Abstract
Human adipose-derived stem cells (hASC) have shown great potential for bone tissue engineering. However, the molecular mechanisms underlying this potential are not yet known, in particular the separate and combined effects of three-dimensional (3D) culture and mechanical loading on hASC osteogenesis. Mechanical stimuli play a pivotal role in bone formation, remodeling, and fracture repair. To further understand hASC osteogenic differentiation and response to mechanical stimuli, gene expression profiles of proliferating or osteogenically induced hASC in 3D collagen I culture in the presence and absence of 10% uniaxial cyclic tensile strain were examined using microarray analysis. About 847 genes and 95 canonical pathways were affected during osteogenesis of hASC in 3D culture. Pathway analysis indicated the potential roles of Wnt/β-catenin signaling, bone morphogenic protein (BMP) signaling, platelet-derived growth factor (PDGF) signaling, and insulin-like growth factor 1 (IGF-1) signaling in hASC during osteogenic differentiation. Application of 10% uniaxial cyclic tensile strain suggested synergistic effects of strain with osteogenic differentiation media on hASC osteogenesis as indicated by significantly increased calcium accretion of hASC. There was no significant further alteration in the four major pathways (Wnt/β-catenin, BMP, PDGF, and IGF-1). However, 184 transcripts were affected by 10% cyclic tensile strain. Function and network analysis of these transcripts suggested that 10% cyclic tensile strain may play a role during hASC osteogenic differentiation by upregulating two crucial factors in bone regeneration: (1) proinflammatory cytokine regulators interleukin 1 receptor antagonist and suppressor of cytokine signaling 3; (2) known angiogenic inductors fibroblast growth factor 2, matrix metalloproteinase 2, and vascular endothelial growth factor A. This is the first study to investigate the effects of both 3D culture and mechanical load on hASC osteogenic differentiation. A complete microarray analysis investigating both the separate effect of soluble osteogenic inductive factors and the combined effects of chemical and mechanical stimulation was performed on hASC undergoing osteogenic differentiation. We have identified specific genes and pathways associated with mechanical response and osteogenic potential of hASC, thus providing significant information toward improved understanding of our use of hASC for functional bone tissue engineering applications.
Introduction
Autologous stem cell-based bone tissue engineering holds great potential for treating bone trauma and pathologies in a patient specific manner. Adult stem cells can be derived from various source tissues such as bone marrow, epidermal tissue, and adipose tissue.1,2 Initially, bone marrow-derived mesenchymal stem cells (MSCs) received the most attention for bone tissue engineering applications given their known osteogenic capability. More recently, however, adipose-derived stem cells (ASC) have received increasing interest for tissue engineering applications owing to their relative ease of harvest, abundance, and multilineage differentiation potential.2–4
When cultured in monolayer in the presence of osteogenic media containing ascorbic acid, β-glycerolphosphate, and dexamethasone, ASC have been shown to undergo osteogenic differentiation, deposit calcium phosphate, and express osteoblast-associated gene markers, osteocalcin, alkaline phosphatase (ALP), and osteopontin in vitro.3,5 To better mimic the in vivo environment, researchers have also utilized three-dimensional (3D) culture conditions. Culture of human ASC (hASC) in 3D collagen gels has been found to promote osteogenic differentiation of hASC by elevating bone marker mRNA expression.6 As with 3D culture, the mechanical environment also plays an important role in stem cell growth, differentiation, and function.
At the tissue level, predominant mechanical stimuli in bone include fluid shear stress and tensile strain.7 In a mechanobiological investigation of mandibular distraction osteogenesis, Loboa et al. found that in vivo bone formation during distraction osteogenesis was stimulated with application of tensile stains at magnitudes of 10%–12.5%. Cyclic tensile strains of these magnitudes have also been found to promote cell proliferation and upregulation of bone marker genes in MSC, osteoblasts, and periosteal cells.8–10 Previous studies in our lab with hMSC and hASC have shown that 10% uniaxial cyclic tensile strain enhances osteogenesis of these stem cells by increasing bone markers and calcium mineral deposition.11–13 However, it is still an ongoing challenge to mimic natural bone and engineer functional, weight-bearing bone tissue from progenitor cells such as ASC. Understanding and elucidating the process of bone formation, along with the molecular biology of osteoprogenitor cells and the osteoinductive environment provided by both physical and chemical stimuli, will help us to optimize use of ASC for functional bone tissue engineering.
In this study, we analyzed differences in gene expression profiles and calcium accretion of hASC in 3D collagen culture during exposure to varying chemical and mechanical stimuli. hASC were cultured in 3D type I collagen gels and maintained in either growth or osteogenic differentiation medium (varied chemical stimulus) in either the presence or absence of 10% cyclic tensile strain (varied mechanical stimulus), a strain magnitude we have previously shown accelerates and increases osteogenic differentiation and calcium accretion of hASC.11 Gene expression profiles were determined and analyzed using advanced microarray analysis, including an evaluation of the canonical pathways affected, to investigate the mechanisms underlying the ability of hASC to undergo osteogenic differentiation, and their response to cyclic tensile strain while undergoing osteogenesis.
Materials and Methods
Cell isolation, culture, and characterization
Excess human adipose tissue from abdominoplasty procedures was obtained from three women (45-year-old African American, 31-year-old Caucasian, and 35-year-old Caucasian) in accordance with an approved Institutional Review Board (IRB) protocol at UNC-Chapel Hill (IRB 04:1622). hASC were isolated from the tissue using a method based on density and differential adhesion, as previously described.3,14 In brief, adipose tissue was digested with 0.075% collagenase type I (Worthington Biochemical Corp., Lakewood, NJ) for 30 min. A dense cell fraction was separated from the adipocytes by centrifugation and resuspended in 160 mM ammonium choloride to lyse the blood cells. Cells were then pelleted by centrifugation for 10 min, and resuspended in growth medium (minimum essential medium Eagle, alpha-modified [α-MEM] supplemented with 10% fetal bovine serum [FBS, lot selected; Atlanta Biologicals, Lawrenceville, GA], 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin). Cells were seeded in T-75 flasks and nonadherent cells were washed out after 24 h. hASC were then characterized via immunohistochemical analysis of surface markers that have been found to be present (CD73, CD105, and CD166) and absent (CD34 and CD45) in MSCs and by their ability to differentiate down osteogenic and adipogenic pathways as previously described.14 All cell culture chemicals and supplies were purchased from Mediatech, Inc. (Herndon, VA) and GIBCO BRL (Grand Island, NY) unless otherwise noted.
Fabrication of collagen gels
hASC were seeded into collagen gels consisting of 70% type I collagen (BD Biosciences, San Jose, CA; pH adjusted to 7.0), 20% 5×α-MEM, and 10% FBS at 60,000 cells/200 μL gel solution. The cell-seeded gel solutions were loaded into Tissue Train® collagen I-coated six-well culture plates (Flexcell International, Hillsborough, NC) to create linear 3D collagen constructs and were allowed to polymerize for 2 h before application of growth media (αMEM supplemented with 10% FBS [lot selected; Atlanta Biologicals], 2 mM L-glutamine, 100 units/mL penicillin, and 100 μg/mL streptomycin).
