SUMMARY
Stress responses in plants are tightly coordinated with developmental processes, but the interaction of these pathways is poorly understood. We used genome-wide assays at high spatio-temporal resolution to understand the processes that link development and stress in the Arabidopsis root. Our meta-analysis finds little evidence for a universal stress response. However, common stress responses appear to exist with many showing cell-type specificity. Common stress responses may be mediated by cell identity regulators, as mutations in these genes resulted in altered responses to stress. Evidence for a direct role for cell identity regulators came from genome-wide binding profiling of the key regulator SCARECROW, which showed binding to regulatory regions of stress-responsive genes. Co-expression in response to stress was used to identify genes involved in specific developmental processes. These results reveal surprising linkages between stress and development at cellular resolution, and show the power of multiple genome-wide datasets to elucidate biological processes.
INTRODUCTION
Plant development is highly plastic and is profoundly influenced by the environment. Surprisingly, little research focuses on the interplay between development and environmental stress, and much of the work that has been done centers on the response of whole organs to a small number of stresses (Kreps et al., 2002; Rabbani et al., 2003; Kilian et al., 2007; Zeller et al., 2009). Previous work has identified several common transcriptional responses to stress (Fujita et al., 2006; Walley et al., 2007; Hirayama and Shinozaki 2010), and has led to the idea that plants have a universal stress response (Walley and Dehesh, 2010; Ma and Bohert, 2007). However, whole organs are a mixture of multiple cell types, and whole organ transcriptional responses often obscure the more complex and subtle changes that occur at cell-type resolution (Birnbaum et al., 2003; Brady et al., 2007; Gifford et al., 2008; Dinneny et al. 2008). Thus, how environmental stress affects the development pathways that regulate individual cell types is largely unknown. To address this issue requires examination of many different stresses at the resolution of individual cell types.
The Arabidopsis root is an excellent model for this purpose. Although 15 cell types have been described, the root can be simplified to a set of concentric cylinders that are radially symmetric around a central axis. From external to internal cell layers, these are the epidermis, cortex, endodermis and stele, with the columella and lateral root cap providing additional layers at the root tip. The longitudinal axis of the root can be viewed as a developmental timeline, with young cells at the root tip in the meristematic zone and older cells distal to the tip in the elongation and differentiation zones.
Each cell type in the root has its own transcriptional profile (Brady et al. 2007), and a recent report demonstrated that cell identity plays an important role in a plant’s response to environmental stress (Dinneny et al. 2008). However, this study was restricted to examining two stimuli, which limited the ability to identify patterns of gene expression within cell types across many stresses. To fully understand the root’s response to stress requires multiple conditions at high resolution using the same cell types and developmental stages. Here we profiled the transcript populations of whole roots, 5 cell types, and 4 developmental stages under two stress conditions, low pH and sulfur deficiency (−S). We combined these with two datasets of similar cell-type resolution describing the root response to high NaCl and iron deficiency (−Fe) at high resolution (Dinneny et al. 2008), and with 10 datasets that describe the whole root response to 10 different stresses (Kilian et al. 2007). This combined dataset allowed us to search for common stress responses in whole roots and cell types, and to identify patterns of gene expression both within a given cell type across multiple stresses and across multiple cell types for a given stress.
Although we find common stress responses in the whole root, we find little evidence for a universal stress response at either whole root or cell-type resolution. We show that common stress responses, such as those mediated by the plant hormone abscisic acid (ABA), exhibit cell-type specificity, and we provide both mutational and genome-wide binding data that suggests this specificity is mediated by cell identity regulators. We show that although different stresses uniquely affect root spatiotemporal transcriptional programs, there is a set of genes enriched in specific cell types regardless of environmental stress, suggesting that cell-cell communication is an important part of the root’s response to stress.
RESULTS
A common stress response in roots
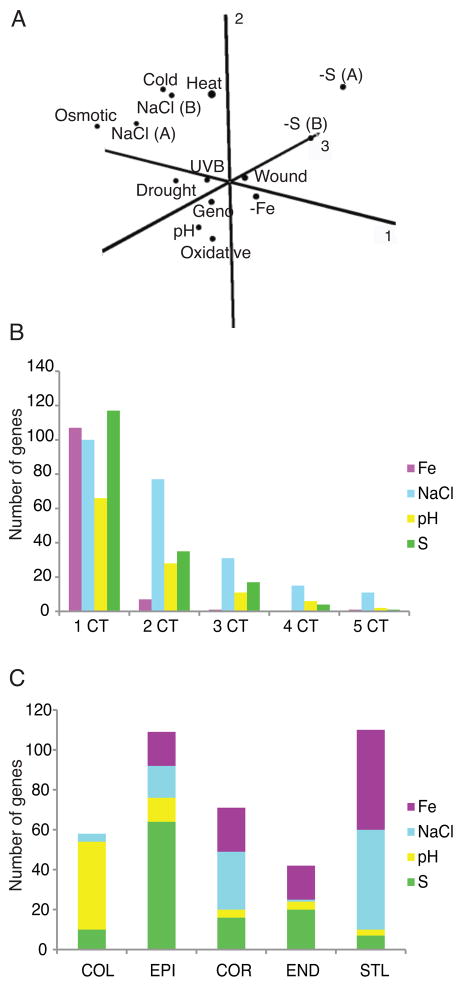
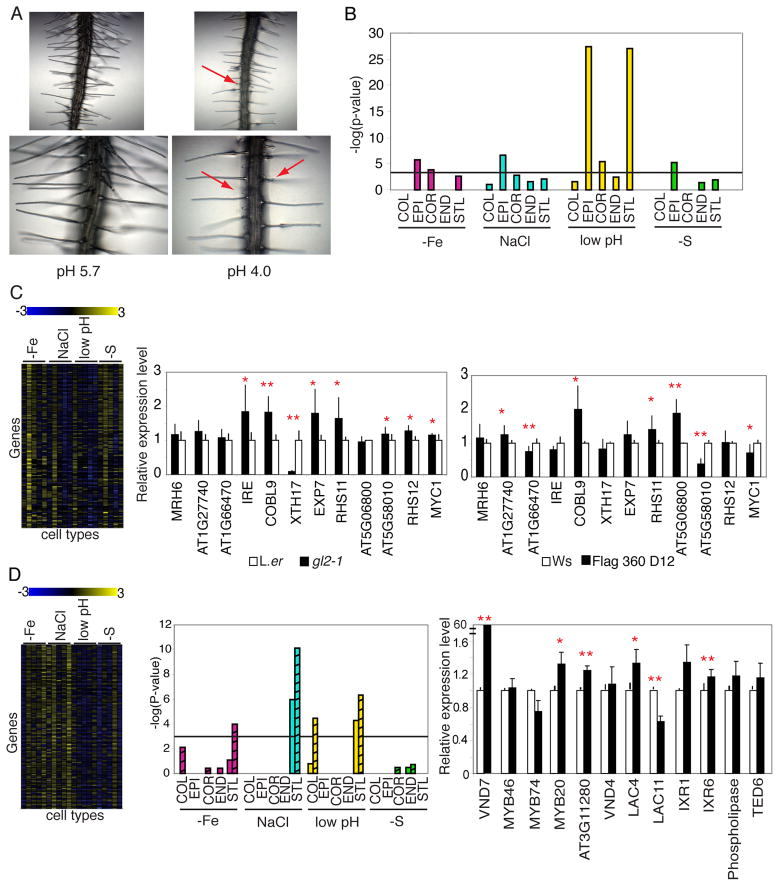
We profiled the whole root transcriptional response to pH 4.6 (low pH) and sulfur deficiency (−S) at different time points after transfer to the stress and combined these data with 12 publically available time-course datasets from whole roots (Table S1) exposed to different environmental stresses to try to identify a “universal stress response” in the Arabidopsis root. To determine whether a universal stress response exists in the root, we first identified differentially expressed genes in each of the 14 treatments using the RankProd package in R (Hong et al., 2006). The resulting P-values were combined using Fisher’s inverse χ2 method and FDR values obtained. We required that significant genes have a combined FDR value of < 0.0001 and, have an FDR < 0.01 in at least 75% (11 of 14) of the conditions (see experimental procedures). Using these criteria we identified 8 genes that were activated and 1 repressed gene (Table S2). The activated genes include a DNAJ heat shock protein and DREB2A, a transcription factor with major roles in drought stress and ABA responses (Sakuma et al., 2006). Consistent with the paucity of universally responsive genes, Principal Component Analysis (PCA) of GO categories significant under each stress showed that each stress elicited distinct functional responses in the root (Figure 1A).
