Abstract
Background
TG-interacting factors (TGIFs) belong to a family of TALE-homeodomain proteins including TGIF1, TGIF2 and TGIFLX/Y in human. Both TGIF1 and TGIF2 act as transcription factors repressing TGF-β signalling. Human TGIFLX and its orthologue, Tex1 in the mouse, are X-linked genes that are only expressed in the adult testis. TGIF2 arose from TGIF1 by duplication, whereas TGIFLX arose by retrotransposition to the X-chromosome. These genes have not been characterised in any non-eutherian mammals. We therefore studied the TGIF family in the tammar wallaby (a marsupial mammal) to investigate their roles in reproduction and how and when these genes may have evolved their functions and chromosomal locations.
Results
Both TGIF1 and TGIF2 were present in the tammar genome on autosomes but TGIFLX was absent. Tammar TGIF1 shared a similar expression pattern during embryogenesis, sexual differentiation and in adult tissues to that of TGIF1 in eutherian mammals, suggesting it has been functionally conserved. Tammar TGIF2 was ubiquitously expressed throughout early development as in the human and mouse, but in the adult, it was expressed only in the gonads and spleen, more like the expression pattern of human TGIFLX and mouse Tex1. Tammar TGIF2 mRNA was specifically detected in round and elongated spermatids. There was no mRNA detected in mature spermatozoa. TGIF2 protein was specifically located in the cytoplasm of spermatids, and in the residual body and the mid-piece of the mature sperm tail. These data suggest that tammar TGIF2 may participate in spermiogenesis, like TGIFLX does in eutherians. TGIF2 was detected for the first time in the ovary with mRNA produced in the granulosa and theca cells, suggesting it may also play a role in folliculogenesis.
Conclusions
The restricted and very similar expression of tammar TGIF2 to X-linked paralogues in eutherians suggests that the evolution of TGIF1, TGIF2 and TGIFLX in eutherians was accompanied by a change from ubiquitous to tissue-specific expression. The distribution and localization of TGIF2 in tammar adult gonads suggest that there has been an ultra-conserved function for the TGIF family in fertility and that TGIF2 already functioned in spermatogenesis and potentially folliculogenesis long before its retrotransposition to the X-chromosome of eutherian mammals. These results also provide further evidence that the eutherian X-chromosome has actively recruited sex and reproductive-related genes during mammalian evolution.
Background
Homeobox genes are characterized by a conserved 180 base pair motif encoding a homeodomain with three structurally conserved helices [1]. The TALE superfamily is distinguished by a three-amino-acid loop extension (TALE) between helix 1 and helix 2 within the homeodomain [2,3]. The third helix is highly conserved playing the major role in DNA binding site recognition [3]. TALE homeodomain proteins are critical transcription factors for embryonic and early development [4,5]. TGIF (named after transforming growth factor-β-induced factor or 5'-TG-3' interacting factor) genes are members of TALE superfamily, containing TGIF1, TGIF2, TGIFLX and TGIFLY [2].
TGIF1 orthologues share a high similarity inside the homeodomain between distantly related vertebrates [6]. TGIF1 is a Smad transcriptional co-repressor that negatively regulates TGF-β-activated gene expression in vertebrates [6,7]. In Drosophila, TGIF1 is a transcriptional activator interacting with Mad and Smad2 [6]. In humans, TGIF1 is highly expressed in the placenta and other adult tissues such as liver, kidney and gonads [3]. Mutations in TGIF1 can cause holoprosencephaly (HPE), a severe disease affecting forebrain and craniofacial development, associated with mental retardation [8]. Two isoforms of Tgif1 were identified by RT-PCR from alternative splicing events that are specific to the mouse [9]. Both alternative splice forms are functional as transcriptional repressors [9]. Mouse Tgif1 mRNA is initially detected at E9.5 with the highest expression in the forebrain, the branchial arches, the otic pit, and the limb buds but not in the heart [10]. Subsequently the expression is maintained at a higher level from then on in the forebrain but declines throughout the whole embryo by E14.5 [10,11]. However, Tgif1 knockout mice do not have any abnormal phenotypes [10], possibly due to a functional redundancy with Tgif2, since both occupy similar spatial and temporal expression domains during embryogenesis [10] and both are co-repressors for TGF-β receptor activated Smads by interacting with histone deacetylases (HDACs) [4].
TGIF2 shares similar DNA binding homeodomains to TGIF1, suggesting both proteins are likely to bind the same DNA sequence [4]. Human TGIF2 has 3 exons which have the highest homology with TGIF1. There are two alternative splicing forms of Tgif2 genes one of which contains a retained intron within the second coding exon that occurs only in mice [9]. Both forms of Tgif2 mRNA transcripts are present and functional as transcriptional repressors in adult and embryonic tissues in mice and have similar expression patterns [9]. TGIF2 is ubiquitously expressed in human tissues, with particularly high expression in the heart, kidney and testis, but the transcript is almost undetectable in the brain and prostate [12]. In the mouse, TGIF2 transcripts are highly expressed in the nervous system at E12.5 and E15.5, indicating this gene has a wide but well controlled expression pattern during early embryo development [10,13]. Both Tgif1 and Tgif2 are required for gastrulation in mice and act to limit Nodal signalling and L-R axis specification [5].
In addition to the autosomal TGIFs described above, orthologues exist on the eutherian mammal × and Y-chromosomes, TGIFLX (transforming growth factor-β-induced factor 2-like, X-linked) and TGIFLY (transforming growth factor-β-induced factor 2-like, Y-linked). TGIFLY lacks the specific C-terminal residues shared by TGIFLX, TGIF1 and TGIF2 [14]. The TGIFLX gene originated from autosomal TGIF2 by retrotransposition [14], characterized by loss of introns, poly A tracts, and flanking short direct repeats [15]. TGIFLX has 2 exons with a 96 base-pair intron [14]. TGIF2 and TGIFLX share high conservation both within the homeodomain and the C-terminus conserved region but show extensive variation outside these domains [14]. TGIFLX is highly conserved between primates. There is an orthologue in mice, Tex1 (testis-expressed homeobox 1), [16], indicating that the TGIFLX retrotransposition event occurred at least 80 million years ago [14]. TGIFLX/Tex1 expression is restricted to the adult testis [14,16]. Tex1 mRNA is detected in the spermatids in the seminiferous tubule and in some residual bodies, suggesting that it may play a critical role in spermatogenesis [16]. Unlike TGIF1 and TGIF2, this gene is not expressed in the brain, so it is unlikely to be involved in brain development [14].
X-linked genes are believed to evolve more rapidly than their autosomal orthologues due to their hemizygosity in males [17,18]. The mammalian sex chromosomes emerged from an ancestral pair of autosomes [19] and have received specific additions and deletions in each lineage. A large number of genes on the X-chromosome have a role in mammalian spermatogenesis [20,21]. Human TGIFLX maps to Xq21.3 in human [14], within a region recently added to the X-chromosome in the eutherian lineage [22] and is expressed in the testis [14]. Homeobox genes expressed in the testis such as TGIFLX and ESX1L are biased on the X-chromosome [17]. Two forms of TGIF genes (Achintya &Vismay) identified in Drosophila are crucial for spermatogenesis, suggesting that there is an ultra-conserved function for these genes in all animals [23,24].
