Abstract
Water-soluble polypeptides adopting α-helical conformations with unprecedented high helicities were obtained by elongating the charge-containing side chains of the constituent amino acids to allow the terminal charges to be situated distally from the peptide backbone. Poly(γ-(4-aminoethylthiopropoxyl)-benzyl-L-glutamate) (PAOBLG-AET) with a charge-peptide backbone distance of 17 σ-bonds exhibited a remarkably high helical content (81%) at a degree of polymerization as low as 10. The helical conformations of these short polypeptides were very stable against various harsh, protein-denaturing conditions, such as extreme pH, high temperature, and high salt or urea concentrations.
Keywords: polypeptides, ring-opening polymerization, amino acid N-carboxyanhydride, NCA, α-helix, thiol-ene click chemistry
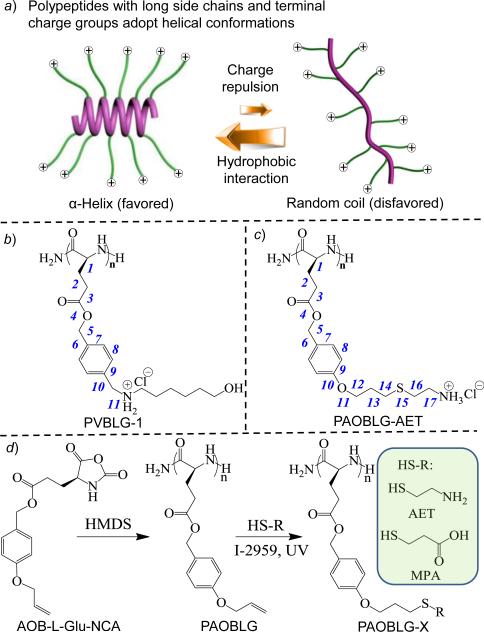
The α-helix is one of the most important functional domains in polypeptides controlling numerous biological activities and functions.1-12 Studies aimed at increasing the overall helicity and stability of helical motifs of proteins and peptides, especially short-chain oligopeptides, have contributed to the fundamental understanding of protein folding/unfolding and have led to improvements in the biological and pharmaceutical activities of peptides.13-19 There is often a drawback in the design of water-soluble, bioactive helical peptides: charged amino acid building blocks provide water solublity but decrease helicity because of disruption of helix due to side-chain charge repulsion.20-23 Increasing the proportion of hydrophobic amino acids tends to increase helicity by increasing side-chain hydrophobic interactions, but the resulting structures show reduced water solubility, which is undesirable for the design of biologically active peptides. It has been a general strategy to integrate both water-soluble and helix-stabilizing motifs in the peptide structure to design water-soluble, helical peptides. Such peptides are often designed to have charged amino acid residues situated on one side of the helix surface and the residues responsible for stabilizing the helix through side-chain hydrophobic interactions,24-26 salt bridges,27-29 or tethering18, 30, 31 situated on the opposite side of the helix surface. These strategies require the design of peptides with specific sequences27 and usually involve tedious chemistries of polypeptide side chains30 that are typically difficult to control. For polypeptides prepared by polymerization instead of through step-wise synthesis, such helix-stabilization strategies mentioned above for the synthesis water-soluble, helical peptides cannot be simply applied.32
Water-soluble, synthetic polypeptides that can adopt stable α-helical conformations have attaracted much attention. Prior efforts have been focused on introducing neutrally charged, hydrophilic functional groups33 or moieties.32 Poly(N-hydroxyalkyl-L-glutamine)33, one of the early design of water-soluble polypeptides derived from aminolysis of poly(L-glutamate) (PLG) with pendant hydroxyl groups, showed excellent water-solubility and fairly high helical contents (up to ca. 65% helicity) in aqueous solution.33 Later, Deming designed PLL containing pendant oligoethyleneglycol moieties.32 The resulting oligoethyleneglycol-graft PLL showed excellent water-solubility and remarkably high helcial content (100% helicity in pH 7 water at 25 °C). Recently, Li and coworkers designed thermo-responsive α-helical polypeptide from peglated poly(L-glutamate), highlighting the recent progress of this class of special polypeptides containing non-charged, water-soluble segments on a α-helical structure.34
