Abstract
BACKGROUND
Mitochondrial DNA (mtDNA) variation has been associated with time to progression to AIDS and adverse effects from antiretroviral therapy (ART). In this study, full mitochondrial DNA (mtDNA) sequence data from U.S.-based adult participants in the AIDS Clinical Trials Group (ACTG) study 384 was used to assess associations between mtDNA variants and CD4 T cell recovery with ART.
METHODS
Full mtDNA sequence was determined using chip-based array sequencing. Sequence and CD4 cell count data was available at baseline and after ART initiation for 423 subjects with HIV RNA levels <400copies/mL plasma. The primary outcome was change in CD4 count of ≥100 cells/mm3 from baseline. Analyses were adjusted for baseline age, CD4 cell count, HIV RNA, and naïve:memory CD4 cell ratio.
RESULTS
Race-stratified analysis of mtDNA variants with a minor allele frequency >1% revealed multiple mtDNA variants marginally associated (P < 0.05 before Bonferroni correction) with CD4 cell recovery. The most significant SNP associations were those tagging the African L2 haplogroup, which was associated with a decreased likelihood of ≥100 cells/mm3 CD4 count increase at week 48 in non-Hispanic blacks (adjusted OR=0.17; 95% CI=0.06–0.53; P=0.002).
CONCLUSIONS
An African mtDNA haplogroup was associated with CD4 cell recovery after ART in this clinical trial population. These initial findings warrant replication and further investigation in order to confirm the role of mtDNA variation in CD4 cell recovery during ART.
Keywords: HIV, CD4 count, Mitochondrial DNA, Pharmacogenetics, Antiretroviral Therapy
Introduction
The CD4+ T-lymphocyte count (CD4 count) is a primary determinant of disease progression and opportunistic infection risk among HIV-infected persons. The CD4 count is also a major factor in the decision to initiate antiretroviral therapy (ART) in asymptomatic HIV-infected individuals.1 There is substantial interindividual variability in the rate and magnitude of CD4 recovery after initiating antiretroviral therapy (ART).2 Studies have demonstrated many ART-treated patients failing to attain substantial increases in CD4 count,2 and poorer CD4 count responses on ART are associated with disease progression even with adequate virologic responses.3,2
Human genetic variation appears to play a role in CD4 count recovery. An ACTG immunogenetics study suggested possible associations between CD4 count recovery ≥200 cells/mm3 after 48 weeks of ART and single nucleotide polymorphisms (SNPs) in several nuclear apoptosis-related genes and cytokine genes.4 These results also highlight the possibility that a potentially important host-related factor that influences HIV-infected CD4 cell turnover is regulation of apoptosis.5,6
Within mitochondrial DNA (mtDNA), combinations of SNPs allow for categorization of individuals into haplogroups.7 The clinical relevance of haplogroups in particular- and mtDNA variation in general- for disease risk is well described.8–14 It is plausible that functional variation in mtDNA would influence T cell dynamics such as apoptosis in response to environmental stressors (e.g. HIV infection and/or ART). We hypothesized that among individuals with control of HIV replication following initiation of ART, variation in mtDNA would influence CD4 count recovery through apoptotic regulatory and/or other mechanisms that modulate the efficiency of CD4cell proliferation. To more fully characterize relationships between mitochondrial genomics and CD4 count recovery on suppressive ART, we examined full mtDNA sequence from ART-naïve, HIV-infected participants from ACTG 384.
Methods
Study Population
ACTG 384 was a muticenter, double-blind, prospective randomized clinical trial comparing the efficacy of ART regimens consisting of three or four drugs in 980 antiretroviral-naïve adults.15 Participants were randomized to one of six treatment arms consisting of either didanosine and stavudine or zidovudine and lamivudine combined with either efavirenz, nelfinavir of both. Median follow-up time was 2.3 years. A secondary end point of ACTG 384 was change in CD4 cell count from baseline. CD4 count was measured for all individuals at 48, 96 and 144 weeks and some individuals also underwent specific flow cytometry for proportions of memory and naïve CD4 cells as part of an immunologic substudy.16 For the purposes of the current study, CD4 count at baseline and weeks 48 and 96 from 530 (54%) participants from ACTG 384 who consented to provide DNA to the ACTG Human DNA Repository (HDR)17 were considered. Previous publications18,19 have reported results from genetic association analysis in the ACTG 384 cohort, and demonstrated no significant differences between ACTG participants who provided DNA as part of the HDR and those who did not.20
Mitochondrial DNA Isolation and Sequencing
Isolation of DNA from study participants was by PUREGENE (Gentra Systems Inc., Minneapolis, MN, USA). Full mtDNA sequencing was performed using the GeneChipR Human Mitochondrial Resequencing Array v2.0 (Affymetrix, Inc., Santa Clara, CA, USA). From full-sequence data, mtDNA variants were identified using the revised Cambridge Reference Sequence (rCRS).21 Haplogroups were assigned using Hernstadt classification,22 and collapsed into higher branch haplogroups for analyses.