Figure 1. Stress responses are stress and cell-type specific.
A) PCA analysis of GO categories significant in each of 14 stresses. Components 1, 2, and 3 explain 25.1, 17.7 and 11.7% of the variation, respectively. A = AtGenExpress; B = Benfey. B) The majority of whole root common stress responses respond in just one cell type, and this cell type differs for different stresses (C). See also Figure S1.
Although the PCA did not suggest the presence of a universal stress response, clustering of some stresses indicated that there may be functions common to a smaller subset of stresses. We therefore examined the set of genes responsive in at least 7 of the 14 conditions, and called these the “common stress response” (CSR; Table S2). The 274 activated genes in the common stress response are enriched for well-known abiotic and biotic stress responses, including “response to abscisic acid stimulus” (P < 10−9) and “response to other organism” (P < 10−5). Four transcription-related categories are also enriched within these genes, including “transcription factor activity” (P < 10−6) and “DNA-dependent regulation of transcription” (P < 10−5). Coincident with this, three families of transcription factors (TFs) are enriched (P < 0.001) in the activated CSR: AP2/EREBP, NAC, and WRKY (Table S2).
Common stress responses show cell-type specificity
We hypothesized that the whole root common stress responses would be present in many, if not all, cell types in the root. Using fluorescently activated cell sorting of plants expressing cell-type specific GFP reporters (Dinneny et al., 2008) coupled with microarray analysis, we profiled 5 different cell types: columella (Col), epidermis (Epi), cortex (Cor), endodermis (End), and stele (Stl) in response to low pH and −S. Plants were grown on standard media and then transferred either to media with a pH of 4.6 (normal is pH 5.7) or sulfur deficient media. Expression of GFP reporters was not altered under stress conditions (Figure S1). We combined these data with that from two additional stresses, high NaCl and Fe deficiency (Dinneny et al., 2008), which covered the same cell types. We defined a stress-regulated gene as one with significantly altered expression (|FC| > 1.5, FDR < 1 × 10−4) under stress conditions compared to standard conditions. Surprisingly, when we examined the distribution of the CSR genes across cell types in −Fe, high NaCl, low pH and −S, we found that the majority of common stress responses show cell-type specificity (Figure 1B). Moreover, these cell-type specific CSR genes do not all respond in the same cell type. For example, some respond in the columella, others in the cortex or endodermis, depending on the stress (Figure1C).
ABA responses are cell-type, developmental-stage, and stress-dependent
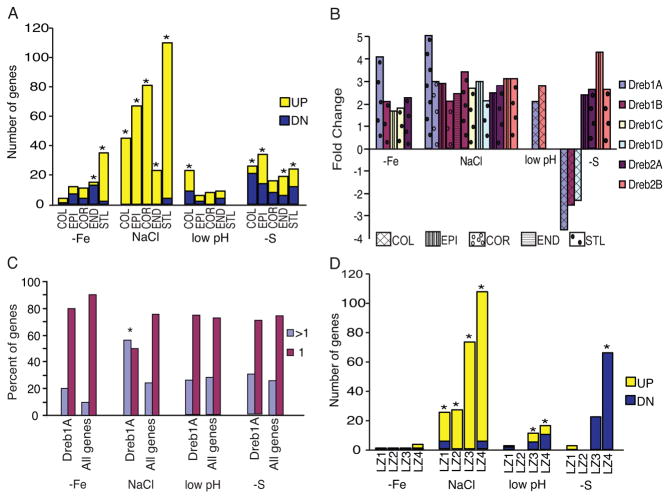
To further examine the cell-type specificity of whole root stress responses we focused on those mediated by the plant hormone abscisic acid (ABA), as it is a well-documented common stress response (Cutler et al., 2010) and ABA responses were enriched in the CSR genes (P = 3.54 × 10−11 in the activated CSR gene set; 22 of the 274 activated CSR genes were ABA marker genes; Table S2). First, we examined the cell-type expression of ABA marker genes (Nemhauser et al., 2006) that significantly responded in at least one of the 14 conditions in our whole root meta-analysis (Table S3, see Table S2 and Figure S2 for only the ABA marker genes that are also CSR genes). We find that ABA regulates responses to stress differentially across stresses and cell types in the root. ABA marker genes are primarily activated and enriched throughout all cell layers in the root under high NaCl, while ABA responses to −Fe, low pH and −S are both activated and repressed and enrichment is restricted to specific cell layers (Figure 2A; Figure S2). Further, the ABA response genes activated and repressed in each cell-type and stress differ (Table S2, 3, Figure S2).
Figure 2. ABA-mediated stress responses are stress and cell-type dependent.
A) ABA responses to salt are enriched in all cell layers, but enrichment is restricted to specific cell-types under −Fe, pH and −S. * = marker gene enrichment P < 0.005. B) Response of the six DREB TFs to different stresses. Multiple bars indicate that the TF responds in more than one cell type; if not shown the TF is not differentially expressed in the stress. C) DREB1A putative targets are enriched (indicated by *; P = 5.3 × 10−5) in genes that respond to salt stress in more than one cell type, but are regulated primarily in a cell-type specific manner under −Fe, low pH and −S. > 1 = more than one cell type; 1 = cell-type specific. D) ABA marker genes are developmental stage and stress specific. LZ1= apical meristem, LZ2= basal meristem, LZ3=elongation zone, LZ4=maturation zone (* significant at P < 0.001). See also Figure S2.
