation, as originally hypothesized. Instead, our findings point to an alternative mechanism involving homo-oligomerization of CALM-AF10 that could increase its binding affinity for interacting proteins, such as DOT1L or other co-activators, that influence chromatin organization and transcriptional regulation, promoting the development of overt leukemia.
Methods
Cloning of CALM and CALM-AF10 expression constructs
cDNAs encoding truncated human CALM and AF10 were obtained by reverse transcriptase-PCR. Recombinatorial PCR was used to clone human CALM-AF10 fusion transcripts representing fusions isolated from patients with breakpoints at nucleotide 1926 or 2091 of CALM, and nucleotide 588 of AF10 (Carlson et al., 2000; Dreyling et al., 1996). For 293T studies, CALM, AF10 or CALM-AF10 cDNAs were cloned upstream and in-frame with the enhanced green fluorescent protein (GFP) using the pEGFP-N1 vector (BD Biosciences Clontech), and transiently-transfected using the Effectene transfection reagent (Qiagen). For retroviral expression, CALM2091AF10 and CALM1926AF10 cDNAs were cloned upstream of the internal ribosomal entry site (IRES) of the MIGR1 (MSCV IRES GFP) vector. Retrovirus was produced by transfection of 293T cells with the pCL ecotropic packaging plasmid and the appropriate retroviral vector. BaF3 cells or E14.5 fetal liver cells were infected by two rounds of spinoculation.
Characterization of Leukemias
Lethally-irradiated BALB/c mice (6-12 weeks old) were infected by retro-orbital injection of 3 ×106 unsorted E14.5 fetal liver cells transduced with CALM-AF10 or control vectors. Transplanted mice were monitored for the onset of disease by performing complete blood counts (Hemavet 850, CDC Technologies) every three weeks or when mice became moribund. All organs were recovered, fixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned at 4 to 5 μm and stained with hematoxylin and eosin (H&E) for histologic examination by a pathologist (J.A.). Peripheral blood, bone marrow aspirates and spleen touch preparations were stained with Wright-Giemsa. Images were obtained using an Olympus microscope (Model BX45) equipped with an Olympus DP12 digital camera. Single cell suspensions of bone marrow and spleen were stained with phycoerythrin (PE)-conjugated antibodies specific for CD71 or Gr-1 and allophycocyanin (APC)-conjugated antibodies specific for Ter119 and Mac-1 (CD11b) (BD BioSciences). Flow cytometry was performed on a FACSCanto (BD Biosciences), and data were analyzed with FlowJo software (Treestar Software). Short-term (24-h) cultures of splenocytes from CALM-AF10+ mice, and spectral karyotyping were performed as described (Le Beau et al., 2002). At least 10 metaphase cells were analyzed per case.
Real-Time PCR Analysis
RNA was isolated using Stat-60 and cDNA was made following standard protocols. Real-time PCR was performed using a Step One Plus Thermocycler (Applied Biosystems). A list of the primers used is shown in supplemental data (Table S5). The relative amounts of each mRNA amplification products were calculated in reference to standard curves, and then normalized to the relative amounts of Gapdh transcripts.
Immunofluorescence and FRET Analysis
Twenty-four hours after 293T cells were transfected with CALM fused to GFP or CALM-AF10 fused to GFP, CFP or YFP, cells were plated on coverslips and grown overnight. An IL-3-dependent cell line was generated by culturing the malignant cells of a CALM2091AF10+ mouse (2616b), and was named CA-2091CL1. These cells were allowed to settle on poly-L-lysine coated coverslips for 30 mins at room temperature. Cells were fixed in 4% paraformaldehyde, quenched with 50 mM NH4Cl, permeabilized with 0.2% Triton X-100, washed, and blocked with PBS plus 5% donkey serum (Jackson ImmunoResearch) and 1% fish gelatin (Sigma) for 30 minutes. For CA2091-CL1 cells, CALM-AF10 was stained with a polyclonal anti-FLAG antibody (Sigma) or anti-CALM antibody (C18, Santa Cruz Biotechnology) followed by a Rhodamine Red-conjugated donkey anti-rabbit IgG or anti-goat IgG, respectively. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). For standard immunofluorescence, cells were viewed with a Zeiss Axioplan epi-fluorescence microscope, and images were processed using Adobe Photoshop software. For FRET analysis, methods were adapted from Cho et al. (Cho and Kehrl, 2007) and Muntau et al. (Muntau et al., 2003), and a detailed description is provided in the supplemental materials.