Osteogenic differentiation and application of 10% cyclic tensile strain
Beginning 24 h after cell seeding, the constructs were cultured for an additional 2 weeks in either growth or osteogenic medium in the presence or absence of 10% uniaxial cyclic tensile strain. Osteogenic medium consisted of growth medium supplemented with 50 μM ascorbic acid, 0.1 μM dexamethasone, and 10 mM β-glycerolphosphate.14–16 Cell-seeded constructs were subjected to 14 days of 10% uniaxial cyclic tensile strain at 1 Hz for 4 h/day using a computer-driven strain device (FX-4000T; Flexcell International). The cell-seeded collagen constructs are fully polymerized around nonwoven anchors at each end, resulting in a 10% uniaxial global tensile strain to the 3D construct.17 Constructs were collected on days 1, 7, and 14 for further analyses.
Cell-mediated calcium accretion was evaluated on day 14 and normalized to cellular DNA content as previously described.18 Briefly, freshly collected constructs on day 14 were rinsed twice with PBS, and cut in half for either calcium or DNA quantification. Constructs for calcium analyses were dissolved in 0.5 N HCl overnight at 4°C and supernatant analyzed with a colorimetric Calcium LiquiColor® assay (Stanbio Laboratory, Boerne, TX). Cellular DNA was extracted from constructs for DNA analyses with DNeasy blood and tissue kit (Qiagen, Valencia, CA) and evaluated using a Nanodrop (Thermo-Fisher Scientific, Wilmington, DE). Data were subjected to a one-way analysis of variance (ANOVA; n=2–3 donors, each with single or duplicate samples per condition).
RNA isolation and real-time reverse transcription–polymerase chain reaction analysis
Constructs were washed twice in PBS, placed in lysis buffer containing β-Mercaptoethanol, and frozen at −80°C until RNA could be isolated. A high cell-seeding density of 60,000 cells/construct was implemented to ensure adequate yield of total RNA. As expected, this resulted in higher contraction of the constructs over time and in some instances construct breakage before day 14.19 Therefore, only one donor had day 14 samples under the strain alone condition (i.e., cultured in complete growth medium, not osteogenic differentiation medium, in the presence of 10% cyclic tensile strain) due to contraction-associated breakage of the constructs by this time; therefore, these samples were only used for reverse transcription–polymerase chain reaction (RT-PCR) validation analysis and not for microarray analysis.
To isolate total RNA, constructs were thawed on ice, ground with a mini-pestle, and homogenized using a QIAshedder (Qiagen). Total RNA was then purified using Eppendorf Perfect RNA mini-columns (Hamburg, Germany) according to the manufacturer's recommended protocol for eukaryotic cells. Total RNA was quantitated using a microplate-based RiboGreen® method (Molecular Probes, Eugene, OR). A single pool of cDNA was reverse transcribed from 3 to 100 ng of each RNA sample using SuperScript™ III (Invitrogen, Carlsbad, CA) with oligo dT primers.
Real-time RT-PCR was performed using an ABI Prism® 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). TaqMan-based PCR Assays-on-Demand™ (Applied Biosystems) were used for gene expression analysis of corin, PDZ and LIM domain 4 (PDLIM4), vascular endothelial growth factor A (VEGF A), and glyceraldehyde-3-phosphate dehydrogenase, the endogenous control. Expression levels were determined using the ΔΔCT method,20 and presented as fold change. Corin expression in hASC was compared with mRNA level in normal human adult bone tissue (BioChain, Hayward, CA), which was set to 1.0; n=2 donors, each with duplicate or triplicate samples per condition. PDLIM4 and VEGF A fold change expression was compared between osteogenic media (set equal to 1), and osteogenic media plus strain; n=2–3 donors each with single or duplicate samples per condition. Data were subjected to a two-tailed Student's t-test to determine significant difference (p<0.05) from control (growth media). Data are presented as mean±standard error.
Biotin labeling, streptavidin antibody staining, scanning, and detection
Gene expression analysis was conducted using Affymetrix Human Genome U133 2.0 Genechip® arrays (Affymetrix, Santa Clara, CA). Total RNA was amplified using the Affymetrix 2-Cycle cDNA Synthesis protocol. Starting with 20 ng of total RNA, cRNA was produced according to the manufacturer's protocol. For each array, 15 μg of amplified cRNAs was fragmented and hybridized to the array for 16 h in a rotating hybridization oven using the Affymetrix Eukaryotic Target Hybridization Controls and protocol. Slides were stained and washed as indicated in the Antibody Amplification Stain for Eukaryotic Targets protocol using the Affymetrix Fluidics Station FS450. Arrays were then scanned with an Affymetrix Scanner 3000. Data were obtained using the Genechip Operating Software (version 1.2.0.037); n=3 donors.
Microarray data analysis
Data preprocessing, normalization, and error modeling was performed with the Rosetta Resolver system (version 7.2; Rosetta Biosoftware, Kirkland, WA). Principal component analysis (PCA) was performed on all samples and all probes to characterize the variability present in the data. Intensity profiles were combined by weight-averaging into Intensity Experiments. When required, Intensity Experiments were built into ratios representing treated/control (osteogenic induction media compared with complete growth media, or osteogenic induction media plus 10% uniaxial cyclic tensile strain compared with osteogenic induction media), as described by Stoughton and Dai (2005).21 An error-weighted ANOVA with Bonferroni test was used to reduce the number of false-positives with p<0.01.
Whole genome expression data were visualized in the context of molecular function, canonical pathways, and biological network using ingenuity pathway analysis (IPA) system (version 8.0; Ingenuity Systems, Mountain View, CA; www.ingenuity.com). Data sets containing gene identifiers and corresponding expression values were uploaded into the application. Each gene identifier was mapped to its corresponding gene. In the case of genes with multiple identifiers, the highest expression value was selected. The function and pathway analysis of IPA was generated based on all currently available published data to identify potential regulatory pathways and molecules.
Results
Osteogenesis of hASC after 14 days in 3D collagen culture in response to osteogenic media and 10% uniaxial cyclic tensile strain
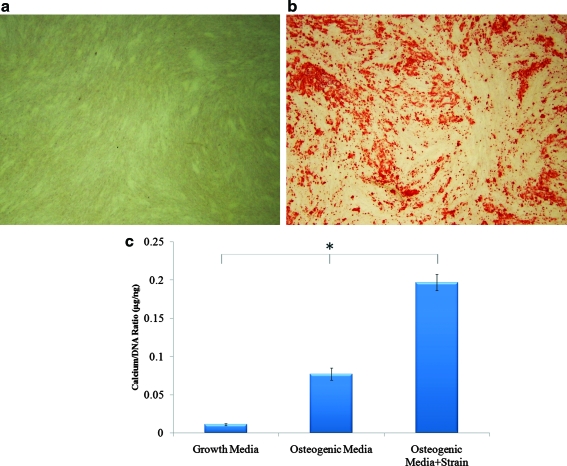
hASC from three donors used in this study had been preselected for positive MSC markers, and capability for adipogenic and osteogenic differentiation in 2D culture.14 All three selected cell lines were also verified to deposit mineral over an area spanning at least 50% of the tissue culture well, determined via Alizarin red staining (Fig. 1a, b).
FIG. 1.
Osteogenic differentiation of hASC. Alizarin red staining for calcium accretion in 2D culture after 14 days (a) in growth media or (b) osteogenic media. (c) Cell-mediated calcium accretion in 3D collagen I culture after 14 days of hASC culture in growth media, osteogenic media, or osteogenic media plus 10% cyclic tensile strain (f=1 Hz, 4 h/day). *p < 0.01. hASC, human adipose-derived stem cells; 3D, three-dimensional. Color images available online at www.liebertonline.com/tea
To evaluate osteogenesis of hASC in 3D collagen culture, accreted calcium and DNA content was quantified. Relative to hASC cultured in complete growth media, calcium content was significantly (p<0.01) increased by osteogenic media induction and further elevated by 10% uniaxial cyclic tensile strain (Fig. 1c).