Exclusion Criteria
Individuals with HIV-1 RNA ≥400 copies/mL at either week 48 (n =77) or 96 (n = 130) were excluded from analyses for the respective time points. Three (<1%) univariate outliers in CD4 cell count change at week 48 were identified with STATA using the minimum volume ellipsoid method described by Hadi23 and removed from analyses of week 48 endpoints. Seven (1.3%) participants lacked 96 week CD4 count data and were removed only from week 96 analyses. For the purposes of genetic data quality control (QC) and subsequent analysis, study participants were stratified by race/ethnicity into self-identified non-Hispanic white, non-Hispanic black, and Hispanic groups to prevent potential confounding. Self-described Asian (n = 11) and Native American participants (n = 2) were excluded due to small sample size. We excluded mitochondrial DNA SNPs with minor allele frequency <1% in the stratified racial subpopulation or genotyping completion rate <95%. We excluded samples with genotyping completion rate <90% (n = 4). Genetic quality control was performed with the Whole-genome Association Study Pipeline (WASP) tool (https://chgr.mc.vanderbilt.edu/plato) inside of the Platform for the Analysis, Translation and Organization of large-scale data (PLATO)24.
Statistical Analysis
For analyses of binary endpoints in this study, a nested case-cohort design was utilized by adjusting for age.25 The primary outcome was the proportion of subjects with an increase of ≥100 CD4 cells/mm3 at week 48. Secondary outcomes included increase of ≥100 CD4 cells/mm3 at week 96, increase of ≥50 CD4 cells/mm3 at week 48 and 96, increase of ≥200 CD4 cells/mm3 at week 48 and 96, and comparison of those participants with an increase of ≥200 CD4 cells/mm3 against participants with an increase of <50 CD4 cells/mm3 at week 48 and 96. CD4 count change was also analyzed as a continuous endpoint.
Analyses of the effect of SNPs on CD4 cell count changes utilized the PLINK tool 26 (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml) to perform logistic and linear regression. Race-stratified regression analyses were performed, adjusting for clinical covariates previously found to be significantly associated with CD4 cell recovery in ACTG 384,16 including age, baseline viral load, baseline CD4 cell count, and baseline ratio of naïve-to-memory CD4 cell count. In the primary analysis of ACTG 384, cART regimen was not found to be a significant predictor of CD4 cell recovery following ART initiation and thus was not adjusted for during analysis.15 Haplogroup analysis was performed in STATA (StataCorp. 2007. Stata Statistical Software: Release 10. College Station, TX: StataCorp LP). Statistical association between haplogroup status and CD4 count change from baseline was analyzed in a race-stratified design using a single indicator variable for each haplogroup. With this design, likelihood of CD4 count increase from baseline ≥100 cells/mm3 at 48 weeks of follow-up within each haplogroup was compared with the likelihood of ≥100 cells/mm3 increase within all other haplogroups. Haplogroup analyses adjusted for the same covariates used in tests of association with individual mtDNA variants.
A Bonferroni multiple-test correction was used to determine the threshold necessary for tests of association to be considered significant. For mtDNA variant analysis, an average of 200 tests were assumed based on the presence of 213, 151 and 216 polymorphisms in the non-Hispanic black, non-Hispanic white and Hispanic self-identified race/ethnicity groups, respectively. For haplogroup analysis, a Bonferroni-corrected threshold was calculated using the 23 haplogroups tested for association with the primary outcome (P threshold<2.2 × 10−3).
Multiple imputation with the mice package27 available in R was used to impute missing values of the naïve-memory ratio conditional on CD4 cell count change, baseline viral load, age and genotype. Imputed values of naïve-memory ratio were not used during analysis due to subsequent discovery of non-random patterns of missingness within the data which violate assumptions of mice.
Results
Baseline Demographics
Baseline demographics of study subjects are shown in Table 1. The median age of individuals remaining after QC and exclusion criteria (n=423) was 36, with 17% being female, 50% self-identifying as non-Hispanic white, 126 (30%) as non-Hispanic black, and 20% as Hispanic. Median baseline CD4 count was 278 cells/mm3 (interquartile range [IQR] 85-434); log10 HIV-1 copies/mL was 5.0 (IQR 4.3–5.5). Baseline naïve-to-memory cell ratio was available for 66% of 423 participants, with a median of 0.59 (IQR 0.28–1.00) among these participants. No demographic data differed significantly before and after QC exclusions.
Table 1.
Baseline demographics for those individuals with DNA in the ACTG Human DNA Repository, for those who passed QC and exclusion criteria, and for case and control groups for the primary analyses.