Preparation of clathrin-coated vesicles
Clathrin-coated vesicles were isolated by a differential centrifugation method adapted from Chakrabarti et al. (Chakrabarti et al., 1993). A detailed description is provided in the supplemental materials.
Transfection of small interfering RNA (siRNA)
All siRNA sequences are provided in the supplementary methods. CA2091-CL1 cells were transfected with 50nM of each hAF10 siRNA, 100 mM of the CLTC pools or 100 nM of the non-targeting siRNA using the Amaxa nucleofection technology.
Western blot analysis
Western blot analysis was performed using standard protocols. Primary antibodies included: FLAG (M2) (Sigma) to detect CALM-AF10, β-tubulin (AA2, Millipore), phospho p44/42 MAPK (Erk1/2) and p44/42 MAPK (Cell Signaling Technologies), H3K79-2me and H3 (Abcam), and clathrin (TD.1, Santa Cruz Biotechnology). To quantitate levels of H3K79-2me, the intensity of the bands was measured using Adobe Photoshop and normalized to H3 loading controls. Clathrin and CALM-A10 levels were normalized to tubulin.
Internalization Assays
CALM-AF10-positive cells or E14.5 fetal liver cells (BALB/c mice) were incubated 15 min on ice with 100 ng/ml SCF or SDF1α (CXCL12) (R&D Systems), then at 37°C for 0-30 minutes (internalization) and washed in ice-cold PBS to remove ligand. Cells were stained with APC-conjugated anti-KIT receptor and PE-conjugated anti-CXCR4 antibodies; receptors remaining on the cell surface were measured by flow cytometric analysis. Internalization was calculated as (MFItime0-MFItimeX)/MFItime0 X 100%, where MFI is the mean fluorescent intensity of KIT or CXCR4 receptors. For cells treated with siRNA and controls, the GFP+ cells were gated for analysis. Transferrin uptake assays are described in the supplementary materials.
Supplementary Material
Acknowledgements
We thank Dr. Vytas Bindokas and the University of Chicago Comprehensive Cancer Center Integrated Microscopy, and Flow Cytometry Facilities for their assistance. We also thank Scott Kogan for advice on classifying myeloid malignancies and Elizabeth Davis for technical assistance and members of the Le Beau laboratory for helpful discussions
This work was supported by a Special Fellow Award and SCOR (7015) from the Leukemia and Lymphoma Society, and grants from the Cancer Research Foundation, Lauri Strauss Leukemia Foundation, and NIH (GM038093).
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- Asnafi V, Radford-Weiss I, Dastugue N, Bayle C, Leboeuf D, Charrin C, et al. CALM-AF10 is a common fusion transcript in T-ALL and is specific to the TCRgammadelta lineage. Blood. 2003;102:1000–6. doi: 10.1182/blood-2002-09-2913. [DOI] [PubMed] [Google Scholar]
- Archangelo LF, Glasner J, Krause A, Bohlander SK. The novel CALM interactor CATS influences the subcellular localization of the leukemogenic fusion protein CALM/AF10. Oncogene. 2006;25:4099–109. doi: 10.1038/sj.onc.1209438. [DOI] [PubMed] [Google Scholar]
- Bernard OA, Mauchauffe M, Mecucci C, Van den Berghe H, Berger R. A novel gene, AF-1p, fused to HRX in t(1;11)(p32;q23), is not related to AF-4, AF-9 nor ENL. Oncogene. 1994;9:1039–45. [PubMed] [Google Scholar]
- Bhattacharya N, Wang Z, Davitt C, McKenzie IF, Xing PX, Magnuson NS. Pim-1 associates with protein complexes necessary for mitosis. Chromosoma. 2002;111:80–95. doi: 10.1007/s00412-002-0192-6. [DOI] [PubMed] [Google Scholar]
- Bohlander SK, Muschinsky V, Schrader K, Siebert R, Schlegelberger B, Harder L, et al. Molecular analysis of the CALM/AF10 fusion: identical rearrangements in acute myeloid leukemia, acute lymphoblastic leukemia and malignant lymphoma patients. Leukemia. 2000;14:93–9. doi: 10.1038/sj.leu.2401614. [DOI] [PubMed] [Google Scholar]
- Bradley SV, Smith MR, Hyun TS, Lucas PC, Li L, Antonuk D, et al. Aberrant Huntingtin interacting protein 1 in lymphoid malignancies. Cancer Res. 2007;67:8923–31. doi: 10.1158/0008-5472.CAN-07-2153. [DOI] [PubMed] [Google Scholar]