Validation of microarray data
To characterize variability present in the microarray data, PCA was performed. PCA results showed discrimination between three treatments: (1) hASC cultured in growth media; (2) hASC cultured in osteogenic media; (3) hASC cultured in osteogenic media in the presence of 10% uniaxial cyclic tensile strain (Fig. 2). As expected, variability was also found between human donors within each treatment. Osteogenic media alone and osteogenic media plus cyclic tensile strain exhibited gene expression profiles closer to each other than hASC cultured in growth media, since both of those populations were undergoing osteogenic differentiation (Fig. 2).
FIG. 2.
Principal component analysis (PCA). (a) PCA results indicated a distinct separation between growth media and osteogenic media, and further between growth media and osteogenic media plus 10% cyclic tensile strain. (b) Rotated (45 degrees) plot for 3D view. Color images available online at www.liebertonline.com/tea
Genes differentially expressed by hASC in response to 3D collagen culture in osteogenic media or osteogenic media plus 10% uniaxial cyclic tensile strain were identified by error-weighted ANOVA. One thousand two hundred eighty-eight gene identifiers were detected as osteogenic sensing and 184 gene identifiers were detected as cyclic tensile strain sensing. Of the previously reported top 20 upregulated genes during osteogenesis of hASC for 14 days in 2D culture,16 we found 43 relevant gene identifiers for 15 genes that were also upregulated during osteogenesis of hASC in 3D collagen I gel culture (Table 1). Four of our top five upregulated genes, alcohol dehydrogenase 1B, glycoprotein M6B (GPM6B), monoamine oxidase A (MAOA), and FK506 binding protein 5 (FKBP5), were also found by Liu et al. as top upregulated genes during osteogenic differentiation of hASC in 2D culture.16 Our fifth top upregulated gene, corin, was not identified by Liu et al. during osteogenesis of hASC in 2D culture.16 Interestingly, corin has also been found to be a top upregulated gene in hMSC during osteogenic differentiation in 2D culture.16 Given this divergence in results for corin by previous investigators during osteogenic differentiation of hMSC versus hASC when analyzing cell response in 2D culture,22 and our current microarray finding that corin was highly upregulated by hASC in 3D culture, we confirmed corin expression using real-time RT-PCR with analysis at a greater number of time points, as described later in the real-time RT-PCR analysis section.
Table 1.
Key Findings from Comparative Analysis of This Study's Findings to the Publicly Available Database
|
Effect of osteogenic induction media |
|
|
Reference |
|
|---|---|---|---|---|
|
Common upregulated genes among top 20 genes upregulated by hASC in 3D collagen culture (present study) and hASC in 2D culture.13 | ||||
| |
|
Fold change |
|
|
| Gene | Entrez gene name | hASC in 3D | hASC in 2D | |
| ADH1B | Alcohol dehydrogenase 1B (class I), beta polypeptide | 100 | 12.1 | Supplementary Table S1 |
| GPM6B | Glycoprotein M6B | 100 | 29.5 | |
| MAOA | Monoamine oxidase A | 100 | 6.4 | |
| FKBP5 | FK506 binding protein 5 zinc finger and BTB | 81.6 | 26.1 | |
| ZBTB16 | Domain containing 16 | 70.6 | 73.6 | |
|
Other top 20 upregulated genes by hASC in 2D culture13 and also upregulated by hASC in 3D collagen culture at day 14 (this study). | ||||
|---|---|---|---|---|
|
Gene | ||||
|
NEBL, CPM, INHBB, RGC32, NEDD9, FMO2, SBLF, B3GNT5, NR2F1 | ||||
|
Common upregulated genes among top 20 genes upregulated by hASC in 3D collagen culture (present study) and hMSC in 2D culture13but not by hASC in 2D culture13at day 14 | ||||
|
Gene |
Entrez gene name |
Fold change |
|
|
| |
|
hASC in 3D |
hMSC in 2D |
|
|
CORIN |
Corin, serine peptidase |
100a |
29.7 |
Figure 6a13 |
| Canonical pathways | Key upregulated genes | Note | ||
| TGF-β signaling | INHBB | Overall downregulation | Figure 2, Supplementary Table S3 | |
| Wnt/β-catenin signaling | SOX13, MMP7, TGFBR3, DKK1, WNT5 B | Upregulation of inhibitors SOX 13, DKK1 |
||
| BMP signaling pathway | NOGGIN, PRKAR2B | Upregulation of inhibitors NOGGIN |
||
| PDGF signaling |
PIK3R1, PDGFD, CAV1 FOXOl, PRKAR2B, |
|||
| IGF signaling | PIK3R1, IRS2, FOXO3 | |||
|
Effect of 10% uniaxial cyclic tensile strain |
|
Reference |
|
|---|---|---|---|
|
Top molecules |
Upregulated genes |
Downregulated genes |
|
| |
PMEPA1, NPTX1, PDLIM4,bIL1RN, KCNG1 |
NAP1L1, LPAR1, NFE2L1, COX11, MKRN1 |
|
|
Canonical pathways |
|
Fold change |
|
| Signaling pathways | Upregulated genes | ||
| Hepatic fibrosis/Hepatic stellate cell activation | FGF, VEGFA,cTGFBR1, MMP2 | 2.7, 1.9, 1.7, 1.4 | Supplementary Table S6 |
| Chondroitin sulfate biosynthesis | HS3ST3A1, HS3ST3B1, CHSY1 | 2.6, 1.7, 1.7 | |
| Endoplasmic reticulum stress pathway | EIF2S1, XBP1 | 1.4, 1.4 | |
| Xenobiotic metabolism signaling | MAP3K4, HS3ST3A, HS3ST3B1 | 2.6, 2.6, 1.7 | |
| Top network | Key upregulated genes | |
|---|---|---|
| Development and function of connective tissue, skeletal, and muscular system | IL1RN, JUND, SOCS3 | Figure 4, Supplementary Table S7 |
| Top function | Upregulated genes | Downregulated genes |
|---|---|---|
| Cardiovascular system development and function | NPTX1, IL1RN, SPHK1, FGF2, SOCS3, PTGES, GEM, ID3, VEGFA, GJA1, ADA, TGFBR1, GBP1, TUBB6, JUND, MMP2 | BTG1, TGFBR2, STX7, NR2F2, CITED2, AGTR1, PTK2B, STC1 |
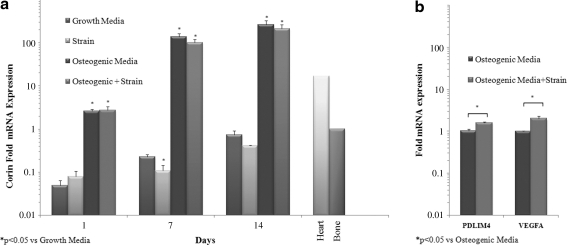
Highly upregulated corin expression was confirmed with RT-PCR at multiple time points (Fig. 6a).
Upregulated PDLIM4 expression was confirmed with RT-PCR (Fig. 6b).
Upregulated VEGFA expression was confirmed with RT-PCR (Fig. 6b).
hASC, human adipose-derived stem cells, 3D, three-dimensional; TGF, transforming growth factors-β; BMP, bone morphogenic protein; PDGF, platelet-derived growth factor; IGF, insulin-like growth factor; RT-PCR, reverse transcription–polymerase chain reaction; PDLIM4, PDZ and LIM domain 4.