To date, TGIFLX and its orthologues have only been studied in eutherian mammals. Since marsupial mammals diverged from eutherian mammals between 130 and 148 MYA [25-27], we investigated whether this gene was present in a marsupial, the tammar wallaby, Macropus eugenii. We characterised TGIF1 and TGIF2 orthologues in the tammar, identified their chromosomal location(s), and examined their expression throughout development and in adult tissues.
Results
The absence of TGIFLX and identification of TGIF1 &TGIF2
Human TGIFLX (ENST00000283891) was used to search all available genome databases to identify TGIFLX-related sequences. An orthologue of TGIFLX was identified from the horse but its chromosomal location is unknown. Horse TGIFLX shared high similarity through the exons and both upstream and downstream flanking sequences. Another orthologue was identified from the mouse known as Tex1 which only shared partial similarity in the homeobox region. No orthologues were identified from other species including the tammar, platypus, chicken and fish (Figure 1A).
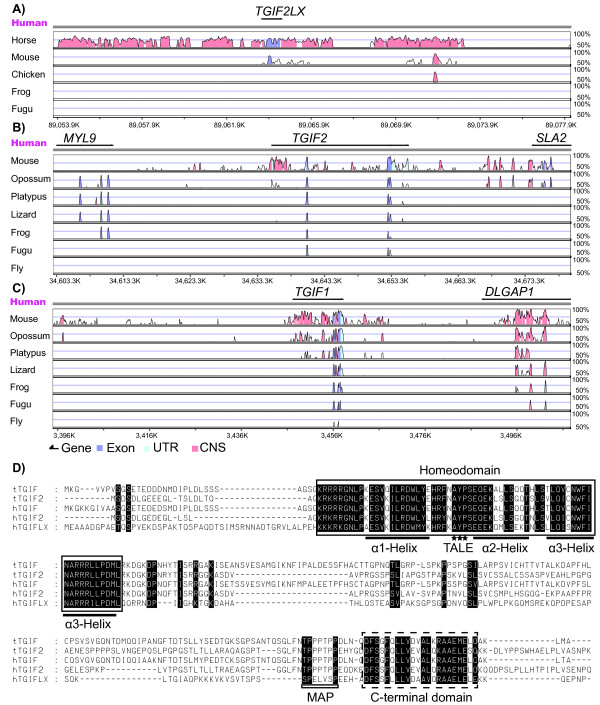
Figure 1.
Comparison of TGIF family. A) TGIFLX orthologues are only found in eutherian mammals including human, horse and mouse, but there are no orthologues in non-mammals including chicken, frog and Fugu with VISTA browser. Human genomic sequence of TGIFLX (chrX: 89,053,736-89,078,217) compared to the horse (contig_22722: 204,584-233,963), mouse (chrX: 115,540,211-115,561,508), chicken (showing only conserved CNS in chr4: 7362172-7362401, no alignment in other region), frog (no alignment and no contigs can be shown) and Fugu (no alignment and no contigs can be shown). B) TGIF2 orthologues are present in all vertebrates, but not in invertebrates. Human genomic sequence encompassing MYL9-TGIF2-SLA2 (chr20: 34,602,792-34,680,072) compared to the mouse (chr2: 156,632,126-156,706,204), opossum (chr1: 389,963,044-390,050,278), platypus (Ultra337: 3,776,448-3,849,788), lizard (scaffold_17: 7,638,665-7,723,584), frog (scaffold_38: 522,992-673,094), Fugu (chrUn: 295,276,805-295,277,868) and fly (chr2R: 8,363,354-8,428,229). C) TGIF orthologues are present in all vertebrates and invertebrates with the conserved downstream and upstream genes in most of species. Human genomic sequence encompassing MYOM1 (partial sequence and not shown)-TGIF1-DLGAP1 (chr18: 3,395,327-3,507,935) compared to the mouse (chr17: 71,147,775-71,255,269), opossum (chr3: 269,037,689-269,144,991), platypus (Contig3116: 1-55,206), lizard (scaffold_70: 2,188,819-2,296,955), frog (scaffold_337: 1,113,822-1,225,651), Fugu (chrUn: 210,908,683-211,016,001) and fly (chr2R: 8,353,314-8,461,279). High similarity over a 100 bp window (showing with a peak region) is seen for exon sequence (slate blue) and untranslated regions (UTR, powder blue) or conserved non-coding sequence (CNS, red). The top double line stands for human genomic sequences. The bottom of each picture showed the human genomic sequence number. D) Amino acids alignment of human (h) TGIF family members and tammar (w) TGIF family members. Identical amino acids between the sequences are shaded black. The homeodomain is boxed and the conserved carboxyl-terminal domain (CCD) is indicated by a dashed box. The position of the three alpha helices (α1, α2, and α3) is indicated by a line below the sequence. The extra 3 residues, AYP, between helix α1 and α2 are highlighted with stars and labeled as TALE. The mitogen-activated protein (MAP) kinase phosphorylation site is indicated with a rectangular box below the sequence. The GenBank accession numbers for these proteins are: hTGIF2, (BAB16424); hTGIF1 (NM_003244) and hTGIFLX (AJ427749). Tammar TGIF1 and TGIF2 sequence have been submitted to GenBank (tTGIF: JF796112; tTGIF2: FJ775183).
Owing to the absence of a TGIFLX orthologue in non-eutherian mammals, the marsupials and monotremes, another BLAST search was performed for its progenitor gene TGIF2 and paralogue TGIF1 from various species including reptiles, amphibians, fish and flies. It showed orthologues of TGIF2 were present in all vertebrates investigated. TGIF2 orthologues shared the same upstream gene MYL9 in most vertebrates except fish and the downstream gene SLA2 that was only present in therians. The TGIF2 orthologues were poorly conserved at the first exon (Figure 1B). No orthologue of human TGIF2 was identified in the fly genome. The TGIF1 orthologues were identified in all species examined and shared the same upstream gene MYOM1 (data not shown) and downstream gene DLGAP1 (Figure 1C).
Characterization of tammar TGIF1 and TGIF2
Tammar TGIF1 and TGIF2 were amplified from adult tammar testis by reverse-transcriptase PCR (RT-PCR) with cross-species primers. The tammar full-length TGIF2 cDNA encodes a predicted protein of 252 amino acids, 15 amino acids longer than human and mouse orthologues. The predicted tammar TGIF1 protein contains 269 amino acids, 3 amino acids shorter than eutherian orthologues. Tammar partial genomic sequences from NCBI http://blast.ncbi.nlm.nih.gov/Blast.cgi were retrieved by alignment with tammar TGIF and used to confirm their genomic structure. Tammar TGIF2 has 3 exons and the open reading frame spanned all exons. The homeobox was located within the second exon. Similarly, tammar TGIF1 also contains 3 exons but the coding region only spanned exons 2 and 3.
Alignment of the predicted amino acid sequences of various orthologues of the TGIF family from human and tammar revealed that they were highly conserved over the homeodomain and carboxyl-terminal conserved domain (CCD) (Figure 1D). Both tammar TGIF1 and TGIF2 contained a three-amino acid loop extension (TALE) between the α1 helix and α2 helix, confirming that tammar TGIF1 and TGIF2 belong to the TALE super-family. The α3 helix was identical between human and tammar TGIF1 and TGIF2. In addition, the region containing the mitogen-activated protein (MAP) kinase phosphorylation sites (TP & PPTP) close to the carboxyl-terminal conserved domain was identical in human and tammar TGIF1/TGIF2. However, the region outside the conserved domain had a very poor similarity and identity, especially between human TGIFLX and other orthologues (Figure 1D).