| Participants with DNA (N=530) | Eligible Participants Passing QC (N= 423) | Increase ≥100 cells at week 48 (N=308) | Increase <100 cells at week 48 (N=115) | P-valuea | |
|---|---|---|---|---|---|
| Median age in years (range) | 36 (17–72) | 36 (17–72) | 35 (17–72) | 40 (20–65) | 0.005 |
| Female sex- N (%) | 93 (18) | 74 (17) | 51 (17) | 23 (20) | 0.47 |
| Race/ethnicity- N (%)b | |||||
| Non-Hispanic White | 258 (49) | 212 (50) | 165 (54) | 47 (41) | 0.06 |
| Non-Hispanic Black | 163 (31) | 126 (30) | 84 (27) | 42 (37) | |
| Hispanic | 96 (18) | 85 (20) | 59 (19) | 26 (23) | |
| Asian and Native American | 13 (2) | 0 | 0 | 0 | |
| Randomized NRTIs-N (%)b | 0.51 | ||||
| ddI+d4T | 280 (53) | 217 (51) | 161 (52) | 56 (49) | |
| ZDV+3TC | 250 (47) | 206 (49) | 147 (48) | 59 (51) | |
| Randomized NNRTI/PI- N (%)b | 0.27 | ||||
| Efavirenz | 166 (31) | 133 (31) | 98 (32) | 35 (30) | |
| Nelfinavir | 173 (33) | 133 (31) | 98 (32) | 35 (30) | |
| Efavirenz+nelfinavir | 191 (36) | 157 (37) | 112 (36) | 45 (39) | |
| CD4 count/mm3-median (IQR) | 275 (85–429) | 278 (85–434) | 261 (67–396) | 342 (190–472) | 0.001 |
| Naïve-to-memory CD4+ lymphocyte ratio- median (IQR) | 0.58 (.26–1.00)c | 0.59 (0.28–1.00)d | 0.62 (0.32–1.09) | 0.46 (0.18–0.72) | 0.003 |
| HIV-1 log10 RNA copies/mL plasma-median (IQR) | 5.00 (4.37–5.55) | 4.95 (4.32–5.53) | 5.13 (4.54–5.64) | 4.57 (4.03–5.15) | <0.001 |
For participants who passed quality control, differences in baseline demographics are compared between those with a CD4 count increase of ≥100 cells/mm3 at 48 weeks (cases for primary endpoint) and those with a CD4 count increase of <100 cells at 48 weeks (controls). P-values from unadjusted Wilcoxon rank-sum, Fisher’s exact or Chi-squared tests.
Totals may not equal 100% due to rounding.
N= 359;
N=281
3TC=l lamivudine; ddI= didanosine; d4T= stavudine; IQR= interquartile range; NRTIs= nucleoside reverse transcripates inhibitors; NNRTI= non-NRTI; PI= protease inhibitor; QC= quality control; ZDV= zidovudine.
Analysis of Change in CD4 Count at Week 48
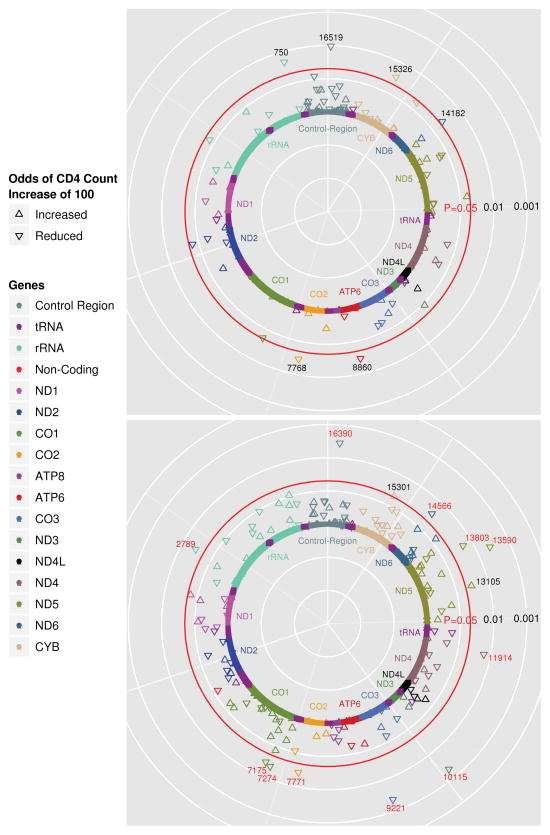
Median CD4 count increase from baseline was 176 cells/mm3 (IQR 94-273) at 48 weeks and 253 cells/mm3 (IQR 138-387) at 96 weeks. The primary study endpoint focused on differences in mtDNA variation between individuals experiencing an increase of ≥100 cells/mm3 (cases) and those with <100 cells/mm3 change (controls) at 48 weeks of follow up, analyzed separately within strata defined by race/ethnicity. Analysis within the non-Hispanic black (42 controls/82 cases), non-Hispanic white (47 controls/164 cases), and Hispanic (26 controls/59 cases) race/ethnicity stratified groups included tests of association with 213, 151, and 216 SNPs respectively due to the differences in genetic variability between the three race/ethnicity groups. All tests used the major allele as the reference. No associations were significant at the P-value threshold determined by Bonferroni correction for multiple comparisons (P<2.5×10−4) in any of the race/ethnicity groups but analysis revealed multiple marginal associations (P<0.05) in analyses of the non-Hispanic white and non-Hispanic black race/ethnicity groups (Table 3). Figures 1a and 1b display negative log-transformed P-values for tests of association with the primary endpoint in individuals self-identifying as non-Hispanic white and non-Hispanic black, respectively. The association test P-values are plotted with respect to base-pair location of the mtDNA variant along the mitochondrial genome. The most significant associations with CD4 count change at 48 weeks were observed within the analysis of non-Hispanic black participants. Polymorphisms at mitochondrial base pair positions 2789, 7175, 7274, 7771, 9221, 10115, 11914, 13590, 13803, 14566 and 16390 (OR range=0.17–0.24, P-value range=0.002–0.027) define haplogroup L2 and were associated with decreased odds of a CD4 count change of ≥100 cells/mm3 in the non-Hispanic black group. Variation at positions 13105 (OR=4.60; 95% CI=1.13–18.70; P=0.033) and 15301 (3.64; 1.02–13.06; P=0.047) were associated with increased odds of a CD4 count change ≥100 cells/mm3 in non-Hispanic blacks. These two polymorphisms define the L2’3’4’6 branch above the L2 haplogroup and associations are to the allele which is prior to this branch.28 As such, the allele which is present within L2 individuals is associated with reduced odds of a CD4 count change ≥100. Polymorphisms at positions 750 (OR=0.04; 95% CI=0.002–0.60; P=0.02), 7768 (0.10; 0.01–0.80; P=0.03), 8860 (0.02; 0.001–0.68; P=0.03), 14182 (0.13; 0.02–0.92; P=0.04), 15326 (0.02; 0.001–0.68; P=0.03) and 16519 (0.24; 0.08–0.71; P=0.01) were associated with reduced odds of CD4 cell recovery in non-Hispanic white individuals. Polymorphism at positions 750, 8860 and 15326 is representative of the H2a2 haplogroup and is typically rare but is likely represented due to the large number of individuals of the H2 haplogroup (N=26) present in the study.