- Broudy VC, Lin NL, Liles WC, Corey SJ, O’Laughlin B, Mou S, et al. Signaling via Src family kinases is required for normal internalization of the receptor c-Kit. Blood. 1999;94:1979–86. [PubMed] [Google Scholar]
- Carlson KM, Vignon C, Bohlander S, Martinez-Climent JA, Le Beau MM, Rowley JD. Identification and molecular characterization of CALM/AF10fusion products in T cell acute lymphoblastic leukemia and acute myeloid leukemia. Leukemia. 2000;14:100–4. doi: 10.1038/sj.leu.2401629. [DOI] [PubMed] [Google Scholar]
- Caudell D, Zhang Z, Chung YJ, Aplan PD. Expression of a CALM-AF10 fusion gene leads to Hoxa cluster overexpression and acute leukemia in transgenic mice. Cancer Res. 2007;67:8022–31. doi: 10.1158/0008-5472.CAN-06-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti R, Joly M, Corvera S. Redistribution of clathrin-coated vesicle adaptor complexes during adipocytic differentiation of 3T3-L1 cells. J Cell Biol. 1993;123:79–87. doi: 10.1083/jcb.123.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KW, Lahad JP, Kuo WL, Lapuk A, Yamada K, Auersperg N, et al. The RAB25 small GTPase determines aggressiveness of ovarian and breast cancers. Nat Med. 2004;10:1251–6. doi: 10.1038/nm1125. [DOI] [PubMed] [Google Scholar]
- Chikatsu N, Kojima H, Suzukawa K, Shinagawa A, Nagasawa T, Ozawa H, et al. ALK+, CD30-, CD20- large B-cell lymphoma containing anaplastic lymphoma kinase (ALK) fused to clathrin heavy chain gene (CLTC) Mod Pathol. 2003;16:828–32. doi: 10.1097/01.MP.0000081729.40230.1F. [DOI] [PubMed] [Google Scholar]
- Cho H, Kehrl JH. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. J Cell Biol. 2007;178:245–55. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Debernardi S, Bassini A, Jones LK, Chaplin T, Linder B, de Bruijn DR, et al. The MLL fusion partner AF10 binds GAS41, a protein that interacts with the human SWI/SNF complex. Blood. 2002;99:275–81. doi: 10.1182/blood.v99.1.275. [DOI] [PubMed] [Google Scholar]
- Deshpande AJ, Cusan M, Rawat VP, Reuter H, Krause A, Pott C, et al. Acute myeloid leukemia is propagated by a leukemic stem cell with lymphoid characteristics in a mouse model of CALM/AF10-positive leukemia. Cancer Cell. 2006;10:363–74. doi: 10.1016/j.ccr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, et al. CALM-AF10+T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19:1948–57. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- Dreyling MH, Martinez-Climent JA, Zheng M, Mao J, Rowley JD, Bohlander SK. The t(10;11)(p13;q14) in the U937 cell line results in the fusion of the AF10 gene and CALM, encoding a new member of the AP-3 clathrin assembly protein family. Proc Natl Acad Sci U S A. 1996;93:4804–9. doi: 10.1073/pnas.93.10.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyling MH, Schrader K, Fonatsch C, Schlegelberger B, Haase D, Schoch C, et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood. 1998;91:4662–7. [PubMed] [Google Scholar]
- Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–8. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Forissier S, Razanajaona D, Ay AS, Martel S, Bartholin L, Rimokh R. AF10-dependent transcription is enhanced by its interaction with FLRG. Biol Cell. 2007;99:563–71. doi: 10.1042/bc20060131. [DOI] [PubMed] [Google Scholar]
- Fu JF, Hsu JJ, Tang TC, Shih LY. Identification of CBL, a proto-oncogene at 11q23.3, as a novel MLL fusion partner in a patient with de novo acute myeloid leukemia. Genes Chromosomes Cancer. 2003;37:214–9. doi: 10.1002/gcc.10204. [DOI] [PubMed] [Google Scholar]
- Harborth J, Weber K, Osborn M. GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. J Biol Chem. 2000;275:31979–85. doi: 10.1074/jbc.M000994200. [DOI] [PubMed] [Google Scholar]
- Hopkins CR. The appearance and internalization of transferrin receptors at the margins of spreading human tumor cells. Cell. 1985;40:199–208. doi: 10.1016/0092-8674(85)90323-x. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–96. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- Kogan SC, Ward JM, Anver MR, Berman JJ, Brayton C, Cardiff RD, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100:238–45. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- Kung AL, Rebel VI, Bronson RT, Ch’ng LE, Sieff CA, Livingston DM, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–7. [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L, Di Fiore PP. Endocytosis and Cancer: an ‘Insider’ Network with Dangerous Liaisons. Traffic. 2008 doi: 10.1111/j.1600-0854.2008.00816.x. [DOI] [PubMed] [Google Scholar]