Next, we examined the expression of master regulators of the ABA signaling pathway within the cell-type data (see Table S4 for all expression values in the cell-type dataset). The AP2 transcription factors DREB1A, 1B, 1D, 2A and 2B control different components of the ABA pathway (Tuteja, 2007). Their expression within the 5 cell types assayed showed that although DREB1A, B, D, and DREB2A and B respond to NaCl in more than one cell-type, they are regulated in a cell-type specific manner in at least one of the other stresses (Figure 2B). Consistent with this, putative targets of DREB1A are significantly enriched among genes that respond to NaCl stress in more than one cell type (P = 5.3 × 10−5) but are cell-type specific under each of the other stresses (Figure 2C).
In addition to the cell-type specificity, ABA-marker genes show developmental-stage specificity. We transcriptionally profiled 4 different developmental stages in the root after transfer to low pH or −S and combined these with similar datasets for high salt and −Fe. Similar to the results at cell-type resolution, we find that ABA marker genes are differentially expressed both in developmental stages and stresses (Figure 2D; see Table S5 for developmental stage dataset expression values).
Together, these results show that although ABA mediates responses to multiple abiotic stresses, these responses exhibit context-dependent cell-type specificity.
Mutations in cell identity regulators lead to altered ABA responses
The cell-type specificity of common stress responses was surprising, and raised the question as to how this is controlled. We hypothesized that this specificity may be due to stress responsiveness of cell identity regulators, which we defined as genes with demonstrated roles in the determination or maintenance of a cell type. As a first test, we examined the expression under stress of a set of cell identity regulators at cell-type resolution. We find that many cell identity regulators are differentially expressed under stress conditions. This occurs both in cell types known to be regulated by them and, occasionally, in cell types in which they have no documented role (Figure 3A, Table S6). For example, CAPRICE (CPC), which is necessary for epidermal patterning, is induced by high NaCl in both the epidermis and stele (Figure 3A).
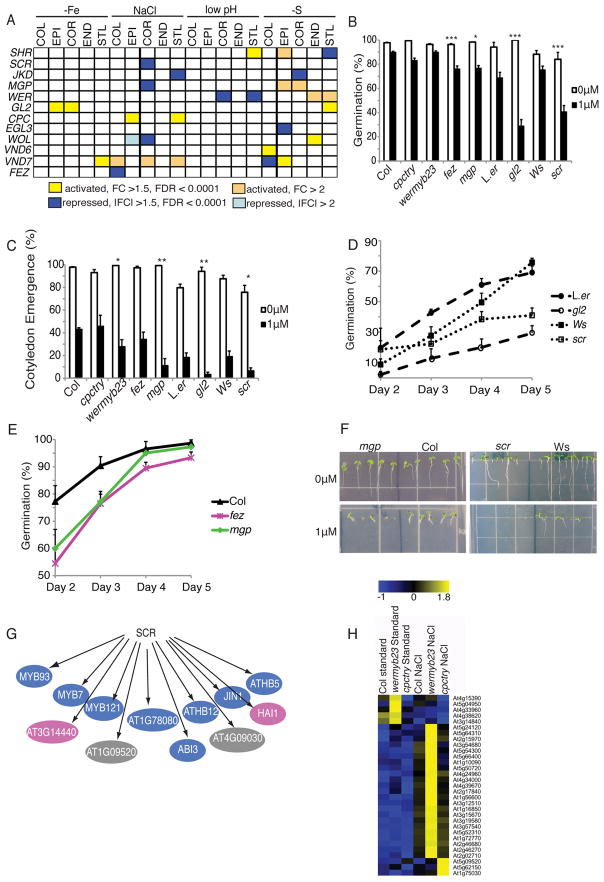
Figure 3. Cell identity regulators interact with the ABA signaling pathway.
A) Cell identity regulators can be stress-regulated both in cell types they regulate and in those they are not known to influence. B – F) Mutants in cell identity regulators result in altered responses to ABA. Error bars show standard error. B) Percent germination of seeds 3 (Col, cpctry, wermyb23, fez, mgp) or 5 (L.er, gl2, Ws, scr) days after transfer to light. L.er is the control for gl2; Ws for scr; Col for remaining mutants. C) Percent fully expanded, green cotyledons 5 (Col, cpctry, wermby23, fez, mgp) or 6 (L.er, gl2, Ws, scr) days after transfer to light. For B) and C), stars denote significant differences between mutant and wild type on 1μM ABA: * = P < 0.05, ** = P < 0.01, *** = P < 0.001. D and E) Germination time-course of hypersensitive mutants on 1μM ABA 2–5 days after transfer to light, D) Ws, scr, L.er, gl2; E) Col, fez, mgp. F) mgp and scr mutants are hypersensitive to ABA. G) SCR CHip-chip reveals that ABA response genes (from Nemhauser et al., 2006 and GO ontologies) are SCR direct targets. Blue = transcription factors, pink = enzymes, gray = other. H) Expression of ABA marker genes under high NaCl in epidermal patterning mutants wermyb23 and cpctry shows that ABA marker gene expression is altered in the ABA hypersensitive wermyb23 mutant.
If cell identity regulators play a role in stress responses such as those mediated by ABA, we hypothesized that mutations in these factors could result in hypersensitivity or resistance to ABA. In addition to its role in stress responses, ABA functions in germination and early seedling development. We examined the germination and cotyledon emergence of six different cell identity regulator mutants, five of which had altered expression patterns under stress (Figure 3A). As shown in Figure 3B – F, mutations in several cell identity regulators result in altered responses to exogenous ABA, either in germination, cotyledon emergence, or both. This is most likely not due to the developmental defects present in some of these mutants, since cpctry, which has no root hairs, is not hypersensitive in either assay.
ABA response genes are direct and indirect targets of cell identity regulators
Many of the cell identity regulators tested above encode transcription factors. We postulated that if these proteins play a role in ABA responses, their direct targets would include known ABA response genes. We tested this using SCR, as the scr mutant is hypersensitive to ABA (Figure 3B – D, F). To determine which genes regulated by SCR are direct targets, we performed chromatin immunoprecipitation of SCR followed by hybridization to an oligonucleotide microarray (ChIP-chip). We identified 181 putative SCR direct target genes (Table S7), several of which are ABA response genes (Figure 3G, Table S7). Indeed, GO category analysis showed that “Response to ABA stimulus” was enriched (P < 0.001) among the putative SCR direct targets (Table S7).
Since wermyb23 and gl2 mutants were also hypersensitive to ABA, we tested whether altered epidermal root hair patterning leads to differential ABA responses under stress. Using publically available microarray data (Dinneny et al., 2008) from the superhairy wermyb23 and hairless cpctry double mutants, we identified stress-regulated hair patterning-dependent ABA marker genes (Figure 3H, Table S7). In line with our phenotypic results above, ABA marker genes were enriched (P = 5.9 × 10−12) among the genes misregulated under stress in the wermyb23 mutant, but not in the cpctry mutant, which is not ABA hypersensitive. Taken together, our results show that cell identity regulators play a role in the root’s response to stress, and suggest that these regulators are entry points for stress and developmental pathway interactions.