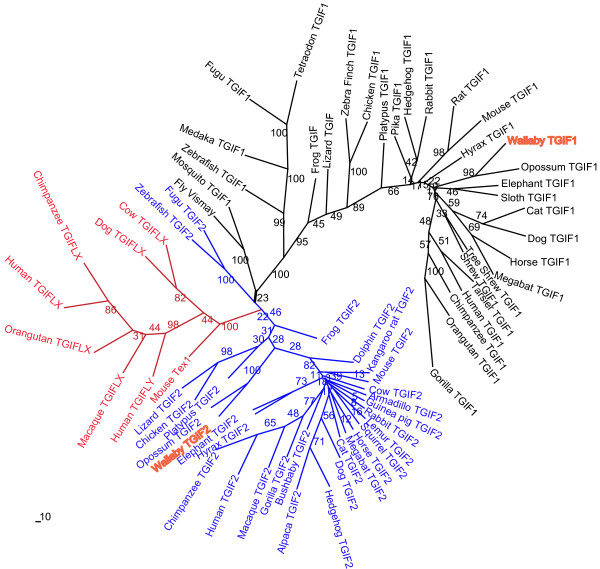
Full length protein sequence or near full-length protein sequence for TGIF1, TGIF2 and TGIFLX/Y homologues were retrieved from NCBI http://www.ncbi.nlm.nih.gov or Ensembl http://www.ensembl.org. There are 30 TGIF1 orthologues from various vertebrates and invertebrates, 28 TGIF2 from various vertebrates and 8 TGIFLX/Y homologues from eutherians. Phylogenetic analysis with PHYLIP program clustered these homologues into two main clusters TGIF1s and TGIF2s (Figure 2). TGIFLX/Y orthologues branched with the TGIF2s. TGIF1 from all mammals including the tammar clustered tightly, with monotremes forming the most distant branch. TGIF1 from reptiles, birds, amphibians and fish formed other branches. Invertebrates including fly and mosquito clustered as a single branch. The biggest branch of the TGIF2 cluster was the eutherian group, followed by the TGIFLX/Y cluster. Tammar and opossum TGIF2 formed a separate branch followed by monotreme, bird and reptile.
Figure 2.
Phylogenetic analyses with neighbour-joining showing evolutionary relationships among members of TGIF1, TGIF2 and TGIFLX/Y family. The phylogenetic tree was constructed with PHYLIP 3.69 program. Tammar TGIF1 and TGIF2 are highlighted in bold. Numbers along branches indicate reliability of each branch with 100 replicates. TGIF1 and fly TGIF formed a branch, TGIF2 produced another branch. The TGIFLX and TGIFLY group is one of the branches of the TGIF2 cluster. GenBank (or Ensembl) accession numbers for these proteins are shown in Additional Table 1.
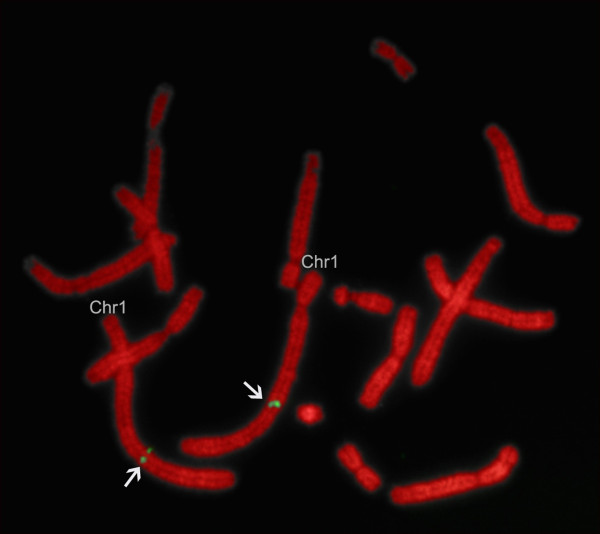
Tammar BAC clones were identified by screening the AGI tammar BAC library with the TGIF2 probe. The BAC clone showing homology to tammar TGIF2 (67G20) was confirmed by PCR amplification. The clone was mapped to the long arm of tammar chromosome 1 by fluorescence in situ hybridization (FISH) (Figure 3). Only one hybridization location was detected.
Figure 3.
Fluorescence in situ hybridization (FISH) of TGIF2. Hybridization signals (green) show that tammar TGIF2 is on the long arm of tammar metaphase chromosome 1 (Chr1; arrow).
Tammar TGIF1 &TGIF2 mRNA distribution in the fetus and developing gonads
RT-PCR was performed to investigate the expression pattern of TGIF1 &TGIF2 in various fetal tissues during organogenesis. Both TGIF1 &TGIF2 mRNA were detected in all the examined tissues (Figure 4A). TGIF2 was ubiquitously expressed during early development. Similarly, TGIF1 was also expressed in all examined fetal tissues. TGIF1 and TGIF2 mRNA transcripts were detected in gonads at all stages examined from the last two days of the 26.5 day gestation to day 44 post partum (Figure 4B and 5).
Figure 4.
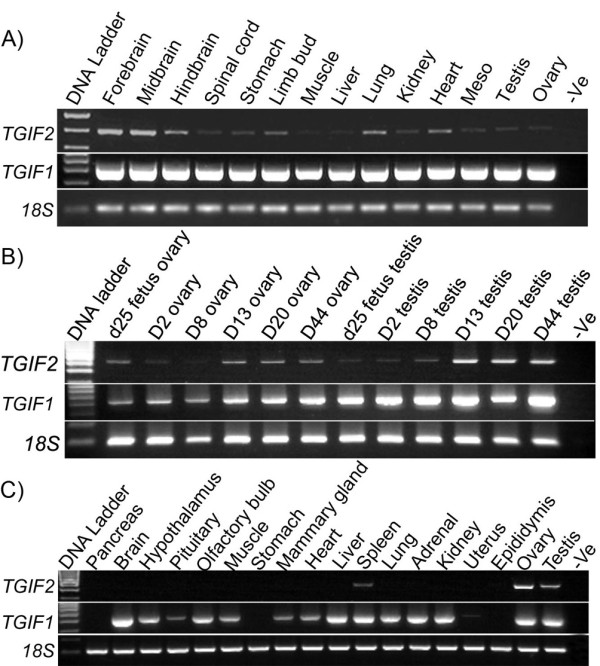
Expression pattern of tammar TGIF1 and TGIF2 in various tissues by RT-PCR. A) Expression of the TGIF1 &TGIF2 gene was assessed in various embryonic tissues. Both TGIF1 and TGIF2 were widely expressed in all examined tissues; B) Expression of the TGIF1 &TGIF2 during gonadal development from the bipotential gonad stage (day 25 of gestation-d25), sexual differentiation stages (days 2-3 post partum for the testis and days 7-8 for the ovary) and at later stages of development (up to day 44 post partum). Tammar TGIF1 and TGIF2 were detected at all stages; C). Expression of TGIF1 &TGIF2 was examined in various adult tissues. TGIF1 was broadly expressed in most tissues and TGIF2 was specifically expressed in the ovary, testis and spleen. PCR for 18S (100 bp) acted as a positive control and "-ve" indicates template free negative control reactions.
Figure 5.