- Le Beau MM, Bitts S, Davis EM, Kogan SC. Recurring chromosomal abnormalities in leukemia in PML-RARA transgenic mice parallel human acute promyelocytic leukemia. Blood. 2002;99:2985–91. doi: 10.1182/blood.v99.8.2985. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Freund C, Conley K, Jones EW. Genetic instability of clathrin-deficient strains of Saccharomyces cerevisiae. Genetics. 1990;124:27–38. doi: 10.1093/genetics/124.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Z, Kluger Y, Greenbaum DS, Tuck D, Gerstein M, Berliner N, et al. Genomic and proteomic analysis of the myeloid differentiation program: global analysis of gene expression during induced differentiation in the MPRO cell line. Blood. 2002;100:3209–20. doi: 10.1182/blood-2002-03-0850. [DOI] [PubMed] [Google Scholar]
- Lin YH, Kakadia PM, Chen Y, Li YQ, Deshpande AJ, Buske C, et al. Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood. 2009;114:651–8. doi: 10.1182/blood-2009-03-209395. [DOI] [PubMed] [Google Scholar]
- Linder B, Newman R, Jones LK, Debernardi S, Young BD, Freemont P, et al. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J Mol Biol. 2000;299:369–78. doi: 10.1006/jmbi.2000.3766. [DOI] [PubMed] [Google Scholar]
- Meyerholz A, Hinrichsen L, Groos S, Esk PC, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–34. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- Muntau AC, Roscher AA, Kunau WH, Dodt G. The interaction between human PEX3 and PEX19 characterized by fluorescence resonance energy transfer (FRET) analysis. Eur J Cell Biol. 2003;82:333–42. doi: 10.1078/0171-9335-00325. [DOI] [PubMed] [Google Scholar]
- Okada Y, Jiang Q, Lemieux M, Jeannotte L, Su L, Zhang Y. Leukaemic transformation by CALM-AF10 involves upregulation of Hoxa5 by hDOT1L. Nat Cell Biol. 2006;8:1017–24. doi: 10.1038/ncb1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle SJ, Bright NA, Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434:1152–7. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selten G, Cuypers HT, Berns A. Proviral activation of the putative oncogene Pim-1 in MuLV induced T-cell lymphomas. EMBO J. 1985;4:1793–8. doi: 10.1002/j.1460-2075.1985.tb03852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signoret N, Oldridge J, Pelchen-Matthews A, Klasse PJ, Tran T, Brass LF, et al. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–64. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So CW, Cleary ML. Dimerization: a versatile switch for oncogenesis. Blood. 2004;104:919–22. doi: 10.1182/blood-2004-03-0992. [DOI] [PubMed] [Google Scholar]
- So CW, Lin M, Ayton PM, Chen EH, Cleary ML. Dimerization contributes to oncogenic activation of MLL chimeras in acute leukemias. Cancer Cell. 2003;4:99–110. doi: 10.1016/s1535-6108(03)00188-0. [DOI] [PubMed] [Google Scholar]
- Tebar F, Bohlander SK, Sorkin A. Clathrin assembly lymphoid myeloid leukemia (CALM) protein: localization in endocytic-coated pits, interactions with clathrin, and the impact of overexpression on clathrin-mediated traffic. Mol Biol Cell. 1999;10:2687–702. doi: 10.1091/mbc.10.8.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–82. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- Vecchi M, Polo S, Poupon V, van de Loo JW, Benmerah A, Di Fiore PP. Nucleocytoplasmic shuttling of endocytic proteins. J Cell Biol. 2001;153:1511–7. doi: 10.1083/jcb.153.7.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler DS, Engstrom LD, Alexander BM, Motto DG, Roulston D. A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes Chromosomes Cancer. 2003;36:26–36. doi: 10.1002/gcc.10136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.