Genes regulated during osteogenic differentiation of hASC in 3D collagen culture for 14 days
Of the 1288 gene identifiers that were significantly and differentially expressed by all three donors' hASC in response to osteogenic differentiation medium, 1218 identifiers were mapped to 847 genes (Supplementary Tables S1 and S2; Supplementary Data are available online at www.liebertonline.com/tea). Eight hundred sixty-four identifiers were eligible for network analysis with IPA; 832 identifiers were eligible for function and pathway analyses.
Ninety-five canonical pathways were mapped as a significant pathway associated with osteogenesis of hASC in 3D collagen culture (p<0.05 calculated by Fisher's exact right-tailed test, determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone; Supplementary Table S3). Transforming growth factors-β (TGF-β) signaling, Wnt/β-catenin signaling, and bone morphogenic protein (BMP) signaling also appeared to play a role in hASC osteogenesis (Fig. 3). Overall, these pathways were downregulated with activation of known inhibitors in Wnt/β-catenin signaling: sex determining region Y-box 13; BMP signaling: dickkopf homolog 1; and noggin (Fig. 3 and Supplementary Table S3). Many pathways related to the triggering of cell cycle, growth, differentiation, and migration, such as axonal guidance signaling, platelet-derived growth factor signaling (PDGF signaling), insulin-like growth factor-1 signaling (IGF-1 signaling), integrin linked kinase signaling, actin cytoskeleton signaling, and sonic hedgehog signaling, were also affected (Fig. 4).
FIG. 3.
Downregulation of TGF-β signaling, Wnt/β-Catenin signaling, and BMP signaling by hASC in 3D collagen culture in response to osteogenic induction media. Red indicates upregulated molecule (also indicated by ↑); green indicates downregulated molecule (also indicated by ↓). TGF-β, tumor growth factor β; BMP, bone morphogenic protein. Color images available online at www.liebertonline.com/tea
FIG. 4.
Canonical pathways affected in hASC by osteogenic induction media. Pathways were categorized based on all published canonical pathway data available at the time of analysis by IPA (Ingenuity® Systems) into (a) organismal growth and development, (b) growth factor signaling, (c) cellular growth, proliferation, and development, and (d) cell cycle regulation. Left y-axis=−log(p-value) from Fisher's exact test right-tailed, at threshold=p-value <0.05 (−log(p-value) > 1.301). Right y-axis=ratio representing the number of differentially expressed genes divided by total number of genes that make up that pathway. Line is connected as a function of the IPA program, does not indicate statistical correlation of the data. AKT, v-akt murine thymoma viral oncogene homolog; AMPK, AMP activated protein kinase; ATM, ataxia telangiectasia mutated protein; IPA, ingenuity pathway analysis; PDGF, platelet-derived growth factor; IGF-1, insulin-like growth factor-1; VEGF, vascular endothelial growth factor; ILK, integrin linked kinase; PI3K, phosphatidyl inositol 3 kinase. Color images available online at www.liebertonline.com/tea
Genes regulated by hASC in 3D collagen culture in response to 10% uniaxial cyclic tensile strain
To identify the effect of 10% uniaxial cyclic tensile strain on hASC undergoing osteogenic differentiation, the gene expression profile of hASC cultured in osteogenic media plus 10% uniaxial cyclic tensile strain were compared with the gene expression profile of hASC cultured in osteogenic media alone (0% strain). One hundred eighty-four transcripts were modulated by 10% uniaxial cyclic tensile strain (Supplementary Tables S4 and S5). Of the 184 transcripts, 176 transcripts were mapped to 147 genes with IPA. One hundred forty-six identifiers were eligible for network analysis and 135 identifiers were eligible for function and pathway analyses.
Cardiovascular system development and function was the top function when categorizing the genes modulated by 10% uniaxial cyclic tensile strain (24 genes, p<10−7–10−3) with upregulation of 16 genes, including fibroblast growth factor 2 (FGF2), interleukin 1 receptor antagonist (IL1RN), matrix metalloproteinase 2 (MMP2), VEGF A, and TGF-β receptor 1 (TGFβR1; Table 1).
The major canonical pathways affected by 10% cyclic tensile strain are shown in Supplementary Table S6 (p<0.05), indicating activation of growth factor-related genes FGF2, VEGF A, and TGFβR 1; extracellular matrix-related genes MMP2, chondroitin sulfate synthase 1, heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1, and heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1; and stress response pathways endoplasmic reticulum (ER) stress and xenobiotic metabolism. The top network identified, development and function of connective tissue, skeletal, and muscular system, was scored at 41 (scores of 2 or higher have at least a 99% confidence of not being generated by random chance alone) with 22 focused molecules and 36 total molecules in the network (Fig. 5 and Supplementary Table S7). Three focused molecules, IL1RN, jun D proto-oncogene, and suppressor of cytokine signaling 3 (SOCS3), were located in the center of the network and linked with 5 or more other molecules.
FIG. 5.
The highest ranked network in response to 10% uniaxial cyclic tensile strain centered with IL1RN, SOCS3, and JUND. Red indicates upregulated molecules; green indicates downregulated molecules. Color intensity indicates the level of expression (see Supplementary Table S7 for full names and expression profile data). Color images available online at www.liebertonline.com/tea
Real-time RT-PCR analysis to validate results for corin, the type II serine protease upregulated during osteogenesis of hASC, PDLIM4, an actin binding protein, and VEGF A, an angiogenic factor, upregulated in response to 10% uniaxial cyclic tensile strain in 3D culture
From the microarray data, corin expression was found to be upregulated 100-fold during osteogenesis of hASC for 14 days in both 3D culture environments: osteogenic media and osteogenic media plus strain (Supplementary Tables S1 and S2). However, the differential expression of corin by strain could not be identified, as 100-fold was the maximum sensitivity of the microarray probes. Therefore, differential corin expression in response to osteogenic media or osteogenic media plus 10% cyclic tensile strain was analyzed in hASC-seeded collagen gels on days 1, 7, and 14 by real-time RT-PCR. On day 1, corin expression of hASC in osteogenic media increased 2.6-fold over hASC in growth media, showing the quick upregulation of corin expression by hASC in osteogenic media (Fig. 6a). By days 7 and 14, corin expression had increased by 139-fold and 270-fold, respectively. The addition of 10% uniaxial cyclic tensile strain did not significantly alter corin expression, except on day 7 of culture in growth media (Fig. 6a). Interestingly, after 14 days of culture in the 3D type I collagen gel without osteogenic induction media, corin was upregulated to approximately its expressed level in bone tissue (Fig. 6a).
FIG. 6.
Real-time reverse transcription–polymerase chain reaction results. (a) Upregulation of corin during hASC osteogenesis. Corin expression in hASC was compared with its expression in bone tissue (expression set to 1). (b) Upregulation of PDZ and LIM domain 4 and VEGF A by hASC in response to 10% uniaxial cyclic tensile strain. Gene expression in osteogenic media set to 1, glyceraldehyde-3-phosphate dehydrogenase normalization.
Two genes upregulated in response to 10% cyclic tensile strain during osteogenesis of hASC were also validated with RT-PCR: (1) PDLIM4, one of the top five upregulated genes that have been shown to have polymorphisms found to associate with bone mineral density regulation23,24; (2) VEGF A, a known angiogenic factor. Real-time RT-PCR results confirmed that expression of both genes was significantly increased by hASC in response to 10% uniaxial cyclic tensile strain (Fig. 6b).