Table 3.
Significant SNP results (P≤0.05) from adjusted logistic regression analysis of discrete outcome ≥ or < 100 cell/mm3 increase in CD4 count at week 48.
| Base position | Gene/Region | MAF | Race/ethnicity | OR | 95% CI | P-value |
|---|---|---|---|---|---|---|
| 2789 | rRNA | 0.194 | NHB | 0.24 | 0.07, 0.85 | 0.03 |
| 7175 | CO1 | 0.194 | NHB | 0.24 | 0.07, 0.85 | 0.03 |
| 7274 | CO1 | 0.187 | NHB | 0.23 | 0.07, 0.82 | 0.02 |
| 7771 | CO2 | 0.194 | NHB | 0.24 | 0.07, 0.85 | 0.03 |
| 9221 | CO3 | 0.315 | NHB | 0.17 | 0.06, 0.53 | 0.002 |
| 10115 | ND3 | 0.306 | NHB | 0.17 | 0.06, 0.52 | 0.002 |
| 11914 | ND4 | 0.268 | NHB | 0.24 | 0.07, 0.76 | 0.02 |
| 13105 | ND5 | 0.333 | NHB | 4.60 | 1.13, 18.70 | 0.03 |
| 13590 | ND5 | 0.312 | NHB | 0.19 | 0.06, 0.58 | 0.004 |
| 13803 | ND5 | 0.187 | NHB | 0.22 | 0.06, 0.78 | 0.02 |
| 14566 | ND6 | 0.194 | NHB | 0.24 | 0.07, 0.85 | 0.03 |
| 15301 | CYB | 0.369 | NHB | 3.64 | 1.02, 13.06 | 0.05 |
| 16390 | CR | 0.311 | NHB | 0.19 | 0.06, 0.58 | 0.004 |
| 750 | rRNA | 0.029 | NHW | 0.037 | 0.002, 0.60 | 0.02 |
| 8860 | ATP6 | 0.053 | NHW | 0.023 | 0.0008, 0.68 | 0.03 |
| 15326 | CYB | 0.024 | NHW | 0.023 | 0.0008, 0.68 | 0.03 |
| 16519 | CR | 0.053 | NHW | 0.24 | 0.08, 0.71 | 0.01 |
CI= confidence interval; CR= control region; MAF= minor allele frequency; NHW= non-Hispanic white; NHB= non-Hispanic black; OR= odds ratio; SNP= single nucleotide polymorphism. See figure legend for gene/region abbreviation listing.
Models adjusted for age, baseline viral load, baseline CD4 cell count, and baseline ratio of naïve-to-memory CD4 cell count.
Fig. 1.
The results of a SNP analysis using the entire distribution of CD4 count change as a continuous outcome variable are shown in Supplemental Digital Content Table S1. The majority of mtDNA variants which were found to be significant in analysis of the primary 48 week endpoint remained significant in this analysis with exception of the variant at position 15301 among Non-Hispanic blacks and variants at positions 750, 8860 and 15326 among non-Hispanic whites, all of which were not significant in analyses of continuous CD4 changes at week 48. There were additional polymorphisms which were found to be marginally significant from the analysis of the continuous CD4 count change at week 48 among non-Hispanic black participants, including positions 2768, 3594 and 14178. In addition, many variants were found to be marginally significant among Hispanic participants, although it appears that most of these SNPs are highly correlated.
CD4 Count Change From Baseline Through 96 weeks
Of the 423 individuals who had data for one or more time points after 48 week exclusion criteria, 370 (87%) were included in secondary 96 week analyses. The decreased sample size between 48 and 96 week time points is due largely to exclusion of those participants with an increase in viral load to above 400 copies/mL on study. Results from analysis of 96 week endpoints were similar to results from analysis of the week 48 primary endpoint and results from the analysis of quantitative CD4 count change (data not shown).