Response centers have developmental defects
The high degree of cell-type specificity within the common stress response genes from whole roots suggested that most genes responsive to stress in the 5 cell types profiled (the cell-type gene set; see Table S4 for expression values) would also be cell-type specific. Indeed, stress-regulation depends both on the cell types and stress examined. The majority of stress-regulated genes in the cell-type gene set respond in just one cell type, regardless of stress (Figure S3) while the majority of responsive genes in this gene set are stress-specific (Figure S3). However, the same gene can respond to different stresses in different cell types (Figure S3). We searched for a universal stress response at cell-type resolution using the same meta-analytic methods as for whole roots. In line with the high degree of cell-type and stress specificity, we found no evidence for such a response.
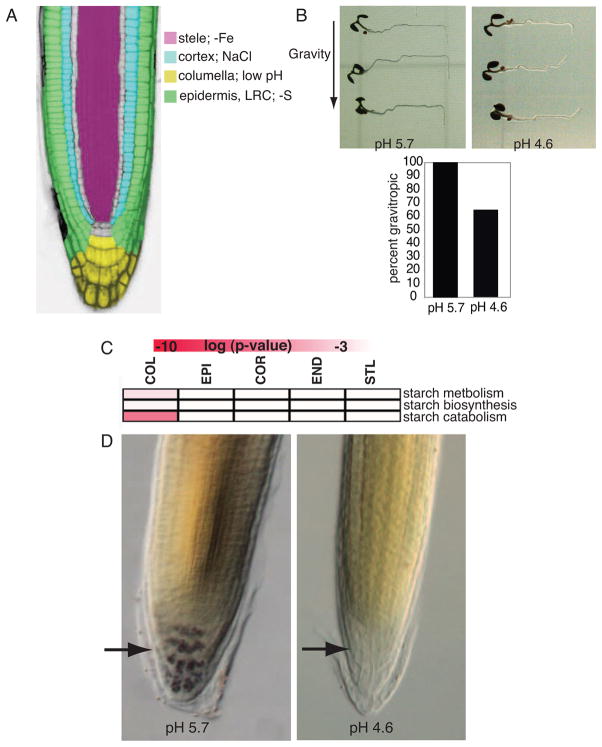
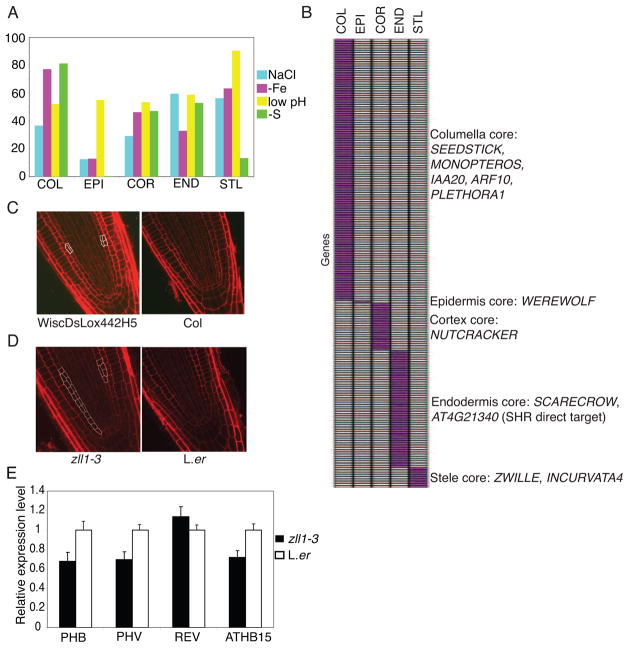
The most responsive cell type differs for each stress examined in the cell type gene set (Figure 4A). These “response centers” often correlate with a phenotypic change. For example, the columella is the most responsive cell type under low pH, and the gravity sensing response is altered under this stress (Figure 4B). Starch-filled plastids in the columella play a role in the gravity response in plants (Chen et al. 2002), and starch catabolism, metabolism and biosynthesis are enriched GO categories in the columella under low pH (Figure 4C). Consistent with this, starch is absent from the columella 24 hours after transfer to low pH (Figure 4D).
Figure 4. The low pH stress response center correlates with developmental changes.
A) The most responsive cell type differs for each of the four stresses examined. B) Defective gravitropic response 24H after transfer to low pH. C) Heat map showing that the GO categories starch catabolism, metabolism and biosynthesis are enriched in the columella under low pH. Red indicates level of enrichment. D) Most starch granules are absent from the columella 24H after transfer to low pH. Arrows point to the columella. See also Figure S3.
Common cell-type stress responses identify transcription modules
Although we did not identify a universal stress response at cell-type resolution, two biological responses, root hair morphogenesis and secondary cell wall biosynthesis, were enriched in at least three of the four stresses. Root hair development is altered under low pH, −Fe and NaCl (Figure 5A and Dinneny et al., 2008) and root hair marker genes are enriched in the epidermis under all four stresses (Figure 5B). We hypothesized that looking for coexpression in response to stresses might identify genes with roles in specific developmental processes, particularly as regulators for each process are stress-regulated (Figure 3A). Using k-means clustering, we grouped the cell-type stress-regulated genes into 35 clusters (Table S8, see supplemental experimental procedures for details). One cluster showed strong enrichment (P = 9.66 × 10−40) for root hair marker genes, including GLABRA2 (GL2), a regulator of epidermal cell identity (Tominaga-Wada et al., 2009). We tested genes in this cluster to determine if they were regulated by GL2 using quantitative RT-PCR (qRT-PCR) in the gl2 mutant (Table S9). Eight of the 12 genes tested were differentially expressed in the mutant (Figure 5C). Many of these, including RHS11 and EXP7, had not previously been shown to be regulated by GL2. We used a similar strategy with the bHLH TF At5G58010, which regulates root hair development both in Arabidopsis and Lotus japonica (Karas et al., 2009) and is also expressed in this cluster. Seven of the 12 genes tested by quantitative RT-PCR (qRT-PCR) in a T-DNA insertion line of At5G58010 were misregulated (Figure 5C). These included known root hair regulators such as COBL9 and RHS11 as well as genes with no known role in root hair development, such as the MYB TF AT5G06800.
Figure 5. Cell-type stress data reveals components of development-associated transcription networks.
A) Root hair development is altered under low pH. Red arrows point to short, swollen root hairs. B) Root hair marker genes are enriched in the epidermis under each stress. Black line marks P < 0.001. C) Root hair cluster with enrichment (P = 9.66 × 10−40) of root hair marker genes. Left, heat map showing fold change of expression for genes in the cluster for all cell types under each stress. Cell types are arranged as COL, EPI, COR, END, STL for each stress. Middle and right, qRT-PCR of selected genes in the root hair cluster in the gl2 mutant (middle) or AT5G58010 T-DNA insertion line (right). D) Secondary cell wall biosynthesis cluster. Left, heat map with fold change of genes under each stress (cell type listed as in C). Middle, secondary cell wall biosynthesis genes are enriched in the STL in three stresses. Striped bars = genes identified from Persson et al. 2005; non-striped from Brown et al. 2005. Right, qRT-PCR of selected genes in this cluster in 35S::VND7. * = P < 0.05; ** = P < 0.01; one-tailed t-test. Error bars denote standard error.
