Abstract
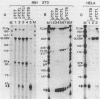
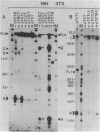
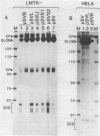
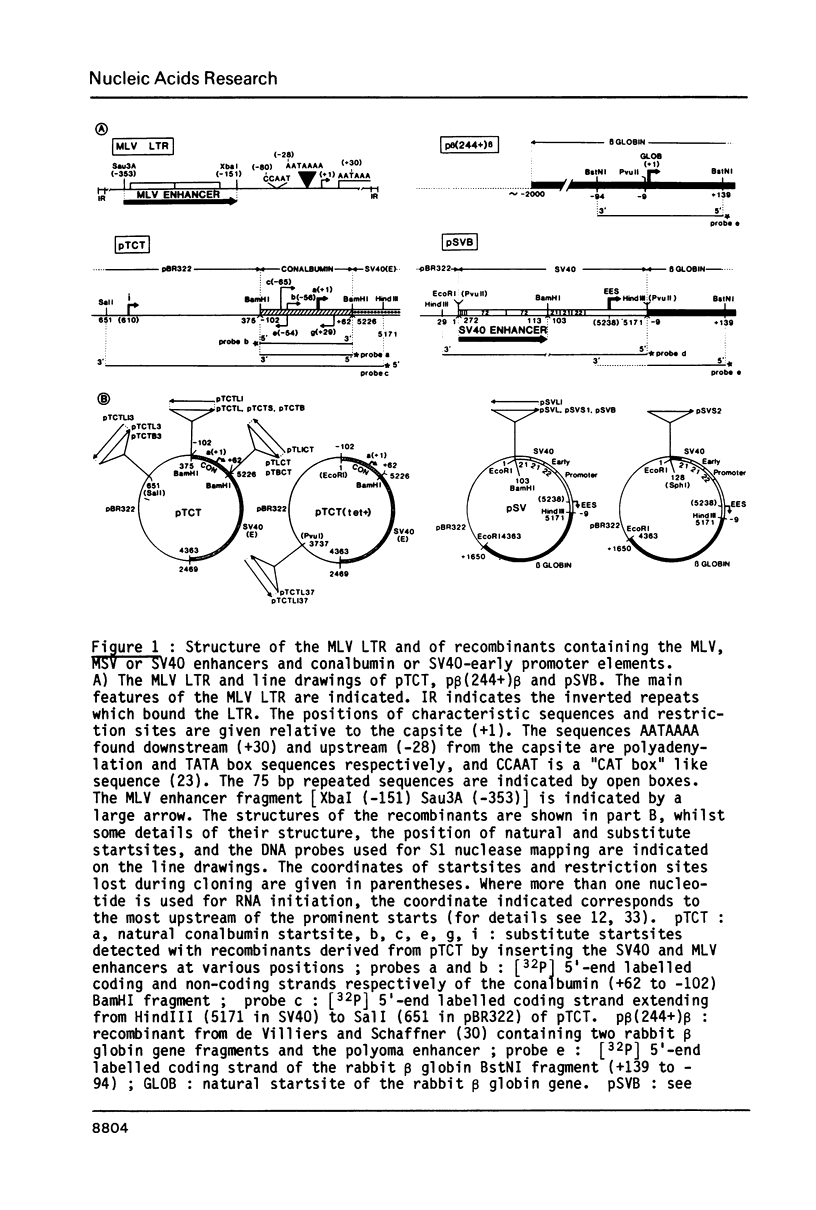
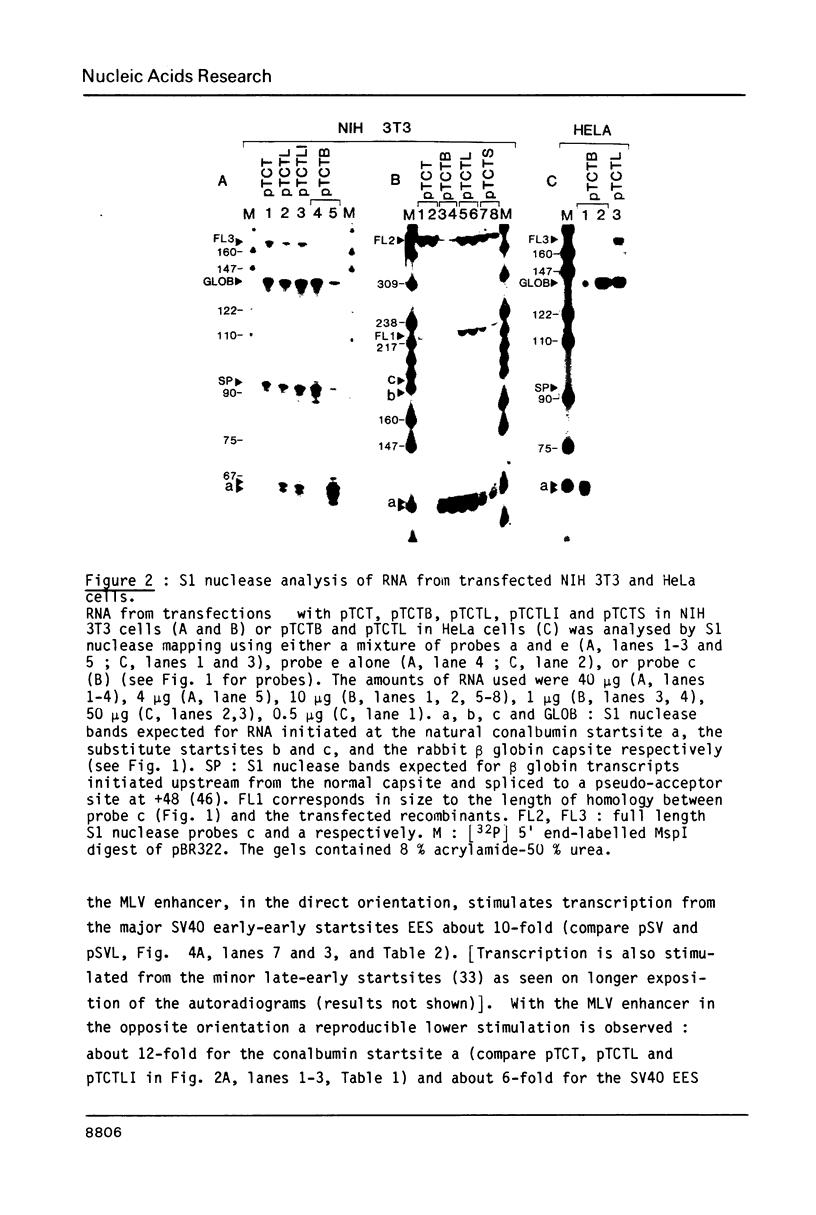
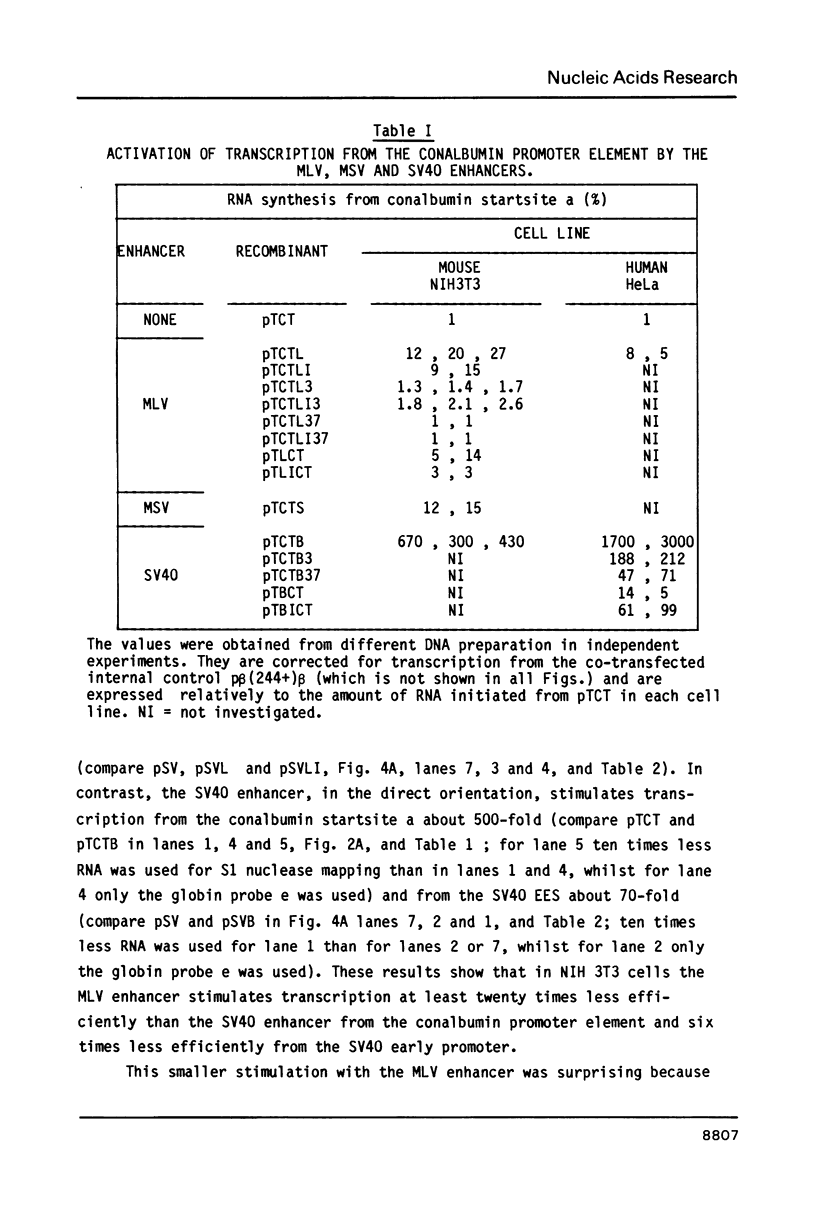
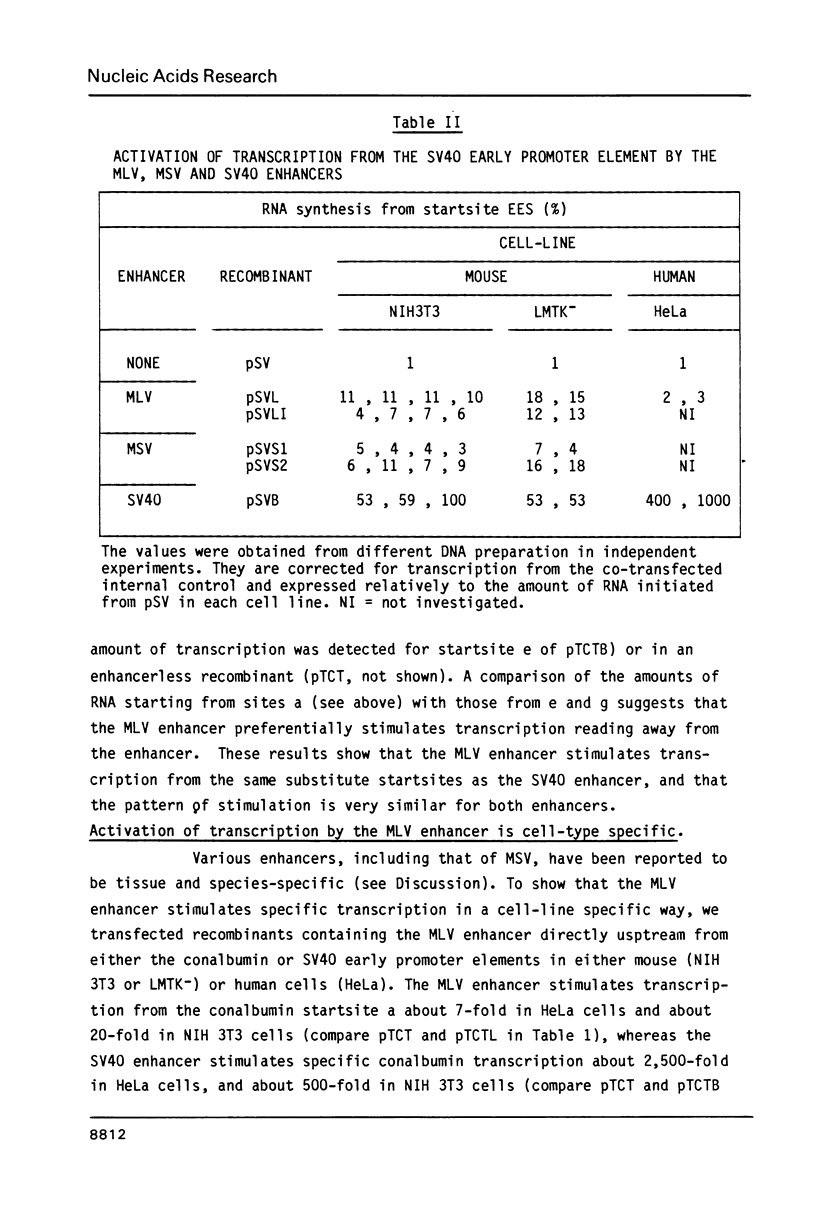
Activation of transcription by the Moloney Murine Leukemia Virus (MLV) and Simian Virus (SV40) enhancers was compared by transfecting recombinants containing these enhancers in either mouse or human cell-lines, and analysing RNA 48 h later by quantitative S1 nuclease mapping. The enhancers share the following properties. They stimulate transcription in an orientation-independent manner from the same startsites on the natural heterologous conalbumin (+62 to -102) or SV40 early promoter elements as well as on substitute promoter elements. The enhancers are most efficient when they are located directly upstream from the conalbumin (+62 to -102) promoter element, but they still stimulate transcription when they are either immediately downstream from the promoter element, or further upstream. Increasing the distance by interposing DNA sequences between the enhancers and the conalbumin promoter fragment results in decreased activation. Both enhancers show some cell-line specificity for activation of transcription. However, in all cell-lines and constructions tested the MLV enhancer was always less efficient than the SV40 enhancer. These results suggest that the MLV and SV40 enhancers stimulate transcription by similar mechanisms.