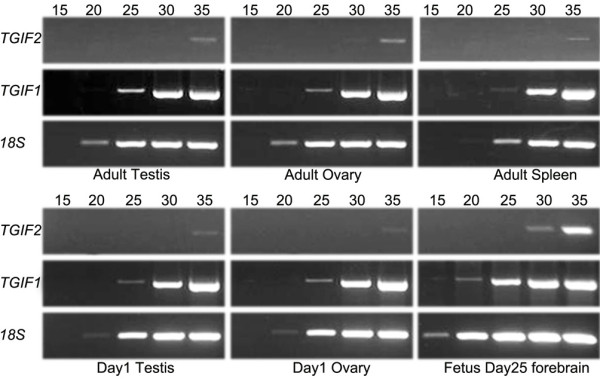
Semi-quantitative PCR. Identical amounts of cDNA template were utilized to identify the exponential (log) phase of the PCR reactions for TGIF1 and TGIF2 genes, while a 50 times cDNA dilution was used for amplification of the housekeeping gene 18S because of its high expression. Six tissues, adult testis, ovary and spleen as well as developing gonads and forebrain, were used for the analyses. PCRs were stopped at various cycles (15, 20, 25, 30, 35) to determine the exponential amplification phase.
Tammar TGIF1 &TGIF2 mRNA distribution in adult tissues
TGIF1 &TGIF2 expression was investigated in adult tissues including the adult testis and ovary. As in the human, tammar TGIF1 was ubiquitously expressed in most adult tissues except the pancreas, stomach and epididymis. In contrast, TGIF2 was only expressed in the adult gonads and was absent in all other tissues except the spleen (Figure 4C and 5). To further confirm our results, we performed semi-quantitative PCR (Figure 5). This further demonstrated that TGIF1 has a broader expression pattern than TGIF2.
Tammar TGIF2 mRNA and protein distribution during spermatogenesis
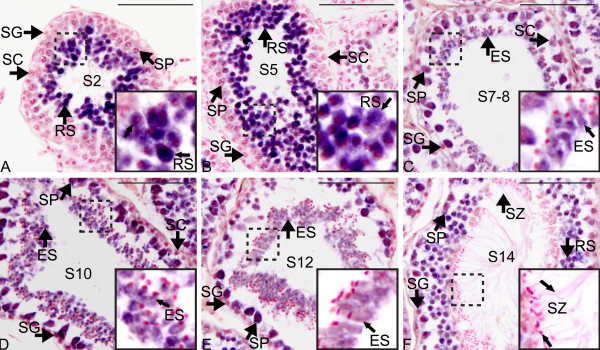
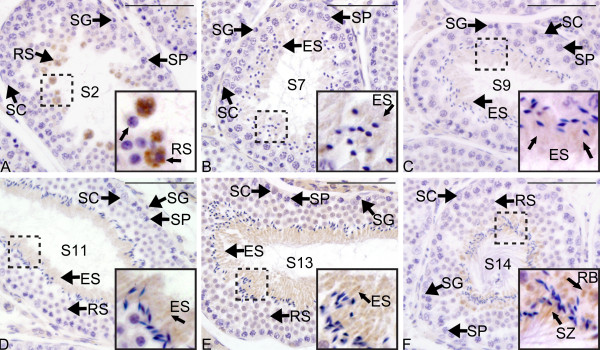
In the adult testis, using in situ hybridization TGIF2 mRNA was detected in the germ cells, predominantly in a layer around the seminiferous cord lumen (Figure 6). No mRNA staining was detectable in the Sertoli cells or interstitial cells. There was strong TGIF2 mRNA staining in round spermatids when they were just newly formed (step 2, [28], Figure 6A) and before transforming into elongated spermatids (step 5, Figure 6B). After round spermatids started to elongate, staining decreased moderately but was still detectable in the cytoplasm of elongating/elongated spermatids and spermatocytes (step 7-8, Figure 6C; step 10, Figure 6D; step 12, Figure 6E). No staining was detected when elongated spermatids transformed into mature spermatozoa and were released into lumen (step 14, Figure 6F).
Figure 6.
mRNA in situ hybridization of TGIF2 during spermatogenesis in tammar adult testis. Each panel shows a seminiferous tubule at different stages of the spermatogenic cycle indicated by the steps 1 to 14 (S1 to S14) [28]. All images shown were hybridized with antisense probe (purple blue) and counterstained with nuclear fast red. Images boxed on the right bottom are the high resolution pictures from the corresponding part highlighted with dashed boxes. TGIF2 mRNA was detected in the cytoplasm of early round spermatids (A, step 2) and round spermatids before transforming to elongated spermatids (B, step 5). Staining was also observed in the cytoplasm of elongated spermatids and spermatocytes (C-E, step7-12). Image F indicates that there was no staining on the mature spermatozoa with arrows showing the sperm head (bottom arrow) and sperm tail (top arrow) (step 14). SG, spermatogonia; SP, spermatocytes; RS, round spermatids; ES, Elongating/elongated spermatids; SZ, spermatozoa; SC, Sertoli cells. Scale bars = 100 μm.
TGIF2 protein was also restricted to the germ cells during spermatogenesis. When round spermatids were newly formed, TGIF2 protein was present and there was strong staining in the cytoplasm of spermatids (step 2, Figure 7A). After that, immunostaining decreased significantly to an almost undetectable level (step 7-11, Figure 7B-D). At late stages of spermatid transformation, TGIF2 was detected to the cytoplasm of germ cells surrounding the lumen (step 13, Figure 7E). When the sperm tail was fully formed and released to the lumen, TGIF2 protein was localized in the residual body and the mid-piece of sperm tail (Figure 7F).
Figure 7.
Immunohistochemistry of TGIF2 protein during spermatogenesis in the tammar adult testis. Each panel shows a seminiferous tubule at different stages of the spermatogenic cycle. Sections were counterstained with haematoxylin (blue). TGIF2 protein (brown staining indicated with arrow) is strongly detected in the cytoplasm of early round spermatids (A, step 2) and was much weaker or undetectable in the later stage round spermatids (B, step 7). As round spermatids transformed into mature spermatozoa, staining decreased dramatically and was almost undetectable in the elongated spermatids (C-E, step 9-13). F shows immunostaining in the residual body (top arrow) and mid-piece (bottom arrow) of the sperm tail (step 14). Images boxed on the right bottom are the high resolution pictures from the corresponding part highlighted with dashed box. SG, spermatogonia; SP, spermatocytes; RS, round spermatids; ES, Elongating/elongated spermatids; SZ, spermatozoa; SC, Sertoli cells; PC, peritubular myoid cells; RB, residual bodies. Scale bars = 100 μm. Steps of spermatogenesis defined in [28].
Tammar TGIF2 mRNA distribution during folliculogenesis
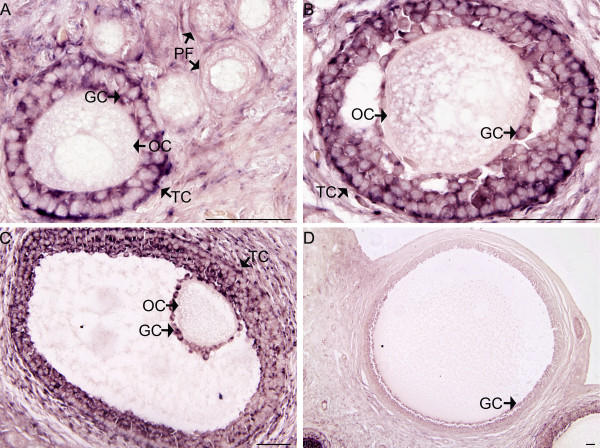
In the adult ovary TGIF2 mRNA was detected in the granulosa cells and theca cells of the secondary follicles and tertiary follicles (Figure 8A-C). There was no mRNA detected in the primary follicles (Figure 8A) and TGIF2 was almost undetectable in the Graafian follicle (Figure 8D). There was no staining seen in the oocyte.