Genes necessary for secondary cell wall biosynthesis (Persson et al., 2005; Brown et al., 2005) are enriched in the stele in −Fe, NaCl and −S (Figure 5D). We identified one cluster (cluster 5, Table S8) with strong enrichment (P = 3.39 × 10−33) of these genes. Cluster 5 contains a transcription module involving the protoxylem cell-identity regulator VND7 (Kubo et al, 2005), which is regulated at the cell-type level in −Fe, high NaCl, low pH and −S. Quantitative RT-PCR showed that multiple genes in this cluster were significantly up- or down-regulated in a 35S::VND7 line (Figure 5D). Several of these have not previously been shown to be regulated by VND7 and may be components of the secondary cell wall transcription network. Coexpression analyses of common cell-type stress responses may be a good predictor for identifying genes downstream of cell identity regulators in the developmental pathways they control.
Core markers suggest cell-cell communication in stress responses
Because cell-cell communication is vital for proper patterning in roots, we examined genes enriched in one cell type relative to all others under the same stress (Figure 6A, Figure S4, see experimental procedures). We identified 199 genes enriched in a cell type under all four stresses and the MS standard condition (Figure 6B, Table S10). Since these genes are enriched in a cell type regardless of environment, we define these as core markers (Dinneny et al., 2008). For example, 9 genes are enriched in the stele compared to all other cell types in all environments tested (Figure 6B).
Figure 6. Stress-enriched genes reveal core markers and cell identity regulators.
A) The epidermis is the most plastic cell type as it shows the least conservation between standard and stress conditions. Percent conservation of enriched genes between standard and stress conditions for each cell type is shown. B) Many cell identity regulators are found within the core markers. C) and D) Mutations in the core markers MYB36 (C) and ZWILLE (D) and identify regulators of radial patterning. E) qRT-PCR showing that HD-ZIP genes PHB, PHV, ATHB15 are repressed in roots of the zll1-3 mutant. Error bars show standard deviation. See also Figure S4.
Since core markers are enriched in a cell type regardless of environment, we hypothesized that these genes may be important for cell identity. Indeed, genes known to regulate cell identity are found among these markers. For example, LONESOME HIGHWAY, INCURVATA4, and ZWILLE (ZLL) are all cell-type markers in the stele, and all play a role in vasculature development (Ohashi-Ito and Bergmann, 2007; Ochando et al., 2006; Tucker et al., 2008). Mutations in the core columella marker PLETHORA1 (PLT1) result in extra columella cells (Aida et al., 2004), while overexpression of the columella marker IAA20 causes the columella not to differentiate and leads to agravitropism (Sato et al., 2008). The TFs, SHORTROOT (SHR) and SCARECROW (SCR) are necessary for the proper patterning of the endodermis and cortex, and two SHR direct targets, an uncharacterized bHLH (At4g21340) (Sozzani et al., 2010) and the transcription factor NUTCRACKER (Sozzani et al., 2010; Levesque et al., 2006; Cui et al., 2007), are cell-type markers in the endodermis and cortex, respectively. Further, eight SHR and SCR direct or indirect targets were identified in the endodermal core marker genes (Sozzani et al., 2010).
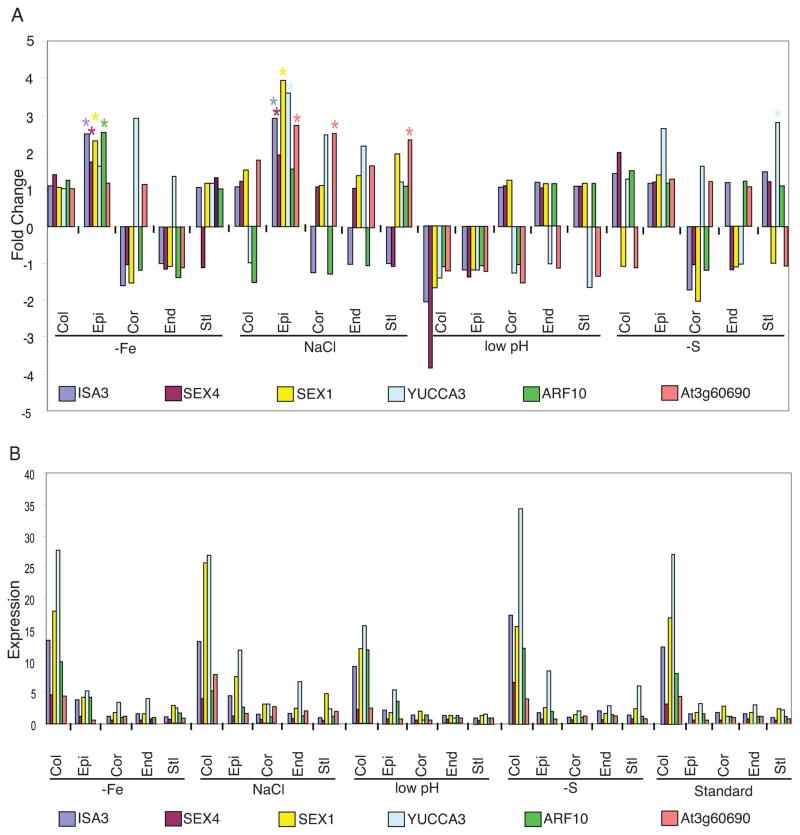
Surprisingly, although core markers are enriched in a cell type under all conditions, they respond to stress in many different cell types (illustrated for selected markers in Figure 7A, and in Table S10 for all markers). However, their expression level is always higher in the core cell type compared to the cell type in which they respond to stress (Figure 7B, Table S10). For example, expression of At4g05170, a bHLH TF and core endodermal marker, is significantly activated in the cortex under −Fe compared to standard MS (FC= 3.8, FDR < 0.0001). However, its absolute expression value in the cortex under −Fe is 2.9, compared to 14.9 in the endodermis under −Fe (Table S10). Thus, although core markers may respond to stress in different cell types, their expression levels are higher in the core cell type, suggesting some mechanism by which relative expression levels are communicated.
Figure 7. Core markers are stress-responsive but are most highly expressed in the core cell type.
Fold change with respect to standard conditions (A) and expression values (B) for all cell types and stresses of six core marker genes in the columella. Several genes significantly (FC > 1.5, FDR < 0.0001) respond to a stress in cell types other than the columella (indicated by a star) (A), but their expression values are highest in the columella compared to all other cell types for a given condition (B). At3g60690 is an auxin-responsive gene. See also Figure S4.