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Baty D., Barrera-Saldana H. A., Everett R. D., Vigneron M., Chambon P. Mutational dissection of the 21 bp repeat region of the SV40 early promoter reveals that it contains overlapping elements of the early-early and late-early promoters. Nucleic Acids Res. 1984 Jan 25;12(2):915–932. doi: 10.1093/nar/12.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss M. A. Enhancer elements in immunoglobulin genes. Nature. 1983 May 26;303(5915):281–282. doi: 10.1038/303281a0. [DOI] [PubMed] [Google Scholar]
- Chen H. R., Barker W. C. Nucleotide sequences of the retroviral long terminal repeats and their adjacent regions. Nucleic Acids Res. 1984 Feb 24;12(4):1767–1778. doi: 10.1093/nar/12.4.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsaro C. M., Pearson M. L. Enhancing the efficiency of DNA-mediated gene transfer in mammalian cells. Somatic Cell Genet. 1981 Sep;7(5):603–616. doi: 10.1007/BF01549662. [DOI] [PubMed] [Google Scholar]
- Dhar R., McClements W. L., Enquist L. W., Vande Woude G. F. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Fromm M., Berg P. Deletion mapping of DNA regions required for SV40 early region promoter function in vivo. J Mol Appl Genet. 1982;1(5):457–481. [PubMed] [Google Scholar]