Haplogroup Analysis
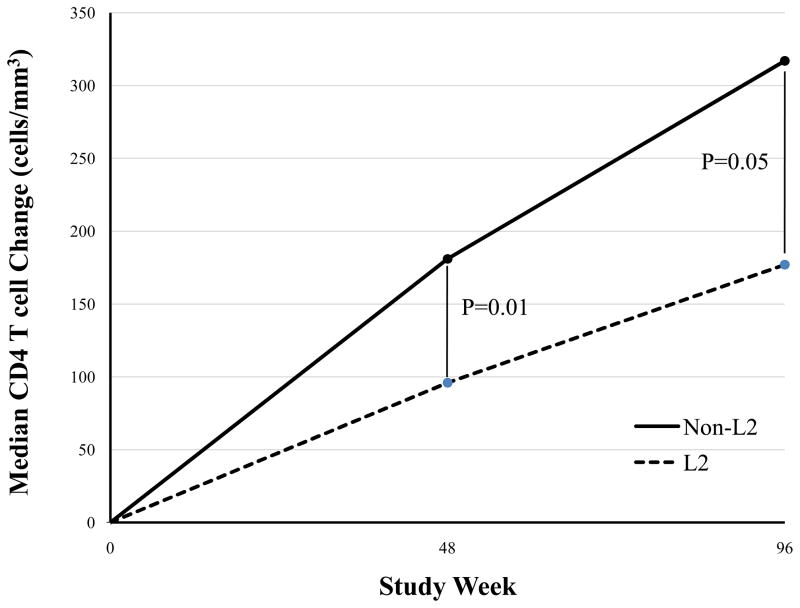
A total of 23 haplogroups were tested for association with a CD4 count increase ≥100 cells/mm3 at 48 weeks of follow-up (A complete listing of haplogroups is shown in Table 2). The Non-Hispanic black participants in this study fall into three major haplogroups, L1 (25%), L2, (32%), and L3 (29%). Analysis of the L2 haplogroup revealed an association with decreased odds (OR=0.17; 95% CI=0.06–0.53; P=0.002) of a CD4 count change of ≥100 cells/mm3. This association remains significant after correcting for the number of haplogroups tested in the association analysis (P<2.2 × 10−3). Among non-Hispanic blacks, those belonging to haplogroup L2 has significantly lower median CD4 T cell increases at study week 48 (96 [IQR 30-252] cells/mm3 vs. 181 [105–285]; P=0.01) and week 96 (177 [120–395] vs. 317 [IQR 173-433]; P=0.05). Haplogroup L2 also remained associated with continuous CD4 count increase at week 48 (adjusted β coefficient= −119.5; 95% CI −187.3,−51.3; P=0.001) after adjustment for covariates noted previously. Haplogroups H1 (13%), H2 (22%), J (13%), K (7%) and U (14%) accounted for the majority of non-Hispanic white participants in this study. Three non-Hispanic white participants were excluded from haplogroup analyses due to inability to assign a haplogroup based on Herrnstadt classification. No tests of association were found to be significant within non-Hispanic whites at the Bonferroni corrected P-value threshold despite the marginal associations observed with polymorphisms indicating the H2a2 sub-haplogroup. Hispanics in our study fell primarily into the A (29%), H2 (11%) and L3 (36%) haplogroups. With such small sample size, statistical power to detect association with Hispanic haplogroups was low and no marginal associations were observed with CD4 count increase.
Table 2.
Haplogroup distributions, overall and by race/ethnicity
| Total (N=423) | Non-Hispanic White (N=212) | Non-Hispanic Black (N=126) | Hispanic (N=85) | |||||
|---|---|---|---|---|---|---|---|---|
| Haplogroupa | N | % | N | % | N | % | N | % |
| A | 31 | 7.33 | 2 | 0.94 | 4 | 3.17 | 25 | 29.41 |
| H | 10 | 2.36 | 9 | 4.25 | 1 | 0.79 | 0 | 0 |
| H1 | 30 | 7.09 | 28 | 13.21 | 0 | 0 | 2 | 2.35 |
| H2 | 59 | 13.95 | 46 | 21.7 | 4 | 3.17 | 9 | 10.59 |
| H3 | 8 | 1.89 | 8 | 3.77 | 0 | 0 | 0 | 0 |
| I | 4 | 0.95 | 3 | 1.42 | 1 | 0.79 | 0 | 0 |
| I1 | 2 | 0.47 | 2 | 0.94 | 0 | 0 | 0 | 0 |
| I2 | 1 | 0.24 | 1 | 0.47 | 0 | 0 | 0 | 0 |
| J1 | 25 | 5.91 | 21 | 9.91 | 2 | 1.59 | 2 | 2.35 |
| J2 | 7 | 1.65 | 6 | 2.83 | 0 | 0 | 1 | 1.18 |
| K | 1 | 0.24 | 1 | 0.47 | 0 | 0 | 0 | 0 |
| K1 | 10 | 2.36 | 8 | 3.77 | 0 | 0 | 2 | 2.35 |
| K1a | 1 | 0.24 | 1 | 0.47 | 0 | 0 | 0 | 0 |
| K2 | 7 | 1.65 | 7 | 3.3 | 0 | 0 | 0 | 0 |
| L1/L2 | 4 | 0.95 | 1 | 0.47 | 2 | 1.59 | 1 | 1.18 |
| L1a | 7 | 1.65 | 0 | 0 | 7 | 5.56 | 0 | 0 |
| L1b | 14 | 3.31 | 1 | 0.47 | 10 | 7.94 | 3 | 3.53 |
| L1c | 15 | 3.55 | 0 | 0 | 14 | 11.11 | 1 | 1.18 |
| L2a | 28 | 6.62 | 0 | 0 | 25 | 19.84 | 3 | 3.53 |
| L2b | 19 | 4.49 | 1 | 0.47 | 15 | 11.9 | 3 | 3.53 |
| L3 | 42 | 9.93 | 3 | 1.42 | 16 | 12.7 | 23 | 27.06 |
| L3b | 20 | 4.73 | 2 | 0.94 | 12 | 9.52 | 6 | 7.06 |
| L3e | 11 | 2.6 | 1 | 0.47 | 8 | 6.35 | 2 | 2.35 |
| T1 | 6 | 1.42 | 5 | 2.36 | 1 | 0.79 | 0 | 0 |
| T2 | 2 | 0.47 | 2 | 0.94 | 0 | 0 | 0 | 0 |
| T2b | 7 | 1.65 | 5 | 2.36 | 1 | 0.79 | 1 | 1.18 |
| U | 1 | 0.24 | 1 | 0.47 | 0 | 0 | 0 | 0 |
| U2 | 3 | 0.71 | 3 | 1.42 | 0 | 0 | 0 | 0 |
| U4 | 7 | 1.65 | 7 | 3.3 | 0 | 0 | 0 | 0 |
| U5 | 4 | 0.95 | 4 | 1.89 | 0 | 0 | 0 | 0 |
| U5a | 6 | 1.42 | 6 | 2.83 | 0 | 0 | 0 | 0 |
| U5a1 | 8 | 1.89 | 8 | 3.77 | 0 | 0 | 0 | 0 |
| U5b | 1 | 0.24 | 1 | 0.47 | 0 | 0 | 0 | 0 |
| U6 | 3 | 0.71 | 0 | 0 | 2 | 1.59 | 1 | 1.18 |
| U9 | 3 | 0.71 | 2 | 0.94 | 1 | 0.79 | 0 | 0 |
| V | 4 | 0.95 | 4 | 1.89 | 0 | 0 | 0 | 0 |
| W | 4 | 0.95 | 4 | 1.89 | 0 | 0 | 0 | 0 |
| W3 | 2 | 0.47 | 2 | 0.94 | 0 | 0 | 0 | 0 |
| X1 | 3 | 0.71 | 3 | 1.42 | 0 | 0 | 0 | 0 |
| No-callsb | 3 | 0.71 | 3 | 1.42 | 0 | 0 | 0 | 0 |