Core markers have functional relevance
Because many of the core marker genes are necessary for root patterning, we reasoned that this dataset may contain previously undescribed root patterning genes. Indeed, we found that mutations in two core markers, MYB36 and ZLL/PINHEAD/ARGONAUTE10 (AGO10), led to deviations from the typical wild type root organization (Figure 6C and D). None of the stresses examined resulted in altered root radial patterning, thus mutants were examined under standard conditions. A T-DNA insertion line (WiscDsLox442H5) of the core endodermal marker MYB36 resulted in increased divisions in the ground tissue, although with low penetrance (6/26 plants; Figure 6C). Although AGO10 is a core marker for the stele, we found extra divisions in the ground tissue of the zll1-3/ago10 mutant compared to wild-type in approximately 54% (14/26) of plants examined (Figure 6D). Mutations in AGO10 result in elevated levels of miR165/166 in stems and leaves and a consequent reduction in their target genes, the HDIII-ZIP TFs PHABULOSA (PHB), PHAVOLUTA (PHV), and REVOLUTA (REV), and ATHB-15 (Liu et al., 2008). We tested whether these target genes were downregulated in roots of zll1-3. Consistent with their expression in stems and leaves, expression levels of PHB, PHV, and ATHB-15 are downregulated in zll1-3 roots (Figure 6E). Together, these results suggest that core markers are important for root function and development.
DISCUSSION
Stress responses in multicellular organisms require the coordination of thousands of genes and regulatory networks. This transcriptional reprogramming differs by cell type, and is required for an organ’s developmental response to changing environmental conditions. In this report we transcriptionally profiled the response to two stresses in 5 cell types, 4 developmental stages, and different timepoints in the Arabidopsis root. We combined these data with two additional high-resolution datasets and 14 conditions in the whole root to form a comprehensive dataset of the effects of abiotic stress on the root. Our results reveal a layer of complex gene regulation within cell types under environmental stress and suggest that cell identity regulators play an important role in stress responses.
A universal stress response in the whole root?
The idea of a universal stress response in plants is compelling, as the identification of such a response could lead to the development of crops able to withstand many types of harsh environmental conditions. Understanding whether the root has a universal stress response depends on the resolution (whole root or cell type) and whether genes or functions are examined. Having explored 14 conditions in the whole root and 4 at cell-type resolution, we found little evidence for a universal stress response in either dataset. However, we identified a set of common stress response (CSR) genes that respond to multiple stimuli, in agreement with other studies (Ma and Bohnert, 2007; Walther et al., 2007; Swindell, 2006). Forty-six of the CSR genes were previously identified as members of a universal stress response cluster that examined 4 abiotic and multiple biotic stresses (Ma and Bohnert, 2007), while 15 of them were found among 26 root stress-general response genes (Swindell, 2006).
Although common stress responses are present in the whole root, these responses cannot be generalized at cell-type resolution, as the majority are cell-type specific. This is in agreement with the different functional responses of cell types to stress and suggests that the mechanisms underlying common stress responses in plants may be fine-tuned at cell-type resolution for each stress. This is analogous to animal systems, in which the expression level of cellular stress response genes and the pathways activated by transcription factors in this response can differ in different cell types (Kültz, 2005), suggesting a convergent evolutionarily theme of context-dependent stress specificity.
Cell identity regulators play a role in the ABA responses
Insight into how common stress responses are regulated at cell-type resolution can be gained from analyzing expression of cell identity regulators under stress. ABA responses are both cell-type and stress-dependent, and we find that several stress-responsive cell identity regulators interact with the ABA response pathway. ABA is not only a stress hormone, but plays a key role in developmental pathways throughout the plant, including senescence, embryogenesis, and lateral root development (Cutler et al., 2010). Since environmental responses often result in morphological changes within the plant, stress and developmental pathways are inextricably linked, yet the interactions between them are poorly understood. However, evidence is building that cell identity regulators have a role in stress responses, and that in turn, stress-responsive transcription factors may regulate developmental patterning. Defects in the columella core marker ARF10 lead to altered root cap formation (Wang et al., 2005) and the cre1 mutant, which has altered xylem organization (Inoue et al., 2001), is hypersensitive to ABA at germination (Tran et al., 2007). Furthermore, the phosphate deficiency induced gene PDR2 is necessary for maintenance of the ground tissue regulator SCR under phosphate deficient conditions (Ticconi et al., 2009), while a putative direct target of the iron-responsive transcription factor POPEYE is the SHORTROOT direct target MGP (Long et al., 2010). In addition, the heat shock protein SCHIZORIA is required for stem cell maintenance (Pernas et al., 2010; ten hove et al., 2010), directly implicating a stress-responsive transcription factor in root patterning. We suggest that one role of cell identity regulators in the root is to interact with different stress response pathways.
Core markers reveal root patterning regulators
We identified a set of genes (core markers) that are always expressed at higher levels in the core cell type relative to all other cell types, regardless of environment. Many of these core markers are cell identity regulators. We have identified a role in radial patterning for the core stele marker ZLL, a member of the AGO protein family. ZLL is required for leaf adaxial identity (Liu et al., 2008) and shoot apical meristem (SAM) maintenance during embryogenesis (Tucker et al., 2008). We find that zll-3 mutants display increased divisions in the ground tissue in the root. Since ZLL is only expressed in the vasculature in the root, this suggests ZLL regulates ground tissue patterning in a non cell-autonomous manner. This is consistent with a previous report demonstrating that ZLL maintains SAM stem cells from the vascular primordium (Tucker et al., 2008). Levels of miR165/166 are elevated in leaves and the SAM of zll mutants. Levels of the HD-III Zip genes PHB, PHV, and REV are correspondingly reduced, suggesting that ZLL genetically represses miR165/166 (Liu et al., 2008). Recently, Carlsbecker et al., 2010, showed that the ground tissue patterning regulators SHR and SCR activate miR165a and miR166b in the endodermis. These microRNAs then move back into the stele, and the resulting microRNA gradient represses HD-ZIP TFs in the endodermis and stele periphery, patterning the cell types of the xylem. ZLL is not affected in either a SHR or SCR induction time course (Sozzani et al., 2010) and thus appears to act in a SHR-and SCR-independent manner. Interestingly, mutations in AGO1, the closest homolog to ZLL in the AGO family, also lead to an increase in ground tissue layers in the root (Miyashima et al., 2009), though the mechanism behind this is not yet established. Recently, ZLL was shown to be a negative regulator of AGO1 at the protein level (Mallory et al., 2008), and the two genes are known to act redundantly in specific pathways. These data suggest that ZLL may regulate ground tissue patterning through a SHR and SCR-independent, non-cell-autonomous, post-transcriptional mechanism.
Cell-cell communication is important for the stress response
Cell-cell communication is a vital part of root development. Since the expression level of core markers is highest in the core cell type regardless of environment, this raises the possibility that cell-cell communication allows cell types in the root to determine the relative concentration of specific genes. As many core markers are cell identity regulators, this may be another point of interaction between root developmental pathways and stress responses.