- Fromm M., Berg P. Transcription in vivo from SV40 early promoter deletion mutants without repression by large T antigen. J Mol Appl Genet. 1983;2(1):127–135. [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Grosveld G. C., de Boer E., Shewmaker C. K., Flavell R. A. DNA sequences necessary for transcription of the rabbit beta-globin gene in vivo. Nature. 1982 Jan 14;295(5845):120–126. doi: 10.1038/295120a0. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly D. J., Esty A. C., Subramani S., Friedmann T., Verma I. M. Elements in the long terminal repeat of murine retroviruses enhance stable transformation by thymidine kinase gene. Nucleic Acids Res. 1983 Mar 25;11(6):1855–1872. doi: 10.1093/nar/11.6.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Gruss P., Pozzatti R., Khoury G. Characterization of enhancer elements in the long terminal repeat of Moloney murine sarcoma virus. J Virol. 1984 Jan;49(1):183–189. doi: 10.1128/jvi.49.1.183-189.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimins L. A., Khoury G., Gorman C., Howard B., Gruss P. Host-specific activation of transcription by tandem repeats from simian virus 40 and Moloney murine sarcoma virus. Proc Natl Acad Sci U S A. 1982 Nov;79(21):6453–6457. doi: 10.1073/pnas.79.21.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson B., Khoury G., Vande Woude G., Gruss P. Activation of SV40 genome by 72-base pair tandem repeats of Moloney sarcoma virus. Nature. 1982 Feb 18;295(5850):568–572. doi: 10.1038/295568a0. [DOI] [PubMed] [Google Scholar]