Figure 8.
mRNA in situ hybridization of TGIF2 in tammar adult ovary. Sections were hybridized with antisense probe without counterstaining. TGIF2 mRNA (purple blue) was predominantly detected in the granulosa cells and theca cells of the secondary follicles (A & B) and tertiary follicles (C). No staining was detected in primary follicles (PF) (A) and TGIF2 mRNA was almost invisible in Graafian follicles (GF) (D). There was no staining detected in the oocytes. Signal was indicated with arrow. PF, primary follicle; GC, granulosa cell; TC, theca cell; OC, oocyte. Scale bars = 50 μm.
Discussion
Conservation of the TGIF family in vertebrates
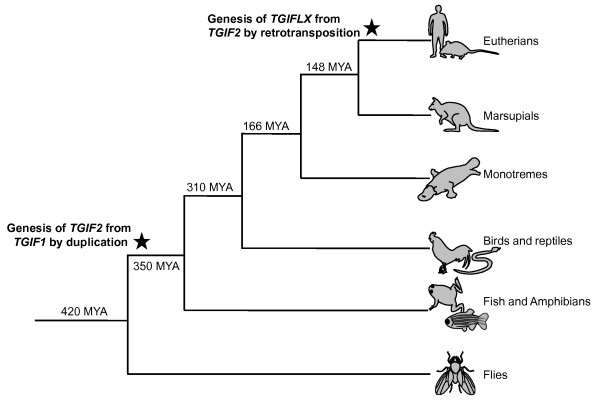
TGIF1 and TGIF2 share the highest degree of homology in the TGIF family. The characterisation of TGIF1 and TGIF2 established that both genes have 3 exons in all species except the orthologue of TGIF1 in Drosophila. The conservation of TGIF1 and TGIF2 throughout the exons, the UTR and flanking regions suggests the functional importance of these genes during evolution. The X-linked TGIFLX is specific to the eutherian lineage and arose after they diverged from marsupials and monotremes around 148 and 166 million years ago respectively.
The entire tammar TGIF2 protein shared 71% amino acid identity with human TGIF2 and only 33% similarity with human TGIFLX. The high degree of conservation over the homeodomain and TALE regions suggests that TGIF1 and TGIF2 interactions via the homeodomain may be similar in marsupial as in human and mouse [4,5]. The third helix within the homeodomain, critical for DNA binding, is the most conserved between eutherian and marsupial orthologues, suggesting they are likely to bind to the same DNA motif as identified in the mouse [4,7]. Phylogenetic analyses showed that the TGIFLX groups tightly with the TGIF2 branch, supporting the suggestion that TGIFLX originated from a TGIF2 retrotransposition event [14,17] (Figure 9).
Figure 9.
Proposed model of TGIFs evolution. TGIF2 emerged from TGIF1 by gene duplication about 420MYA while TGIFLX was generated from TGIF2 by a retrotransposon after eutherians diverged from marsupials about 148MYA.
Both TGIF1 &TGIF2 are important for embryonic and gonadal development
Marsupial TGIF1 and TGIF2 mRNA was expressed in all embryonic tissues examined, as in mice [11,13]. Tammar TGIF2 mRNA was abundant in embryonic brain, consistent with the high level seen in the murine embryonic nervous system [13]. Tammar TGIF1 was observed in all examined tissues, again consistent with previous studies in mice, and there was abundant mRNA expression in the brain [10,11]. In contrast, mouse Tgif1 mRNA transcripts were restricted in the brain from E15 indicating that Tgif1 may be involved in cerebellum development and maturation [11]. The overlapping expression of these two genes and the phenotype of the Tgif1 knockout mouse suggest that Tgif2 can compensate for the loss of Tgif1 during embryogenesis [10]. The ubiquitous expression of TGIF1 and TGIF2 in the tammar similarly suggests that both TGIF1 and TGIF2 may be functionally redundantly during marsupial embryogenesis.
Differential function of tammar TGIF1 and TGIF2 in adult tissues
Tammar TGIF1 was broadly expressed in adult tissues including the testis, in similar locations to those described for the human and mouse [3,11]. Mouse Tgif1 is found in the nuclei of peripheral regions of tubules such as spermatogonia and primary spermatocytes in the adult testis [23]. Murine Tgif2 mRNA transcripts are present and functional as transcriptional repressors in adult and embryonic tissues and have similar expression patterns [9]. Similarly, TGIF2 is ubiquitously expressed in human tissues [12]. In contrast, tammar TGIF2 was exclusively expressed in the adult testis, ovary and spleen. The expression of tammar TGIF2 in adult gonadal tissues is instead similar to that of human TGIFLX and mouse Tex1 that are expressed only in adult testis and are thought to be involved in spermatogenesis [14,16,17]. Marsupial TGIF2 was strongly expressed in round spermatids but appeared to be down-regulated as they transformed into elongated spermatids, identical to the Tex1 expression pattern in mice [16]. Tammar TGIF2 protein was strongly detected in early round spermatids and in mature spermatids, and also detected in the residual bodies, in line with Tex1 mRNA distribution in mice [16]. Since TGIFLX is undetectable in some humans with abnormal spermatogenesis, TGIFLX is likely to have a function in male reproduction and development [29]. Therefore, the expression pattern of marsupial TGIF2 and its specific localisation in the germ cells during spermatogenesis suggests that TGIF2 has developed a specialised function in marsupial germ cells.
The mRNA of tammar TGIF2 was examined in the adult ovary and was detected primarily in the granulosa cells in developing ovarian follicles, predominantly in secondary and tertiary follicles, suggesting that TGIF2 may also function in folliculogenesis and the development of oocyte. This is the first examination of TGIF2 in ovaries of any mammal. TGIF2 was also detected in the thecal cells, known to be critical for follicular function [30,31].
Evolution of the TGIF family from fly to human: specialised function in sex and reproduction?