Discussion
Differentiation of mesenchymal progenitor cells into the osteoblastic lineage has been studied extensively in transgenic animal models.25,26 While animal models provide an in vivo system to identify the pivotal role of specific factors, there are possibilities of interspecies differences. In vitro studies on human cell lines are thus required to fully develop our knowledge for clinical application. Research on osteoblastic differentiation of human MSCs from multiple source tissues has been performed,27,28 but they have lacked incorporation of a 3D, mechanically loaded culture system that provides bone ECM-mimetic 3D culture and mechanical loading, and the global analysis to obtain insight into associated regulatory mechanisms of osteogenesis.
In the present study, collagen type I was used to mimic the organic ECM of bone and 10% uniaxial cyclic tensile strain was applied as an appropriate mechanical load to promote greater osteogenic differentiation and calcium accretion of hASC.11,13,29,30 Calcium accretion data indicated that hASC differentiated down an osteogenic pathway by accreting calcium in 3D collagen culture when in the presence of soluble osteogenic inductive factors. Ten percent uniaxial cyclic tensile strain further increased calcium accretion of hASC (Fig. 1c), as we have previously shown in 2D culture.11 Global analysis of over 47,000 gene identifiers from microarray data revealed the canonical pathways involved with osteogenic differentiation of hASC in 3D culture and their response to 10% cyclic tensile strain.
As expected, many genes that were modulated in response to soluble osteogenic inductive factors were associated with cellular functions that are important for the process of progenitor cell differentiation into specific cell lineages, as this process requires the exit of cells from cell cycle or self-renewal toward cell maturation with associated changes in cell growth, cell development, and cell morphology. Many canonical pathways affected by osteogenic media were also reported as signaling required for bone formation. After 14 days, common bone marker genes ALP, runt-related transcription factor-2 (RUNX2), and osteocalcin/bone gamma-carboxyglutamate (gla) protein were not significantly upregulated relative to culture in 3D type I collagen gels without soluble osteogenic inductive factors. The time point and culture system chosen likely caused this result, as we have found that endogenous ALP activity highly increases in hASC without chemical or mechanical stimuli after culture in linear 3D type I collagen gels for 10 days (Supplementary Fig. S1). Further, it has been previously shown that when soluble osteogenic inductive factors are included in the culture media, 3D collagen culture of hASC causes increased mRNA expression of bone markers RUNX2, ALP, osteonectin, osteopontin, Collagen I, and mitogen-activated protein kinase 9 (MAPK9) compared with hASC cultured in 2D monolayer on tissue culture plastic.6
Factors influencing embryonic osteogenesis,31 PDGF, IGFs, and TGF-β, were also involved with osteogenesis of hASC, either directly or through their signaling cascades. PDGFs are known to play a role in bone regeneration by acting as a chemotactic agent, thus increasing the pool of osteogenic cells at an injury site.32 A previous study on hMSC expression profiles has shown that PDGF signaling is essential for hMSC growth and tri-lineage differentiation (adipogenic, chondrogenic, and osteogenic lineages).33 Our data indicate that PDGF signaling might play a role in osteogenic differentiation of hASC via the direct induced modulation of PDGF-D and PDGF receptor, alpha polypeptide (PDGFRA). PDGF-D is a newly found member of the PDGF family.34 A recent study on PDGF-D function in transgenic mice showed that PDGF-D induces macrophage recruitment and blood vessel maturation during angiogenesis.34
Our data on IGF-1 mRNA expression indicated that neither the secreted factor (IGF-1) nor its receptor (IGF1R) was directly affected during osteogenic differentiation of hASC. However, IGF-1 signaling was affected with the activation of insulin receptor substrate 1 and 2 (IRS-1 and IRS-2) and its downstream cascades, including phosphatidyl inositol 3 kinase (PI3K). Secreted IGF-1 from osteoblasts has been reported to act as a chemotactic factor with its function potentially inhibited by PI3K inhibitor.35 Both IRS-1 and IRS-2 are expressed in bone. IRS-1 is important for maintaining bone turnover (bone formation and bone resorption). IRS-1 expression is also required for skeletal growth and fracture healing.36–38 IRS-2 is not required for bone healing but plays a critical role in the coupling of bone resorption to bone formation.39,40 These findings suggest that IGF-1 signaling, even without the modulation of mRNA expression of IGF-1 itself, may play a critical role for osteogenic differentiation of hASC.
TGF-β signaling and Wnt/ß-catenin signaling pathways, known to stimulate proliferation and early osteoblast differentiation, have been reported to inhibit osteoblast maturation with continuing activation.41,42 Our results also indicate downregulation of these pathways at day 14 in hASC, suggesting that hASC might also require the downregulation of these two pathways for terminal osteogenic differentiation.
Interestingly, another pathway found to be downregulated was the antigen presentation pathway with the suppressed expression of major histocompatibility complex-A,-B,-C,-G (HLA-A,-B,-C,-G), and calnexin. Donor-specific HLA antibodies have been associated with graft dysfunction and failure.43 A study on immunological properties of hASC and hMSC has shown low immunogenecity and suggested the potential application of these cell types for allogenic transplantation.44 Our data suggest that osteogenic induction also suppresses the immune characteristics of hASC further, indicating their potential use for allograft bone tissue engineering.
Of the canonical pathways involved with hASC osteogenesis, molecular mechanisms of cancer were ranked first. These mechanisms were comprised of downregulation of membrane receptors (WNT, BMP, and TGF-β); upregulation of cytoplasmic signals such as PI3K and IRS-1; repression of apoptosis and cell cycle arrest through the activation of forkhead box O1 and cyclin D3; and the deactivation of cyclin-dependent kinase inhibitor 1A (p21 and Cip1), and cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4). Senescence suppression of hMSC by addition of FGF-2 has been found to be regulated by a similar mechanism by which TGF-β and cyclin-dependent kinase inhibitor (p53, p21, and p16) are downregulated, resulting in the upregulation of cyclin D.45 However, self-renewal of stem cells and immortality of cancer are intricately associated. Many signals such as Notch, Hedgehog, and Wnt have been found to regulate both self-renewal of stem cells and also to control the development of cancer.46 Further analyses are required to distinguish the two processes.
In addition to the pathway analysis, of interest was the gene corin, which was found to be upregulated by 100-fold but was not matched with any canonical pathways. Corin is a serine protease normally associated with the heart, where it has been found to help maintain blood pressure levels.47 Corin has also been found to be expressed in developing bone near hypertrophic chondrocytes and in perichondrocytes.48 This latter report suggested that corin may be associated with chondrogenesis. Recently, corin was found to be relatively highly upregulated compared with other genes during osteogenic differentiation of hMSC, but not during osteogenic differentiation of hASC in 2D culture at days 3 and 14.16 Corin has not previously been studied in the context of its potential role in osteogenesis or chondrogenesis. In the present study, we have shown that corin may be involved with osteogenesis of hASC in 3D culture in response to soluble osteogenic inductive factors but not by 10% uniaxial cyclic tensile strain. Taken together with its presence in vivo, more studies on the role of corin in both chondrogenesis and osteogenesis might lead to a better understanding of its role in endochondral and potentially intramembranous bone formation.