Haplogroups assigned using methods described by Herrnstadt, et al.22
Subjects unable to be classified.
Discussion
In this study, we examined the effect of mtDNA variation on CD4 cell recovery during ART. Race-stratified analysis of mtDNA variants suggested possible associations between the African L2 haplogroup and reduced magnitude of CD4 cell recovery during ART in non-Hispanic black participants, although no single SNP association withstood correction for multiple testing. Significant correlation structure exists between mtDNA variants however, forming the basis for mitochondrial haplogroups, and an association with the L2 haplogroup was found which is significant after considering the number of haplogroups analyzed. The association was seen in analyses of a discrete outcome of 100 cell/mm3 increase at 48 weeks, and with CD4 count increase as a continuous outcome. It is unlikely that this haplogroup association is the result of an effect of race/ethnicity, as race/ethnicity was not found to be associated with CD4 cell count change in our study or in that of the original immunologic study of ACTG 384.16 It is also unlikely that associations were related to ART toxicity, virologic failure of treatment, and/or non-adherence, as we only included participants with suppressed HIV-1 RNA at the time point used for analyses.
Associations with other mtDNA SNPs implicate a role for variation not directly related to haplogroup. The role of these variations is not immediately apparent, particularly because the majority of mtDNA variants observed to have a significant association with CD4 count recovery are synonymous (i.e. do not change amino acid sequence). The exception is the 13105 variant, which causes a conservative isoleucine-to-valine change in the ND5 complex I protein. It will be necessary to assess CD4 recovery and mtDNA variation in larger cohorts to replicate these associations, and in model systems to characterize their functional effects.
Limitations of our data include small sample sizes which may have impaired our ability to detect associations in other racial/ethnic populations, use of self-identified race/ethnicity instead of genetic ancestry for stratification, missing data on naïve-to-memory cell ratio, and an incomplete understanding of the biologic rationale for the observed genomic associations. There are multiple potential mechanisms through which variation in mtDNA could alter CD4 cell count. The role of the mitochondrion as an apoptotic regulator has been increasingly recognized, and its specific importance in HIV-infected CD4+ lymphocytes has been studied.29 In a recent study, PBMCs from HIV-infected long-term non-progressors demonstrated less mitochondrial dysfunction and mitochondrially-mediated apoptosis.30 Published data on the effects of specific mtDNA variation on apoptosis include a murine model of mtDNA depletion31 and small studies of patients with mtDNA tRNA point mutations showing increases32 or no increases33 in TUNEL-positive staining in muscle fibers. An additional study in fibroblasts demonstrated massive ROS-induced apoptosis in the presence of a mtDNA point mutation.34
Hendrickson, et al. reported associations between European haplogroups J and U5a and increased pre-ART progression to AIDS and CD4 count <200 cells/mm3 among a Caucasian, HIV-infected cohort; 97% of whom were male.35 These findings are also consistent with a role for mtDNA variation in CD4 T cell dynamics, but different associations in our study may be due to a smaller population of persons of European descent and/or different phenotypes: disease progression prior to ART and CD4 count recovery during ART may differ, even if similar mechanisms (e.g. apoptosis regulation) play a role in both. The current study provides insight into the contribution of mitochondrial genomics to CD4 count recovery in individuals of non-European descent. Further study in larger populations is necessary to more definitively determine the role of mtDNA variation in CD4 T cell dynamics.
Supplementary Material
Fig. 2.