The mammalian X-chromosome is known to contain a disproportionate number of genes involved in testis and brain function [19,32]. This is thought to have evolved due to direct selection of male advantage genes on the hemizygous × [17]. Whether X-linked genes are more likely to evolve testis functions or testicular genes are more likely to be relocated to the × remains to be tested. The marsupial X-chromosome is the smallest × in mammals and is most similar to the X-chromosome of the therian ancestor [32,33]. In the eutherian lineage the × has been expanded, mainly by the addition of two large autosomal blocks from the therian ancestor. These two large additions are autosomal in the marsupial and non-mammalian vertebrate lineages [33-36]. TGIFLX maps to the conserved region of the X-chromosome, present in all therian mammals, but was absent on the marsupial X-chromosome. Eutherian TGIFLX was not part of the two large autosomal additions but arose via retrotransposition, presumably from TGIF2 [14]. Once on the eutherian X-chromosome it was exposed to positive selection that favours the evolution of male reproductive advantage genes [17,21], possibly leading to its specialisation in the adult testis. The accelerated rate of TGIFLX evolution therefore appears to be due its location on the × and its potentially redundant function with its progenitor TGIF2 [17,37]. Despite the autosomal location of tammar TGIF2, it too has a specialised role in the adult marsupial in spermatogenesis and possibly folliculogenesis. The conflicting expression profiles of TGIF2 in adult marsupials and eutherians make it impossible to determine if the ancestral TGIF2 orthologue was a broadly expressed gene in the adult or was gonad-specific. There is good evidence that the × selectively retains translocated testis-specific genes that may confer a male advantage [21,32,33]. Thus the ancestral TGIF2 may have already had a function in mammalian spermatogenesis before a translocated copy moved to the × in eutherians. Due to the new TGIFX orthologue now fulfilling the testicular function, autosomal TGIF2 was able to diversify and take on broader functions in the adult. This suggestion is supported by the similar degree of functional specialisation in reproduction from the autosomal copy of TGIF2 in marsupials, demonstrating that a location on the sex chromosome is not the only the driving force behind the rapid specialisation of this gene in spermatogenesis.
Conclusions
This study demonstrates that tammar TGIF1 had a similar expression pattern to that of eutherians [11], but TGIF2 has a restricted expression pattern, very similar to that of eutherian TGIFLX/Tex1. Since TGIFLX and its orthologues are only present in the eutherian lineage, the retrotransposition event resulting in its creation must have occurred after the marsupial-eutherian divergence between 130 and 148MYA [25] but before ~80 MYA when the primate-rodent lineages split [14,17]. The germ cell specific expression of marsupial TGIF2 in adult testes suggests it may have a role in spermatogenesis, similar to that of TGIFLX/Tex1 in eutherian mammals and of the TGIF genes in Drosophila. TGIF2 expression was shown for the first time in follicles and oocytes so it is likely that it also has a role in folliculogenesis. Together these data suggest that there has been an ultra-conserved function for the TGIF family in fertility and that TGIF2 already had specialised role in the adult, in spermatogenesis and folliculogenesis long before its retrotransposition in eutherian mammals.
Methods
Gene cloning and structure
Both TGIF1 and TGIF2 were initially cloned by RT-PCR using cross-species primers (MeTGIFF1 and MeTGIFR1 for TGIF1, TF1 and TR1 for TGIF2) based on conserved regions in the human, mouse and opossum genomes. The resulting 554 bp PCR product (TGIF2) was sequenced and then used to design tammar specific primers for 3' and 5' RACE to clone the full TGIF2 mRNA transcript [GenBank Accession No. FJ775183]. Sequencing primers are listed in Table 1.
Table 1.
Primers designed for analysis of TGIF1 &TGIF2 expression by RT-PCR
| Primers | Sequence (5'→3') | Function |
|---|---|---|
| meTGIFF1 | CGAGACTGGCTCTATGAA | RT-PCR |
| meTGIFR1 | AGATTTACCCGTGTCCTC | RT-PCR |
| TF1 | TGAAGAT(C/T)CTCCGAGACTGG | Cross species cloning & RT-PCR |
| TR1 | CCACCAGCAG(G/C/T)TGGAAGC | Cross species cloning & RT-PCR |
| TF2 | ATGGCAAGGACCCTAACCG | 3' RACE and RT-PCR |
| TR2 | TGGAAGAAGCCGCCGTCGTG | 5' RACE |
| SMART IV | AAGCAGTGGTATCAACGCAGAGTGGCCATTACGGCCGGG | 5' RACE |
| CDS III | ATTCTAGAGGCCGAGGCGGCCGACATG-d(T)30N-1N (N = A, G, C, or T; N-1 = A, G, or C) | 3' RACE |
| 5'PCR primer | AAGCAGTGGTATCAACGCAGAGT | 5' RACE |
| 18S F | GATCCATTGGAGGGCAAGTCT | RT-PCR |
| 18S R | CCAAGATCCAACTACGAGCTTTTT | RT-PCR |
F denotes forward primers, R denotes reverse primers. All primers are shown in the 5'→3' direction; CDSIII, Smart IV and 50 PCR were synthesized by SIGMA (Genosys) according to the manual of the SMART cDNA library construction kit from Clontech.
cDNA was reverse-transcribed from total RNA isolated from an adult tammar wallaby testis, using the SMART cDNA library construction kit (Clontech, Mountain View, California, USA). 5' RACE was performed using primer 5' PCR and primer TR1. Due to an incomplete coding sequence at the 5' end, we designed a new primer TR2 as a nested primer and repeated 5' RACE. 3' RACE was performed using primer TF1 and CDS III, nested PCR was performed using the TF2 and CDS III primers. PCR cycling conditions were: 35 cycles of 30s, 95°C; 60s, 55°C; 120s, 72°C, in a 25 μl reaction with GoTaq Green Master Mix (Promega, Wisconsin, USA; Primers listed in Table 1).
Genomic sequence of tammar TGIF1 and TGIF2 were retrieved from the tammar wallaby genome trace archives http://www.ncbi.nlm.nih.gov/. These reads were aligned with the full length cDNA sequence to determine the gene structure of TGIF2. This confirmed the number and the length of exons.
Genomic sequence analysis and phylogenetic tree
Human genomic sequences for TGIFLX, TGIF2 and TGIF1 were obtained from the UCSC genome browser http://genome.ucsc.edu/. The conserved block of genes surrounding these loci were identified in amniote species using the VISTA genome browser program http://pipeline.lbl.gov/cgi-bin/gateway2. Blast was then performed in mouse, opossum, platypus, lizard, frog, fugu and fly. Genomic sequence from these orthologous blocks was also obtained from UCSC and aligned using the LAGAN algorithm available on the mVISTA website with default parameters http://genome.lbl.gov/vista/mvista/submit.shtml except that TGIFLX orthologues were aligned with PROLAGAN algorithm. In an attempt to identify any TGIFX orthologues in marsupials or platypus we performed a low stringency BLAST using the individual human TGIFX exons as well as a protein BLAT. In each case no homology was found.
Searches for homologues of TGIF1, TGIF2 and TGIFLX/Y were performed at NCBI http://www.ncbi.nlm.nih.gov and Ensembl http://www.ensembl.org and orthologues were found in eutherians, marsupials, monotreme (platypus), birds and reptiles, fish and invertebrates (fruit fly and mosquito) (Additional file 1, Table S1). Phylogenetic analysis was performed with PHYLIP 3.69 program with default parameters (University of Washington) using neighbour-joining analysis with 100 replicates, and viewed with TREE-view 1.66 http://taxonomy.zoology.gla.ac.uk/rod/treeview.html.
RT-PCR
RT-PCR was used to check the mRNA expression pattern of TGIF1 and TGIF2 in embryonic tissues (fetal stage at day 25 in 26.5 days gestation), developing gonads (from fetal stage before birth to pouch young stage of day 44 after birth) and adult tissues (Pancreas, brain, hypothalamus, pituitary, olfactory bulb, muscle, stomach, heart, liver, spleen, lung, adrenal, kidney, uterus, epididymis, ovary and testis). Total RNA was isolated using RNAwiz (Ambion Inc., Austin, Texas, USA) according to the manufacturer's instructions. The quality and quantity of total RNA was verified by gel electrophoresis and optical density reading with a Nanodrop (ND-1000 Spectrophotometer, Wilmington, USA). 2 μg of total RNA was DNase-treated with DNase I (Ambion Inc., Austin, Texas, USA) for 30 minutes. 1 μg of DNased RNA was reverse-transcribed with oligodT primer using the SuperScript III kit (Invitrogen, California, USA).