Other genes found to be highly expressed during osteogenic differentiation of hASC in 3D collagen culture were GPM6B, MAOA, FKBP5, and zinc finger and BTB domain containing 16 (ZBTB16), which have also been found to be upregulated during osteogenic differentiation of hASC and hMSC after 14 days in 2D culture.16 Likewise, alcohol dehydrogenases may be involved in retinoic acid synthesis,49 which has been found to induce osteogenic differentiation in mouse ASC.50
Finally, mechanical load, a critical factor in bone formation and resorption, was also found to significantly affect hASC during osteogenic differentiation. Application of 10% uniaxial cyclic tensile strain to hASC induced upregulation of chondroitin sulfate biosynthesis and ER stress pathways. Depletion of heparan/chondroitin sulfate in MSC has been reported to result in altered BMP and Wnt activity.51 Further, XBP1 and ATF4 from the ER stress pathway have been found to express and regulate the onset of osteoblast differentiation.52–54 This suggests that chondroitin sulfate biosynthesis and the ER stress pathway may play key roles in hASC response to 10% cyclic tensile strain.
Since many affected genes have not been fully studied, such as the mechanosensing genes of hASC in this study, our analysis had to be based on known function and canonical pathways that could conceivably bias the focus to some of the extensively studied genes. The network analysis was generated to identify key regulatory genes. The network analysis of genes differentially expressed by 10% uniaxial cyclic tensile strain showed that IL1RN and SOCS3 centered in the first rank network. Interestingly, both of these genes are inhibitors of inflammatory signaling. IL1RN is a competitive inhibitor of IL-1, and has been shown to reduce osteoclast formation, bone loss, and bone resorption in estrogen deficient (ovariectomized) mice.55–57 SOCS3 negatively regulates IL-6, and mRNA expression of SOCS3 is stabilized by tumor necrosis factor α (TNF-α).56,58,59 IL-6 and TNF-α are known to initiate the bone healing process by promoting extracellular matrix synthesis, stimulating angiogenesis, and promoting osteoclastogenesis and osteoclast function.60,61 Although we found the expression of IL-1 and IL-6 inhibitors, there was no differential expression of the IL-1 and IL-6 genes. A previous study on the role of proinflammatory cytokines during bone fracture healing showed that the expression of IL-1 and IL-6 is temporal, with peaks at 24 h and 7 days.61 As we also found that one of the top classified biofunctions was cardiovascular development with the upregulation of many genes that promoted angiogenesis such as FGF2, MMP2, and VEGF A, this suggests that 10% uniaxial cyclic tensile strain may induce angiogenesis in hASC, possibly through the activation of IL-1 and IL-6, thus requiring IL1RN and SOCS3 to negatively control the temporal expression of IL-1 and IL-6.
In summary, our data suggest that after 14 days of osteogenic differentiation in 3D collagen I gels in the absence of cyclic tensile strain, hASC downregulated TGF-β signaling, Wnt/β-Catenin signaling, and BMP signaling through some of their inhibitor molecules. Further, osteogenic differentiation of hASC in 3D collagen culture elevated the expression of some genes, for example, corin, in a distinctly different response from a previous analysis of hASC osteogenic differentiation in 2D culture.16 Pathway analysis with nonoverlapping results by the addition of 10% cyclic tensile strain relative to the effects of osteogenic differentiation media alone suggested that increased calcium accretion by hASC in response to mechanical loading is associated with a different set of genes than those affected by soluble osteogenic inductive factors. Of particular interest was the upregulation of PDLIM4, one of the top five upregulated genes that have been shown to have polymorphisms found to associate with bone mineral density regulation.23,24 The addition of 10% uniaxial cyclic tensile strain also resulted in hASC upregulation of two crucial factors in bone regeneration: (1) proinflammatory cytokine regulators IL1RN and SOCS3; (2) angiogenic inductors FGF2, MMP2, and VEGF A.
In conclusion, this is the first study to investigate the effects of both 3D collagen culture and 10% uniaxial cyclic tensile strain on hASC osteogenic differentiation. A complete microarray analysis investigating both the separate effect of soluble osteogenic inductive factors, and the combined effects of chemical and mechanical stimulation was performed on hASC undergoing osteogenic differentiation. We have identified specific genes and pathways associated with mechanical response and osteogenic potential of hASC, thus providing significant information toward improved understanding of our use of hASC for functional bone tissue engineering applications.
Supplementary Material
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, by National Center for Research Resources grant 10KR51023 (EGL), and by NIH/NIBIB grant R03EB008790-01A2 (EGL).
Disclosure Statement
No competing financial interests exist.
References
- 1.Toma J.G. Akhavan M. Fernandes K.J.L. Barnabé-Heider F. Sadikot A. Kaplan D.R. Miller F.D. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat Cell Biol. 2001;3:778. doi: 10.1038/ncb0901-778. [DOI] [PubMed] [Google Scholar]
- 2.Kern S. Eichler H. Stoeve J. Kluter H. Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 3.Zuk P.A. Zhu M. Ashjian P. De Ugarte D.A. Huang J.I. Mizuno H. Alfonso Z.C. Fraser J.K. Benhaim P. Hedrick M.H. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hattori H. Sato M. Masuoka K. Ishihara M. Kikuchi T. Matsui T. Takase B. Ishizuka T. Kikuchi M. Fujikawa K. Osteogenic potential of human adipose tissue-derived stromal cells as an alternative stem cell source. Cells Tissues Organs. 2004;178:2. doi: 10.1159/000081088. [DOI] [PubMed] [Google Scholar]
- 5.Halvorsen Y.D.C. Franklin D. Bond A.L. Hitt D.C. Auchter C. Boskey A.L. Paschalis E.P. Wilkison W.O. Gimble J.M. Extracellular matrix mineralization and osteoblast gene expression by human adipose tissue-derived stromal cells. Tissue Eng. 2001;7:729. doi: 10.1089/107632701753337681. [DOI] [PubMed] [Google Scholar]
- 6.Gabbay J.S. Heller J.B. Mitchell S.A. Zuk P.A. Spoon D.B. Wasson K.L. Jarrahy R. Benhaim P. Bradley J.P. Osteogenic potentiation of human adipose-derived stem cells in a 3-dimensional matrix. Ann Plast Surg. 2006;57:89. doi: 10.1097/01.sap.0000205378.89052.d3. [DOI] [PubMed] [Google Scholar]
- 7.Carter D.R. Beaupré G.S. Giori N.J. Helms J.A. Mechanobiology of skeletal regeneration. Clin Orthop. 1998;355:S41. doi: 10.1097/00003086-199810001-00006. [DOI] [PubMed] [Google Scholar]