Acknowledgments
The authors gratefully acknowledge the many HIV-infected patients who participated in ACTG study 384 and protocol A5128. We also acknowledge Laura Smeaton, Diana Ventura, and Roy Matining (Harvard School of Public Health) for invaluable assistance with clinical data from ACTG study 384, and Melanie Robinson in the Vanderbilt Microarray Shared Resource Core Facility.
Additional members of the ACTG 384 study leadership team included: Victor De Gruttola (Harvard School of Public Health), Sally Snyder, Thomas Nevin (Social & Scientific Systems), Carla Pettinelli (National Institutes of Health), Michael Dube (Indiana University), Margaret Fischl (Miami University), Richard Pollard (Univ. California-Davis), Robert Delaphna (Howard University), Linda Gideon (Frontier Science and Technology Foundation), Charles van der Horst (Univ. of North Carolin-Chapel Hill), Robert Murphy (Northwestern University), Mark Becker (Agouron), Richard D’Aquila (Vanderbilt University), Stefano Vella (Istituto Superiore de Sanita), Thomas Merigan (Stanford University) and Martin Hirsch (Harvard Medical School).
Funding support:
Funding for the genomic sequencing was provided by the National Institutes of Health (NIH)/National Institute of Neurological Diseases and Stroke grant NS059330. The project described was also supported by Award Number U01AI068636 from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. AIDS Clinical Trials Group (ACTG) Study 384 was also supported by grant AI38858, and by Agouron/Pfizer, Bristol Myers Squibb, and GlaxoSmithKline. The ACTG sites contributing DNA for these analyses were funded by NIH grants AI069513, AI34835, AI069432, AI069423, AI069477, AI069501, AI069474, AI069428, AI69467, AI069415, Al32782, AI27661, AI25859, AI069495, AI069471, AI069532, AI069452, AI069450, AI069556, AI069484, AI069472, AI34853, AI069465, AI069511, AI38844, AI069424, AI069434, AI46370, AI069502, and AI069419. The ACTG Human DNA Repository is also supported in part by the Vanderbilt CTSA grant UL1 RR024975 from NCRR/NIH. Additional NIH grant support included: AI077505, AI54999, HL087726, AI69495, 5T32GM80178 and AI062435.
Footnotes
These data were presented at the 16th Annual Meeting of the International Genetic Epidemiology Society, Boston, MA, October 2010 [Abstract #183] and at the 60th Annual Meeting of the American Society for Human Genetics, Washington D.C., November 2010 [Abstract #2412].
Potential conflicts of interest:
G.K.R. has received research support from Gilead Sciences, Schering-Plough, and served as a consultant for Abbott Laboratories and Boehringer-Ingelheim.
D.W.H. has received research grants from Boehringer-Ingelheim, Bristol-Myers Squibb, Gilead Sciences, and Merck, and has served as a consultant for Boehringer-Ingelheim.
T.H. has received research funding from Merck.
M.D.R. has served as a consultant for Boehringer-Ingelheim.
B.J.G., D.C.S., D.S., J.A.C., R.P., R.S., S.A.K., D.G.M.: No conflicts.
References
- 1.Kitahata MM, Gange SJ, Abraham AG, et al. Effect of early versus deferred antiretroviral therapy for HIV on survival. N Engl J Med. 2009;360:1815–1826. doi: 10.1056/NEJMoa0807252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutierrez F, Padilla S, Masia M, et al. Clinical outcome of HIV-infected patients with sustained virologic response to antiretroviral therapy: long-term follow-up of a multicenter cohort. PLoS One. 2006;1:e89. doi: 10.1371/journal.pone.0000089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grabar S, Le MV, Goujard C, et al. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann Intern Med. 2000;133:401–410. doi: 10.7326/0003-4819-133-6-200009190-00007. [DOI] [PubMed] [Google Scholar]
- 4.Haas DW, Geraghty DE, Andersen J, et al. Immunogenetics of CD4 Lymphocyte Count Recovery during Antiretroviral Therapy: An AIDS Clinical Trials Group Study. J Infect Dis. 2006;194:1098–1107. doi: 10.1086/507313. [DOI] [PubMed] [Google Scholar]
- 5.Petit F, Arnoult D, Viollet L, et al. Intrinsic and extrinsic pathways signaling during HIV-1 mediated cell death. Biochimie. 2003;85:795–811. doi: 10.1016/j.biochi.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Varbanov M, Espert L, Biard-Piechaczyk M. Mechanisms of CD4 T-cell depletion triggered by HIV-1 viral proteins. AIDS Rev. 2006;8:221–236. [PubMed] [Google Scholar]
- 7.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–230. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 8.van der Walt J, Nicodemus KK, Martin E, et al. Mitochondrial Polymorphisms Significantly Reduce the Risk of Parkinson Disease. Am J Hum Genet. 2003 doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Autere J, Moilanen JS, Finnila S, et al. Mitochondrial DNA polymorphisms as risk factors for Parkinson’s disease and Parkinson’s disease dementia. Hum Genet. 2004;115:29–35. doi: 10.1007/s00439-004-1123-9. [DOI] [PubMed] [Google Scholar]