PCR was performed in a 25 μl reaction with GoTaq Green Master Mix, primers (meTGIF1F and meTGIF1R for TGIF1, TF2 and TR1 for TGIF2) and first-strand cDNA products. Amplification conditions were: 95°C 30s; 49.2°C 30s; 72°C, 30s for 35 cycles (TGIF1) and 95°C 30s; 57°C 30s; 72°C, 30s for 35 cycles (TGIF2), or 25 cycles (18S). Samples were analyzed on a 2% agarose gel. Furthermore, adult tissues (testis, ovary and spleen), developing gonads (testis and ovary at day 1 after birth) and embryonic tissue (forebrain at day 25 of gestation) expressing both TGIF1 and TGIF2 were selected to validate that our PCR results were within the exponential amplification phase of the PCR [38]. Briefly, same amount of cDNA template was used for TGIF1 and TGIF2 amplifications and 50 times dilution of cDNA template was used for 18S amplification. PCRs were stopped after a variable number of cycles (15, 20, 25, 30 and 35) to determine the exponential phase. For the TGIF2 primers, 35 cycles was within the exponential phase; for TGIF1 primers, 35 cycles almost reached the plateau phase, but was still within the exponential phase; for 18S primers, amplification had plateaued by 30 cycles, but was in the exponential phase after 25 cycles.
mRNA in situ hybridization
Antisense and sense RNA probes were prepared separately from a region including TGIF2 domain (~500 bp, spanning nucleotides 178-732) of the tammar wallaby transcript using SP6 and T7 polymerase respectively and labeled with digoxigenin-UTP (Roche, Castle Hill, NSW, Australia, Cat#10999644001). Tissues were fixed in 4% paraformaldehyde overnight at 4°C rinsed several times in 1X PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH = 7.4), embedded in paraffin, sectioned at 8 um and placed onto polysine coated slides (Menzel-Gläser, Germany). After dewax and rehydration, the sections were washed several times with 1X PBS, 0.1 M Glycine, 0.1% Triton X-100 and freshly made triethanolamine (TEA) buffer (100 mM triethanolamine, 0.25% (v/v) acetic anhydride, pH = 8.0), and then were immediately pre-hybridized in prehybridisation buffer containing 10% Dextran sulfate, 1× Denhardt's solution, 4× SSC, 50% Deionized formamide, 2 mM EDTA and 500 μg/ml herring sperm DNA for 1 hours at 37°C. Hybridization was performed on each section with 50-100 μl hybridization buffer containing 100 ng/ml of DIG-labeled RNA probe for 16-18 hours at 42°C. Hybridization signals were detected immunologically with alkaline phosphatase-conjugated anti-DIG antibody (Roche, Castle Hill, NSW, Australia, Cat# 11093274910) and visualized with NBT/BCIP (Roche, Castle Hill, NSW, Australia, Cat# 11681451001) according to the manufacturer's instructions. Tissues were counterstained with 0.1% nuclear Fast Red (Aldrich Chemical Corp., Milwaukee, WI, USA).
Immunohistochemistry
Fresh tissues were collected and fixed in 4% paraformaldehyde and treated as described above. Then tissue sections were dewaxed in histolene (Grale Scientific, VIC, Australia) and rehydrated in a series of ethanol/water solutions. Antigen retrieval was performed in boiling sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 20 minutes, sections were then treated with 5% hydrogen peroxide in methanol for 20 minutes. The primary antibody (TGIF2 purified MaxPab rabbit polyclonal antibody, Cat# H00060436-D01P, Abnova, USA) was applied to tammar adult gonad tissue sections at 1:150 dilutions and incubated at 4°C overnight. Signal was amplified using the streptavidin/HRP kit (DAKO, Australia, Cat# P0397), visualized with DAB (DAKO, Australia, Cat# 1967), and counterstained with hematoxylin.
BAC library screening and chromosomal mapping
To establish the chromosomal localization of TGIF2 in the tammar, a BAC library was screened with a TGIF2 554 bp PCR product as a probe. Membranes were pre-hybridized for 2 hours in Church buffer (0.25 M NaHPO4, 1 mM EDTA, 1% BSA and 7% SDS). 25 ng of probe PCR product was labeled with the Amersham Megaprime DNA Labeling System (GE Healthcare, Rydalmere, NSW, Australia, Cat# RPN1606) with [α-32P] dCTP (10 mCi/ml, PerkinElmer, Melbourne, VIC, Australia) following the manufacturer's instructions. Hybridization was performed at 65°C overnight. Filters were washed with 2XSSC/0.1%SDS, 1XSSC/0.1%SDS, and 0.1XSSC/0.1%SDS for 10 minutes each at 65°C and autoradiographed at -70°C for 2-5 days [39]. BACs were identified based on a duplicate spot pattern according to the Arizona Genomics Institute protocol http://www2.genome.arizona.edu/research/protocols.
Chromosome preparations were made from pouch young testis cells according to standard methods with minor modifications [40]. Chromosome fluorescence in situ hybridization (FISH) was performed as previously described with minor modifications [41]. The BAC genomic DNA was labeled with dUTP-digoxygenin (DIG) by nick translation at 14°C for one hour and pre-blocked with tammar wallaby Cot-1 DNA prior to hybridization. The probe was hybridized to tammar metaphase chromosome spreads at 37°C overnight. Hybridization was detected using mouse anti-DIG-FITC antibody (Roche, Castle Hill, NSW, Australia, Cat#. 11207741910; 1/200). After hybridization, the chromosome preparations were stained with DAPI (4, 6-diamidino-2-phenylindole) (Vector lab. Inc., Burlgamine, CA, USA, Cat# H-1200) to visualize the chromosomes. Images were taken using a Zeiss microscope.
Authors' contributions
All authors participated in the design of the study and collected the tissue samples. YH performed all the experiments, HY participated in FISH and ISH, AJP also participated in FISH. All authors analyzed the results. YH and HY drafted the manuscript. All authors read, modified and approved the final manuscript.
Supplementary Material
Table S1. Homologues of TGIF family from various species.
Contributor Information
Yanqiu Hu, Email: y.hu@zoology.unimelb.edu.au.
Hongshi Yu, Email: hongshiy@unimelb.edu.au.
Geoff Shaw, Email: g.shaw@unimelb.edu.au.
Marilyn B Renfree, Email: m.renfree@unimelb.edu.au.
Andrew J Pask, Email: andrew.pask@uconn.edu.
Acknowledgements
We thank all members of the wallaby research group and in particular Kerry Martin and Scott Brownlees for assistance with the animals. This study was supported by the Australian Research Council Centre of Excellence in Kangaroo Genomics; a Federation Fellowship to MBR, and a National Health and Medical Research Council R D Wright Fellowship to AJP. There is no financial or other potential conflict of interest. We also acknowledge the Albert Shimmins Fund (The University of Melbourne) for providing financial support to YH during preparation of the manuscript.