- 8.Kanno T. Takahashi T. Ariyoshi W. Tsujisawa T. Haga M. Nishihara T. Tensile mechanical strain up-regulates Runx2 and osteogenic factor expression in human periosteal cells: implications for distraction osteogenesis. J Oral Maxillofac Surg. 2005;63:499. doi: 10.1016/j.joms.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Fong K.D. Nacamuli R.P. Loboa E.G. Henderson J.H. Fang T.D. Song H.M. Cowan C.M. Warren S.M. Carter D.R. Longaker M.T. Equibiaxial tensile strain affects calvarial osteoblast biology. J Craniofac Surg. 2003;14:348. doi: 10.1097/00001665-200305000-00013. [DOI] [PubMed] [Google Scholar]
- 10.Koike M. Shimokawa H. Kanno Z. Ohya K. Soma K. Effects of mechanical strain on proliferation and differentiation of bone marrow stromal cell line ST2. J Bone Miner Metab. 2005;23:219. doi: 10.1007/s00774-004-0587-y. [DOI] [PubMed] [Google Scholar]
- 11.Hanson A.D. Marvel S.W. Bernacki S.H. Banes A.J. van Aalst J. Loboa E.G. Osteogenic effects of rest inserted and continuous cyclic tensile strain on hASC lines with disparate osteodifferentiation capabilities. Ann Biomed Eng. 2009;37:955. doi: 10.1007/s10439-009-9648-7. [DOI] [PubMed] [Google Scholar]
- 12.Sumanasinghe R.D. Bernacki S.H. Loboa E.G. Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP-2) mRNA expression. Tissue Eng. 2006;12:3459. doi: 10.1089/ten.2006.12.3459. [DOI] [PubMed] [Google Scholar]
- 13.Wall M.E. Rachlin A. Otey C.A. Loboa E.G. Human adipose-derived adult stem cells upregulatepalladin during osteogenesis and in response to cyclic tensile strain. Am J Physiol Cell Physiol. 2007;293:C1532. doi: 10.1152/ajpcell.00065.2007. [DOI] [PubMed] [Google Scholar]
- 14.Wall M.E. Bernacki S.H. Loboa E.G. Effects of serial passaging on the adipogenic and osteogenic differentiation potential of adipose-derived human mesenchymal stem cells. Tissue Eng. 2007;13:1291. doi: 10.1089/ten.2006.0275. [DOI] [PubMed] [Google Scholar]
- 15.Halleux C. Sottile V. Gasser J. Seuwen K. Multi-lineage potential of human mesenchymal stem cells following clonal expansion. J Musculoskelet Neuronal Interact. 2001;2:71. [PubMed] [Google Scholar]
- 16.Liu T.M. Martina M. Hutmacher D.W. Po Hui J.H. Lee E.H. Lim B. Identification of common pathways mediating differentiation of bone marrow- and adipose tissue-derived human mesenchymal stem cells into three mesenchymal lineages. Stem Cells. 2007;25:750. doi: 10.1634/stemcells.2006-0394. [DOI] [PubMed] [Google Scholar]
- 17.Pfeiler T.W. Sumanasinghe R.D. Loboa E.G. Finite element modeling of 3D human mesenchymal stem cell-seeded collagen matrices exposed to tensile strain. J Biomech. 2008;41:2289. doi: 10.1016/j.jbiomech.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCullen S. Zhu Y. Bernacki S. Narayan R. Pourdeyhimi B. Gorga R. Loboa E. Electrospun composite poly (L-lactic acid)/tricalcium phosphate scaffolds induce proliferation and osteogenic differentiation of human adipose-derived stem cells. Biomed Mater. 2009;4:035002. doi: 10.1088/1748-6041/4/3/035002. [DOI] [PubMed] [Google Scholar]
- 19.Sumanasinghe R.D. Osborne J.A. Loboa E.G. Mesenchymal stem cell-seeded collagen matrices for bone repair: effects of cyclic tensile strain, cell density, and media conditions on matrix contraction in vitro. J Biomed Mater Res Part A. 2009;88:778. doi: 10.1002/jbm.a.31913. [DOI] [PubMed] [Google Scholar]
- 20.Livak K.J. Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 CT method. Methods. 2001;25:402. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Stoughton R. Dai H. Statistical Combining of Cell Expression Profiles, 2005. US patent No. 6351712
- 22.Liu T.M. Martina M. Hutmacher D.W. Hui J.H. Lee E.H. Lim B. Identification of common pathways mediating differentiation of bone marrow and adipose tissues derived human mesenchymal stem cells (MSCs) into three mesenchymal lineages. Stem Cells. 2006;25:750. doi: 10.1634/stemcells.2006-0394. [DOI] [PubMed] [Google Scholar]
- 23.Omasu F. Ezura Y. Kajita M. Ishida R. Kodaira M. Yoshida H. Suzuki T. Hosoi T. Inoue S. Shiraki M. Association of genetic variation of the RIL gene, encoding a PDZ-LIM domain protein and localized in 5q31. 1, with low bone mineral density in adult Japanese women. J Human Genet. 2003;48:342. doi: 10.1007/s10038-003-0035-1. [DOI] [PubMed] [Google Scholar]
- 24.Xiong Q. Jiao Y. Hasty K.A. Canale S.T. Stuart J.M. Beamer W.G. Deng H.W. Baylink D. Gu W. Quantitative trait loci, genes, and polymorphisms that regulate bone mineral density in mouse. Genomics. 2009;93:401. doi: 10.1016/j.ygeno.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uutela M. Wirzenius M. Paavonen K. Rajantie I. He Y. Karpanen T. Lohela M. Wiig H. Salven P. Pajusola K. PDGF-D induces macrophage recruitment, increased interstitial pressure, and blood vessel maturation during angiogenesis. Blood. 2004;104:3198. doi: 10.1182/blood-2004-04-1485. [DOI] [PubMed] [Google Scholar]
- 26.Ducy P. Cbfa1: a molecular switch in osteoblast biology. Dev Dynamics. 2000;219:461. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1074>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 27.Kern S. Eichler H. Stoeve J. Klüter H. Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 28.Caton D. Roche S. Bony C. Lehmann S. Casteilla L. Jorgensen C. Cousin B. Cell specific differences between human adipose-derived and mesenchymal-stromal cells despite similar differentiation potentials. Exp Cell Res. 2008;314:1575. doi: 10.1016/j.yexcr.2007.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Sumanasinghe R.D. Pfeiler T.W. Monteiro-Riviere N.A. Loboa E.G. Expression of proinflammatory cytokines by human mesenchymal stem cells in response to cyclic tensile strain. J Cell Physiol. 2009;219:77–83. doi: 10.1002/jcp.21653. [DOI] [PubMed] [Google Scholar]
- 30.Sumanasinghe R.D. Bernacki S.H. Loboa E.G. Osteogenic differentiation of human mesenchymal stem cells in collagen matrices: effect of uniaxial cyclic tensile strain on bone morphogenetic protein (BMP-2) mRNA expression. Tissue Eng. 2006;12:3459. doi: 10.1089/ten.2006.12.3459. [DOI] [PubMed] [Google Scholar]
- 31.Rosen V. Thies R.S. New York: Springer; 1995. The Cellular and Molecular Basis of Bone Formation and Repair. [Google Scholar]
- 32.Hollinger J.O. Hart C.E. Hirsch S.N. Lynch S. Friedlaender G.E. Recombinant human platelet-derived growth factor: biology and clinical applications. J Bone Joint Surg. 2008;90:48. doi: 10.2106/JBJS.G.01231. [DOI] [PubMed] [Google Scholar]
- 33.Ng F. Boucher S. Koh S. Sastry K.S.R. Chase L. Lakshmipathy U. Choong C. Yang Z. Vemuri M.C. Rao M.S. PDGF, TGF-{beta}, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood. 2008;112:295. doi: 10.1182/blood-2007-07-103697. [DOI] [PubMed] [Google Scholar]
- 34.Uutela M. Wirzenius M. Paavonen K. Rajantie I. He Y. Karpanen T. Lohela M. Wiig H. Salven P. Pajusola K. PDGF-D induces macrophage recruitment, increased interstitial pressure, and blood vessel maturation during angiogenesis. Blood. 2004;104:3198. doi: 10.1182/blood-2004-04-1485. [DOI] [PubMed] [Google Scholar]