Together, our results highlight the complexity of gene regulation within cell types under stress and demonstrate the power of multiple genome-wide analyses from different environmental stimuli to uncover root patterning factors and regulatory associations within cell types. Our data suggest that cell identity regulators play dual roles in stress and development. We speculate that this may contribute to the enormous phenotypic plasticity observed in the plant kingdom.
EXPERIMENTAL PROCEDURES
Plant materials and growth conditions for microarrays
Columbia-0 (Col-0) was used for all microarray experiments. Seeds were surface sterilized with 50% Bleach and 0.1% Tween for 5 minutes and then rinsed 3 times with sterile water. Seeds were stratified at 4°C for 48 hours before sowing and plated on nylon mesh on agar for all experiments as described (Dinneny et al. 2008). High NaCl, −Fe, and MS standard media are as described (Dinneny et al. 2008). Sulfur deficient media has a similar concentration of nutrients (with the exception of S) as the MS standard (full description in Supplemental Experimental Procedures). Low pH media is 1X concentration MS salt mixture (Caisson laboratories), 3mM DMG (Sigma), 1% sucrose, 1% agar and adjusted to pH 4.6 with KOH. A separate standard media (pH standard) was used to compare to low pH for all experiments. The pH standard is equivalent to the low pH media with the exception that the pH was adjusted to 5.7.
ABA assays
For radicle and cotyledon emergence assays, seeds of Col-0, Ws, L.er, scr4 (Ws control), mgp (Col-0 control), fez-2 (Col-0 control), wermyb23 (Col-0 control), cpctry (Col-0 control), and gl2 (L.er control) were cold-treated for 48 hours, sterilized as above and plated on 0μM and 1μM ABA plates. 1μM ABA plates were 1X MS salt mixture, 0.05% MES, 1% agar and 1μM ABA (Sigma). 0μM ABA plates were the same except an equivalent volume of ethanol was added in place of the ABA. Radicle emergence was scored using a Leica 6SE dissecting scope; Cotyledons were scored as fully emerged if they were green and open past 90 degrees. Experiments were repeated three times for fez, wermyb23, cpctry, and scr; and twice for mgp, and gl2 (n ≈ 60/genotype/concentration/experiment, except gl2 and scr in which n ≈ 30.). The average of experiments is shown ± standard error (SE). Significance was tested using a one-tailed t-test with equal variance.
FACS sorting and GFP reporter lines
FACS and GFP reporter lines used for cell sorting are as described (Brady et al., 2007; Dinneny et al. 2008).
Sample preparation for time-course (TC), cell-type and longitudinal datasets
Sample preparation for the TC, cell-type and longitudinal datasets is as described (Dinneny et al. 2008) except that seedlings were transferred to either low pH and the pH standard or −S and fresh MS. A full description is in Supplemental Experimental Procedures. All low pH and −S micorarray datasets have been submitted to GEO (SuperSeries GSE30166).
Sorting effect sample preparation
Low pH samples for the effect of sorting were prepared as described (Dinneny et al. 2008). Cell sorting affected probesets are listed in Table S9.
Microarray and ChIP-Chip Analysis
Meta-analysis
All computations for the meta-analysis were done using R. . CEL files for AtGenExpress datasets were downloaded from TAIR. All arrays from each condition were background corrected and normalized together using RMA with the affy package. The RankProd package (Hong et al., 2006) with two classes was used for identifying differentially expressed genes within each time-course. P-values from RankProd were used to generate a combined p-value from 14 treatments for each probeset. Combined p-values were generated using Fisher’s method (Fisher, 1932; Burguillo et al., 2010) in the survcomp package. FDR values were determined using the q-value package (Storey and Tibshirani, 2003) with the default settings. Since Fisher’s method is asymmetric in the weight it gives to small numbers of significant values (Zaykin et al., 2002) significant probesets had to meet two criteria: First, a combined FDR value < 0.0001 and second, in at least ¾ of the treatments, a FDR < 0.01. Thus to be considered significant among 14 conditions in the whole root, a probeset with a combined FDR<0.0001 also had to have an individual stress FDR < 0.01 in at least 11 of 14 stresses. For the meta-analysis at cell-type resolution, a probeset had to have a combined FDR < 0.0001 and an individual treatment FDR < 0.01 in at least 15 of 20 conditions (5 cell types, 4 stresses = 20 conditions). All probesets on the array were used in the meta-analysis.
Normalization and identification of differentially expressed probesets for stress-regulated genes
All arrays were normalized and differentially expressed probesets identified using a mixed-model ANOVA Perl script as described (Levesque et al., 2006). Arrays for each stress were normalized separately. See Table S4 and S5 for expression values for cell type and developmental stage datasets. The Pearson correlation product was calculated for all replicates. A cut-off value of 0.91 was used for the low pH dataset and 0.88 for the −S dataset.
A list of 20,385 singleton probesets was generated by eliminating both the Affymetrix probesets that were predicted to hybridize to more than one locus, and loci that were predicted to have multiple probesets that detected expression (Table S9). This was based on the lookup table from Affymetrix published on May 29, 2008. Only probesets that hybridized to nuclear genes were used for analysis. Differentially expressed probesets for all time-course and stress-regulated lists were identified using an |1.5| fold change cut-off and a FDR of 1 × 10−4. Differentially expressed probesets for the stress-enriched lists were identified using a 1.5 fold change cut-off and a FDR of 1 × 10−4. The exception was the −S enriched probesets for the stele and epidermis, for which a 2-fold change cut-off value was used.
Stress-regulated probesets for the cell-type and developmental stage datasets were identified by comparing each cell type or developmental stage under stress to the same cell type or developmental stage under standard conditions. Stress-enriched probesets were identified by comparing each cell type to all other cell types under the same condition (See Figure S4 for description of significant vs enriched probesets). Heatmaps were created using TMV microarray software (www.tm4.org). For all cell-type analyses except k-means clustering, sorting affected probesets were removed from the analysis for high NaCl, −Fe and low pH (listed in Table S9). In the text, “responsive” always refers to those genes that meet the significance cut-off.
DREB1A target analysis
DREB1A target analysis was as described (Dinneny et al. 2008). A full description is in Supplemental Experimental Procedures.
Gene Ontology (GO) enrichment analysis
GO analysis was preformed using the ChipEnrich program as described in (Orlando et al. 2010). Statistical significance for each GO category was determined using the hyper-geometric distribution as described (Brady et al., 2007; Dinneny et al., 2008; Orlando et al., 2009).
Stress-regulated hair patterning dependent ABA marker genes
From results in (Dinneny et al., 2008) we obtained a list of genes that are differentially expressed (FDR <0.0001, |FC| > 1.5) under NaCl and dependent on hair patterning. This list was then used to query the ABA marker genes from (Nemhauser et al., 2006).
Quantitative RT-PCR
Procedures for quantitative RT-PCR are as described (Dinneny et al. 2008, Tsukagoshi et al. 2010). A full description is in Supplemental Experimental Procedures.