References
- Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annual review of biochemistry. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270(52):31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- Melhuish TA, Gallo CM, Wotton D. TGIF2 interacts with histone deacetylase 1 and represses transcription. J Biol Chem. 2001;276(34):32109–32114. doi: 10.1074/jbc.M103377200. [DOI] [PubMed] [Google Scholar]
- Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137(2):249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman CA, Bartholin L, Newfeld SJ, Wotton D. Drosophila TGIF proteins are transcriptional activators. Mol Cell Biol. 2003;23(24):9262–9274. doi: 10.1128/MCB.23.24.9262-9274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massague J. A Smad transcriptional corepressor. Cell. 1999;97(1):29–39. doi: 10.1016/S0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massague J, Muenke M. et al. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat Genet. 2000;25(2):205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- Melhuish TA, Wotton D. The Tgif2 gene contains a retained intron within the coding sequence. BMC Mol Biol. 2006;7:2. doi: 10.1186/1471-2199-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Walsh CA. Targeted disruption of Tgif, the mouse ortholog of a human holoprosencephaly gene, does not result in holoprosencephaly in mice. Mol Cell Biol. 2005;25(9):3639–3647. doi: 10.1128/MCB.25.9.3639-3647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino E, Wildt S, Richards G, Clerc RG. Expression of a novel murine homeobox gene in the developing cerebellar external granular layer during its proliferation. Dev Dyn. 1996;205(4):410–420. doi: 10.1002/(SICI)1097-0177(199604)205:4<410::AID-AJA5>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Imoto I, Pimkhaokham A, Watanabe T, Saito-Ohara F, Soeda E, Inazawa J. Amplification and overexpression of TGIF2, a novel homeobox gene of the TALE superclass, in ovarian cancer cell lines. Biochem Biophys Res Commun. 2000;276(1):264–270. doi: 10.1006/bbrc.2000.3449. [DOI] [PubMed] [Google Scholar]
- Jin L, Zhou Y, Kuang C, Lin L, Chen Y. Expression pattern of TG-interacting factor 2 during mouse development. Gene Expr Patterns. 2005;5(4):457–462. doi: 10.1016/j.modgep.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Blanco-Arias P, Sargent CA, Affara NA. The human-specific Yp11.2/Xq21.3 homology block encodes a potentially functional testis-specific TGIF-like retroposon. Mamm Genome. 2002;13(8):463–468. doi: 10.1007/s00335-002-3010-9. [DOI] [PubMed] [Google Scholar]
- Rogers JH. The origin and evolution of retroposons. International review of cytology. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Lai YL, Li H, Chiang HS, Hsieh-Li HM. Expression of a novel TGIF subclass homeobox gene, Tex1, in the spermatids of mouse testis during spermatogenesis. Mechanisms of development. 2002;113(2):185–187. doi: 10.1016/S0925-4773(02)00026-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J. Rapid evolution of mammalian X-linked testis-expressed homeobox genes. Genetics. 2004;167(2):879–888. doi: 10.1534/genetics.103.025072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhang J. Rapid evolution of primate ESX1, an X-linked placenta- and testis-expressed homeobox gene. Hum Mol Genet. 2007;16(17):2053–2060. doi: 10.1093/hmg/ddm153. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex Chromosomes and Sex Linked Genes. Berlin: Springer; 1967. [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet. 2005;14(19):2911–2918. doi: 10.1093/hmg/ddi322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27(4):422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Sargent CA, Boucher CA, Blanco P, Chalmers IJ, Highet L, Hall N, Ross N, Crow T, Affara NA. Characterization of the human Xq21.3/Yp11 homology block and conservation of organization in primates. Genomics. 2001;73(1):77–85. doi: 10.1006/geno.2000.6491. [DOI] [PubMed] [Google Scholar]
- Wang Z, Mann RS. Requirement for two nearly identical TGIF-related homeobox genes in Drosophila spermatogenesis. Development. 2003;130(13):2853–2865. doi: 10.1242/dev.00510. [DOI] [PubMed] [Google Scholar]
- Ayyar S, Jiang J, Collu A, White-Cooper H, White RA. Drosophila TGIF is essential for developmentally regulated transcription in spermatogenesis. Development. 2003;130(13):2841–2852. doi: 10.1242/dev.00513. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Luo ZX, Ji Q, Wible JR, Yuan CX. An Early Cretaceous tribosphenic mammal and metatherian evolution. Science. 2003;302(5652):1934–1940. doi: 10.1126/science.1090718. [DOI] [PubMed] [Google Scholar]
- Ji Q, Luo ZX, Yuan CX, Tabrum AR. A swimming mammaliaform from the Middle Jurassic and ecomorphological diversification of early mammals. Science. 2006;311(5764):1123–1127. doi: 10.1126/science.1123026. [DOI] [PubMed] [Google Scholar]
- Lin M, Harman A, Rodger JC. Spermiogenesis and spermiation in a marsupial, the tammar wallaby (Macropus eugenii) J Anat. 1997;190(Pt 3):377–395. doi: 10.1046/j.1469-7580.1997.19030377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi M, Ousati-Ashtiani Z, Nazarian A, Modarressi MH, Heidari M. Association of TGIFLX/Y mRNA expression with azoospermia in infertile men. Mol Reprod Dev. 2008;75(12):1761–1766. doi: 10.1002/mrd.20906. [DOI] [PubMed] [Google Scholar]
- Peters H, McNatty KP. The Ovary A Correlation of Structure and Function in Mammals. Berkeley and Los Angeles, California: University of California Press; 1980. Ovarian Hormones; pp. 44–59. [Google Scholar]
- Tajima K, Orisaka M, Mori T, Kotsuji F. Ovarian theca cells in follicular function. Reprod Biomed Online. 2007;15(5):591–609. doi: 10.1016/S1472-6483(10)60392-6. [DOI] [PubMed] [Google Scholar]
- Graves JA. Review: Sex chromosome evolution and the expression of sex-specific genes in the placenta. Placenta. 2010;31(Suppl):S27–32. doi: 10.1016/j.placenta.2009.12.029. [DOI] [PubMed] [Google Scholar]
- Graves JA. Weird animal genomes and the evolution of vertebrate sex and sex chromosomes. Annual review of genetics. 2008;42:565–586. doi: 10.1146/annurev.genet.42.110807.091714. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C. et al. Physical map of two tammar wallaby chromosomes: a strategy for mapping in non-model mammals. Chromosome Res. 2008;16(8):1159–1175. doi: 10.1007/s10577-008-1266-y. [DOI] [PubMed] [Google Scholar]
- Kohn M, Kehrer-Sawatzki H, Vogel W, Graves JA, Hameister H. Wide genome comparisons reveal the origins of the human × chromosome. Trends Genet. 2004;20(12):598–603. doi: 10.1016/j.tig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human × chromosome. Science. 1999;286(5441):964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Hurst LD. Evolutionary genomics. Sex and the X. Nature. 2001;411(6834):149–150. doi: 10.1038/35075697. [DOI] [PubMed] [Google Scholar]
- Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biological procedures online. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- Schempp W, Meer B. Cytologic evidence for three human X-chromosomal segments escaping inactivation. Hum Genet. 1983;63(2):171–174. doi: 10.1007/BF00291539. [DOI] [PubMed] [Google Scholar]
- Wilcox SA, Toder R, Foster JW. Rapid isolation of recombinant lambda phage DNA for use in fluorescence in situ hybridization. Chromosome Res. 1996;4(5):397–398. doi: 10.1007/BF02257276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Homologues of TGIF family from various species.