- Marx J. L. Immunoglobulin genes have enhancers. Science. 1983 Aug 19;221(4612):735–737. doi: 10.1126/science.6410507. [DOI] [PubMed] [Google Scholar]
- Mercola M., Wang X. F., Olsen J., Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983 Aug 12;221(4611):663–665. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. S. Expression and regulation of immunoglobulin heavy chain gene transfected into lymphoid cells. EMBO J. 1983;2(8):1373–1378. doi: 10.1002/j.1460-2075.1983.tb01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D., Schaffner W. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 1984 Jan 5;307(5946):80–82. doi: 10.1038/307080a0. [DOI] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Srinivasan A., Reddy E. P., Dunn C. Y., Aaronson S. A. Molecular dissection of transcriptional control elements within the long terminal repeat of the retrovirus. Science. 1984 Jan 20;223(4633):286–289. doi: 10.1126/science.6322296. [DOI] [PubMed] [Google Scholar]
- Van Beveren C., Rands E., Chattopadhyay S. K., Lowy D. R., Verma I. M. Long terminal repeat of murine retroviral DNAs: sequence analysis, host-proviral junctions, and preintegration site. J Virol. 1982 Feb;41(2):542–556. doi: 10.1128/jvi.41.2.542-556.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. E. Form and function of retroviral proviruses. Science. 1982 May 21;216(4548):812–820. doi: 10.1126/science.6177038. [DOI] [PubMed] [Google Scholar]
- Wartell R. M., Reznikoff W. S. Cloning DNA restriction endonuclease fragments with protruding single-stranded ends. Gene. 1980 May;9(3-4):307–319. doi: 10.1016/0378-1119(90)90329-p. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Chambon P. Short and long range activation by the SV40 enhancer. Nucleic Acids Res. 1984 Jul 25;12(14):5589–5608. doi: 10.1093/nar/12.14.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Matthes H., Wintzerith M., Chambon P. Transcription from the SV40 early-early and late-early overlapping promoters in the absence of DNA replication. EMBO J. 1983;2(9):1605–1611. doi: 10.1002/j.1460-2075.1983.tb01631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Yaniv M. Enhancing elements for activation of eukaryotic promoters. Nature. 1982 May 6;297(5861):17–18. doi: 10.1038/297017a0. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Banerji J., Schaffner W. Analysis of the transcriptional enhancer effect. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):911–919. doi: 10.1101/sqb.1983.047.01.105. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Tyndall C., Schaffner W. Transcriptional 'enhancers' from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982 Dec 20;10(24):7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]