ChIP-chip procedure and analysis
Procedures for ChIP-chip and analysis were as described (Sozzani et al., 2010; Long et al., 2010; Tsukagoshi et al., 2010; Busch et al., 2010). Two biological replicates each of homozygous pSCR:SCR:GFP scr-4 and Columbia (Lehle) (control) were processed. A full description is in Supplemental Experimental Procedures. ChIP-chip data has been submitted to GEO as part of SuperSeries GSE30166.
Supplementary Material
HIGHLIGHTS.
A common, but not universal, stress response in roots
Common stress responses are mediated by cell identity regulators
SCARECROW transcription factor directly regulates abscisic acid responses
Genes co-expressed in response to stress tend to share specific developmental roles
Acknowledgments
This work was funded by grants to PNB from the NSF Arabidopsis 2010 program and from the NIH (R01-GM043778). AIP was supported by a National Institutes of Health Ruth Kirschstein National Research Service Award postdoctoral fellowship. We thank Terri Long and Jose Dinneny for helpful discussions, as well as Pete Pascuzzi and members of the Benfey lab for comments on the manuscript. We are grateful to T. Demura for the 35S::VND7:GFP line.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science. 2007;318:801–806. doi: 10.1126/science.1146265. [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LA, Ellis J, Goodacre R, Turner SR. Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell. 2005;17:2281–2295. doi: 10.1105/tpc.105.031542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burguillo FJ, Martin J, Barrera I, Bardsley WG. Meta-analysis fo microarray data; the case of imatinib resistance in chronic myelogenous leukemia. Comput Biol Chem. 2010;34:184–192. doi: 10.1016/j.compbiolchem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. Transcriptional control of a plant stem cell niche. Dev Cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Guan C, Boonsirichai K, Masson PH. Complex physiological and molecular processes underlying root gravitropism. Plant Mol Biol. 2002;49:305–317. [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–425. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Scheifelein J, Benfey PN. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science. 2008;320:942–945. doi: 10.1126/science.1153795. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Statistical Methods for Research Workers. Oliver and Boyd; Edinburgh: 1932. [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc Natl Acad Sci U S A. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CM, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K. Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol. 2009;151:1175–1185. doi: 10.1104/pp.109.143867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T. Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 2005;19:1855–1860. doi: 10.1101/gad.1331305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kültz D. Molecular and Evolutionary Basis of the Cellular Stress Response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann J, Scheres B, Benfey PN. Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 2006;4:e143. doi: 10.1371/journal.pbio.0040143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Yao X, Pi L, Wang H, Cui X, Huang H. The ARGONAUTE10 gene modulates shoot apical meristem maintenance and leaf polarity establishment by repressing miR165/166 in Arabidopsis. Plant J. 2008;58:27–40. doi: 10.1111/j.1365-313X.2008.03757.x. [DOI] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN. The BHLH transcription factor POPEYE regulates response to iron deficiencey in Arabidopsis roots. Plant Cell. 2010;22:2219–2236. doi: 10.1105/tpc.110.074096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 2007;8:R49. doi: 10.1186/gb-2007-8-4-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory AC, Elmayan T, Vaucheret H. MicroRNA maturation and action--the expanding roles of ARGONAUTEs. Curr Opin Plant Biol. 2008;11:560–566. doi: 10.1016/j.pbi.2008.06.008. [DOI] [PubMed] [Google Scholar]
- Miyashima S, Hashimoto T, Nakajima K. ARGONAUTE1 acts in Arabidopsis root radial pattern formation independently of the SHR/SCR pathway. Plant Cell Physiol. 2009;50:626–634. doi: 10.1093/pcp/pcp020. [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, Martinez-Laborda A, Micol JL. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol. 2006;141:607–619. doi: 10.1104/pp.106.077149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development. 2007;134:2959–2968. doi: 10.1242/dev.006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando DA, Brady SM, Koch JD, Dinneny JR, Benfey PN. Manipulating large-scale Arabidopsis microarray expression data: Identifying dominant expression patterns and biological process enrichment. Methods Mol Biol. 2009;553:57–77. doi: 10.1007/978-1-60327-563-7_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernas M, Ryan E, Dolan L. SCHIZORIZA controls tissue system complexity in plants. Curr Biol. 2010;20:818–823. doi: 10.1016/j.cub.2010.02.062. [DOI] [PubMed] [Google Scholar]
- Persson S, Wei H, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci U S A. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133:1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Yamamoto KT. Overexpression of the non-canonical Aux/IAA genes causes auxin-related aberrant phenotypes in Arabidopsis. Physiol Plant. 2008;133:397–405. doi: 10.1111/j.1399-3054.2008.01055.x. [DOI] [PubMed] [Google Scholar]
- Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, Brady SM, Dewitte W, Murray JA, Benfey PN. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–132. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Swindell WR. The association among gene expression responses to nine abiotic stress treatments in Arabidopsis thaliana. Genetics. 2006;174:1811–1824. doi: 10.1534/genetics.106.061374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove CA, Willemsen V, de Vries WJ, van Dijken A, Scheres B, Heidstra R. SCHIZORIZA encodes a nuclear factor regulating asymmetry of stem cell divisions in the Arabidopsis root. Curr Biol. 2010;20:452–7. doi: 10.1016/j.cub.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S. ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristerms to phosphate availability. Proc Natl Acad Sci USA. 2009;106:14174–14179. doi: 10.1073/pnas.0901778106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga-Wada R, Iwata M, Sugiyama J, Kotake T, Ishida T, Yokoyama R, Nishitani K, Okada K, Wada T. The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J. 2009;60:564–574. doi: 10.1111/j.1365-313X.2009.03976.x. [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought and salt stress in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukagoshi H, Busch W, Benfey PN. Transcriptional regulation of ROS controls the transition from cellular proliferation to differentiation in the root. Cell. 2010;143:606–616. doi: 10.1016/j.cell.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Tucker MR, Hinze A, Tucker EJ, Takada S, Jurgens G, Laux T. Vascular signalling mediated by ZWILLE potentiates WUSCHEL function during shoot meristem stem cell development in the Arabidopsis embryo. Development. 2008;135:2839–2843. doi: 10.1242/dev.023648. [DOI] [PubMed] [Google Scholar]
- Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walley JW, Dehesh K. Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J Integr Plant Biol. 2010;52:354–359. doi: 10.1111/j.1744-7909.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- Walley JW, Coughlan S, Hudson ME, Covington MF, Kaspi R, Banu G, Harmer SL, Dehesh K. Mechanical stress induces biotic and abiotic stress responses via a novel cis-element. PLoS Genet. 2007;3:1800–1812. doi: 10.1371/journal.pgen.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther D, Brunnemann R, Selbig J. The regulatory code for transcriptional response diversity and its relation to genome structural properties in A. thaliana PLoS Genet. 2007;3:e11. doi: 10.1371/journal.pgen.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Zue HW, Chen XY. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- Zeller G, Henz SR, Widmer CK, Sachsenberg T, Ratsch G, Weigel D, Laubinger S. Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J. 2009;58:1068–1082. doi: 10.1111/j.1365-313X.2009.03835.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.