- 35.Nakasaki M. Yoshioka K. Miyamoto Y. Sasaki T. Yoshikawa H. Itoh K. IGF-I secreted by osteoblasts acts as a potent chemotactic factor for osteoblasts. Bone. 2008;43:869. doi: 10.1016/j.bone.2008.07.241. [DOI] [PubMed] [Google Scholar]
- 36.Ogata N. Kawaguchi H. Involvement of insulin and IGF-1 signaling molecules in bone metabolism. Clin Calcium. 2008;18:614. [PubMed] [Google Scholar]
- 37.Shimoaka T. Kamekura S. Chikuda H. Hoshi K. Chung U. Akune T. Maruyama Z. Komori T. Matsumoto M. Ogawa W. Impairment of bone healing by insulin receptor substrate-1 deficiency. J Biol Chem. 2004;279:15314. doi: 10.1074/jbc.M312525200. [DOI] [PubMed] [Google Scholar]
- 38.Ogata N. Insulin receptor substrate-1 in osteoblast is indispensable for maintaining bone turnover. J Clin Investig. 2000;105:935. doi: 10.1172/JCI9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawaguchi H. Molecular backgrounds of age-related osteoporosis from mouse genetics approaches. Rev Endocr Metab Disord. 2007;7:17. doi: 10.1007/s11154-006-9011-3. [DOI] [PubMed] [Google Scholar]
- 40.Akune T. Ogata N. Hoshi K. Kubota N. Terauchi Y. Tobe K. Takagi H. Azuma Y. Kadowaki T. Nakamura K. Insulin receptor substrate-2 maintains predominance of anabolic function over catabolic function of osteoblasts. J Cell Biol. 2002;159:147. doi: 10.1083/jcb.200204046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodda S.J. McMahon A.P. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 42.Ryoo H.M. Lee M.H. Kim Y.J. Critical molecular switches involved in BMP-2-induced osteogenic differentiation of mesenchymal cells. Gene. 2006;366:51. doi: 10.1016/j.gene.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Zeevi A. Lunz J.G., III Shapiro R. Randhawa P. Mazariegos G. Webber S. Girnita A. Emerging role of donor-specific anti–human leukocyte antigen antibody determination for clinical management after solid organ transplantation. Hum Immunol. 2009;70:645. doi: 10.1016/j.humimm.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Niemeyer P. Kornacker M. Mehlhorn A. Seckinger A. Vohrer J. Schmal H. Kasten P. Eckstein V. Südkamp N.P. Krause U. Comparison of immunological properties of bone marrow stromal cells and adipose tissue-derived stem cells before and after osteogenic differentiation in vitro. Tissue Eng. 2007;13:111. doi: 10.1089/ten.2006.0114. [DOI] [PubMed] [Google Scholar]
- 45.Ito T. Sawada R. Fujiwara Y. Tsuchiya T. FGF-2 increases osteogenic and chondrogenic differentiation potentials of human mesenchymal stem cells by inactivation of TGF-β signaling. Cytotechnology. 2008;56:1. doi: 10.1007/s10616-007-9092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reya T. Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 47.Wu Q. Kuo H.C. Deng G.G. Serine proteases and cardiac function. BBA-Proteins Proteomics. 2005;1751:82. doi: 10.1016/j.bbapap.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 48.Yan W. Sheng N. Seto M. Morser J. Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274:14926. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 49.Duester G. Alcohol dehydrogenase as a critical mediator of retinoic acid synthesis from vitamin A in the mouse embryo. J Nutrition. 1998;128:459S. doi: 10.1093/jn/128.2.459S. [DOI] [PubMed] [Google Scholar]
- 50.Wan D.C. Shi Y.Y. Nacamuli R.P. Quarto N. Lyons K.M. Longaker M.T. Osteogenic differentiation of mouse adipose-derived adult stromal cells requires retinoic acid and bone morphogenetic protein receptor type IB signaling. Proc Natl Acad Sci. 2006;103:12335. doi: 10.1073/pnas.0604849103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manton K.J. Leong D.F.M. Cool S.M. Nurcombe V. Disruption of heparan and chondroitin sulfate signaling enhances mesenchymal stem cell-derived osteogenic differentiation via bone morphogenetic protein signaling pathways. Stem Cells. 2007;25:2845. doi: 10.1634/stemcells.2007-0065. [DOI] [PubMed] [Google Scholar]
- 52.Zambelli A. Mongiardini E. Villegas S. Carri N. Boot-Handford R. Wallis G. Transcription factor XBP-1 is expressed during osteoblast differentiation and is transcriptionally regulated by parathyroid hormone (PTH) Cell Biol Int. 2005;29:647. doi: 10.1016/j.cellbi.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 53.Wu J. Kaufman R. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Diff. 2006;13:374. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- 54.Roybal C.N. Yang S. Sun C.W. Hurtado D. Vander Jagt D.L. Townes T.M. Abcouwer S.F. Homocysteine increases the expression of VEGF by a mechanism involving endoplasmic reticulum stress and transcription factor ATF4. J Biol Chem. 2004;279:14844. doi: 10.1074/jbc.M312948200. [DOI] [PubMed] [Google Scholar]
- 55.Kimble R. Vannice J. Bloedow D. Thompson R. Hopfer W. Kung V. Brownfield C. Pacifici R. Interleukin-1 receptor antagonist decreases bone loss and bone resorption in ovariectomized rats. J Clin Investig. 1994;93:1959. doi: 10.1172/JCI117187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasukawa H. Ohishi M. Mori H. Murakami M. Chinen T. Aki D. Hanada T. Takeda K. Akira S. Hoshijima M. Hirano T. Chien K.R. Yoshimura A. IL-6 induces an anti-inflammatory response in the absence of SOCS3 in macrophages. Nat Immunol. 2003;4:551. doi: 10.1038/ni938. [DOI] [PubMed] [Google Scholar]
- 57.Kitazawa R. Kimble R. Vannice J. Kung V. Pacifici R. Interleukin-1 receptor antagonist and tumor necrosis factor binding protein decrease osteoclast formation and bone resorption in ovariectomized mice. J Clin Investig. 1994;94:2397. doi: 10.1172/JCI117606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Croker B.A. Krebs D.L. Zhang J. Wormald S. Willson T.A. Stanley E.G. Robb L. Greenhalgh C.J. Forster I. Clausen B.E. Nicola N.A. Metcalf D. Hilton D.J. Roberts A.W. Alexander W.S. SOCS3 negatively regulates IL-6 signaling in vivo. Nat Immunol. 2003;4:540. doi: 10.1038/ni931. [DOI] [PubMed] [Google Scholar]
- 59.Ehlting C. Lai W.S. Schaper F. Brenndorfer E.D. Matthes R.J. Heinrich P.C. Ludwig S. Blackshear P.J. Gaestel M. Haussinger D. Regulation of suppressor of cytokine signaling 3 (SOCS3) mRNA stability by TNF-{alpha} involves activation of the MKK6/p38MAPK/MK2 cascade. J Immunol. 2007;178:2813. doi: 10.4049/jimmunol.178.5.2813. [DOI] [PubMed] [Google Scholar]
- 60.Logar D.B. Komadina R. Preželj J. Ostanek B. Trošt Z. Marc J. Expression of bone resorption genes in osteoarthritis and in osteoporosis. J Bone Mineral Metab. 2007;25:219. doi: 10.1007/s00774-007-0753-0. [DOI] [PubMed] [Google Scholar]
- 61.Kon T. Cho T.J. Aizawa T. Yamazaki M. Nooh N. Graves D. Gerstenfeld L.C. Einhorn T.A. Expression of osteoprotegerin, receptor activator of NF-κB ligand (osteoprotegerin ligand) and related proinflammatory cytokines during fracture healing. J Bone Mineral Res. 2001;16:1004. doi: 10.1359/jbmr.2001.16.6.1004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.