- 10.Castro MG, Huerta C, Reguero JR, et al. Mitochondrial DNA haplogroups in Spanish patients with hypertrophic cardiomyopathy. Int J Cardiol. 2006;112:202–206. doi: 10.1016/j.ijcard.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 11.De BG, Rose G, Carrieri G, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 12.Niemi AK, Hervonen A, Hurme M, et al. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 13.Canter JA, Robbins GK, Selph D, et al. African mitochondrial DNA subhaplogroups and peripheral neuropathy during antiretroviral therapy. J Infect Dis. 2010;201:1703–1707. doi: 10.1086/652419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulgan T, Haubrich R, Riddler SA, et al. European mitochondrial DNA haplogroups and metabolic changes during antiretroviral therapy in AIDS Clinical Trials Group Study A5142. AIDS. 2011;25:37–47. doi: 10.1097/QAD.0b013e32833f9d02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins GK, De GV, Shafer RW, et al. Comparison of sequential three-drug regimens as initial therapy for HIV-1 infection. N Engl J Med. 2003;349:2293–2303. doi: 10.1056/NEJMoa030264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi RT, Spritzler J, Chan E, et al. Effect of baseline- and treatment-related factors on immunologic recovery after initiation of antiretroviral therapy in HIV-1-positive subjects: results from ACTG 384. J Acquir Immune Defic Syndr. 2006;42:426–434. doi: 10.1097/01.qai.0000226789.51992.3f. [DOI] [PubMed] [Google Scholar]
- 17.Haas DW, Wilkinson GR, Kuritzkes DR, et al. A multi-investigator/institutional DNA bank for AIDS-related human genetic studies: AACTG Protocol A5128. HIV Clin Trials. 2003;4:287–300. doi: 10.1310/MUQC-QXBC-8118-BPM5. [DOI] [PubMed] [Google Scholar]
- 18.Kallianpur AR, Hulgan T, Canter JA, et al. Hemochromatosis (HFE) gene mutations and peripheral neuropathy during antiretroviral therapy. AIDS. 2006;20:1503–1513. doi: 10.1097/01.aids.0000237366.56864.3c. [DOI] [PubMed] [Google Scholar]
- 19.Motsinger AA, Ritchie MD, Shafer RW, et al. Multilocus genetic interactions and response to efavirenz-containing regimens: an adult AIDS clinical trials group study. Pharmacogenet Genomics. 2006;16:837–845. doi: 10.1097/01.fpc.0000230413.97596.fa. [DOI] [PubMed] [Google Scholar]
- 20.Hulgan T, Haas DW, Haines JL, et al. Mitochondrial haplogroups and peripheral neuropathy during antiretroviral therapy: an adult AIDS clinical trials group study. AIDS. 2005;19:1341–1349. doi: 10.1097/01.aids.0000180786.02930.a1. [DOI] [PubMed] [Google Scholar]
- 21.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 22.Herrnstadt C, Elson JL, Fahy E, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hadi AS. Identifying Multiple Outliers in Multivariate Data. Journal of the Royal Statistical Society. 1992;54:761–771. [Google Scholar]
- 24.Grady BJ, Torstenson E, Dudek SM, et al. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac Symp Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 25.Little J, Sharp L, Khoury MJ, et al. The epidemiologic approach to pharmacogenomics. Am J Pharmacogenomics. 2005;5:1–20. doi: 10.2165/00129785-200505010-00001. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Buuren S, Oudshoorn C. mice: Multivariate Imputation by Chained Equations library. 2010. [Google Scholar]
- 28.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 29.Garg H, Blumenthal R. HIV gp41-induced apoptosis is mediated by caspase-3-dependent mitochondrial depolarization, which is inhibited by HIV protease inhibitor nelfinavir. J Leukoc Biol. 2006;79:351–362. doi: 10.1189/jlb.0805430. [DOI] [PubMed] [Google Scholar]
- 30.Peraire J, Miro O, Saumoy M, et al. HIV-1-infected long-term non-progressors have milder mitochondrial impairment and lower mitochondrially-driven apoptosis in peripheral blood mononuclear cells than typical progressors. Curr HIV Res. 2007;5:467–473. doi: 10.2174/157016207781662452. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, Silva JP, Gustafsson CM, et al. Increased in vivo apoptosis in cells lacking mitochondrial DNA gene expression. Proc Natl Acad Sci U S A. 2001;98:4038–4043. doi: 10.1073/pnas.061038798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mirabella M, Di GS, Silvestri G, et al. Apoptosis in mitochondrial encephalomyopathies with mitochondrial DNA mutations: a potential pathogenic mechanism. Brain. 2000;123 (Pt 1):93–104. doi: 10.1093/brain/123.1.93. [DOI] [PubMed] [Google Scholar]
- 33.Sciacco M, Fagiolari G, Lamperti C, et al. Lack of apoptosis in mitochondrial encephalomyopathies. Neurology. 2001;56:1070–1074. doi: 10.1212/wnl.56.8.1070. [DOI] [PubMed] [Google Scholar]
- 34.Geromel V, Kadhom N, Cebalos-Picot I, et al. Superoxide-induced massive apoptosis in cultured skin fibroblasts harboring the neurogenic ataxia retinitis pigmentosa (NARP) mutation in the ATPase-6 gene of the mitochondrial DNA. Hum Mol Genet. 2001;10:1221–1228. doi: 10.1093/hmg/10.11.1221. [DOI] [PubMed] [Google Scholar]
- 35.Hendrickson SL, Hutcheson HB, Ruiz-Pesini E, et al. Mitochondrial DNA haplogroups influence AIDS progression. AIDS. 2008;22:2429–2439. doi: 10.1097/QAD.0b013e32831940bb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.