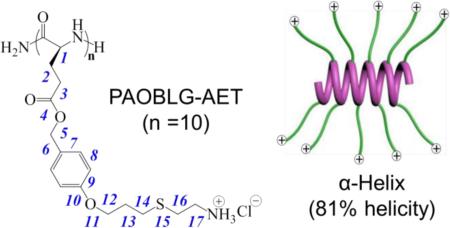
In a separate effort, we designed charged, water-soluble polypeptides that adopt stable α-helical conformations (i.e., α-helical polypeptide electrolytes; αHPEs), by using polypeptide containing charged side chains but elongating the charge-containing amino acid side chains to place the charges distally from the polypeptide backbone (Figure 1a).35 When the charges are 11 σ-bonds away from the peptide backbone, as in poly(γ-(4-(1-hexanol-6-aminomethyl))benzyl-L-glutamate) (PVBLG-1; Figure 1b), the resulting polypeptide with a degree of polymerization (DP) of 60 ((PVBLG-1)60) maintains a stable α-helical conformation with 91% helicity.35 PVBLG-1’s with very low DPs, such as (PVBLG-1)10 with a DP value of 10, however, have mixed conformations containing both β-sheets and α-helices, with a helicity of only 26% for (PVBLG-1)10.35 Because both the charge-backbone distance and the hydrophobicity of the side chains in αHPEs have significant effect on the stability of α-helix,35 we reasoned that further elongating the side chain will not only further reduce side chain charge repulsion by increasing the charge-backbone distance but also increase the side-chain hydrophobicity. By doing so, it is possible to obtain a water-soluble αHPE with ultra-stable α-helix and high helicity even at a very low DP. Here, we report the design and synthesis of a water-soluble αHPE, (poly(γ-(4-aminoethylthiopropoxyl)benzyl-L-glutamate) (PAOBLG-AET, Figure 1c), with side chain charge situated 17 σ-bonds away from the peptide backbone, adopts an unprecedented, remarkably high helicity (81%) with a DP of 10 at pH 2 aqueous solution.
Figure 1.
(a) Polypeptide with charged side chains and the postulated helix–coil transition in response to the length of the side chains. Chemical structures of (b) PVBLG-1 and (c) PAOBLG-AET. (d) Synthesis of PAOBLG-AET and PAOBLG-MPA.
The synthesis of PAOBLG-AET is illustrated in Figure 1d. γ-(4-Allyloxylbenzyl)-L-glutamate N-carboxyanhydride (AOB-l-Glu-NCA) can be easily prepared in multi-gram scale (see Supporting Information). The ring-opening polymerization of AOB-l-Glu-NCA initiated by hexamethyldisilazane (HMDS)36-38 yielded PAOBLGs with controlled molecular weights (MWs) and narrow molecular-weight distributions (MWDs) that were determined by gel permeation chromatography (GPC) (Table S1). For example, at a monomer/initiator (M/I) ratio of 10 with expected Mn of 3.0 × 103 g·mol-1, the resulting PAOBLG had an Mn of 2.8 × 103 g·mol-1 with a narrow MWD of 1.22 (entry 1, Table S1). The MW and MWD of PAOBLG10 obtained by matrix-assisted laser desorption ionization mass spectrometry (MALDI-TOF MS, Figure S3) agreed well with the values obtained by GPC. To accelerate the polymerization of AOB-l-Glu-NCA and synthesize higher MW PAOBLGs, we used 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD) as a co-catalyst,39, 40 which gave faster yet controlled NCA polymerization (Figure S4).41 In the presence of a small amount of TBD (HMDS/TBD = 1/0.1), the polymerizations yielded corresponding PAOBLGs with the expected MWs and narrow MWDs (entries 3 and 4, Table S1).
The PAOBLGs were then treated with 2-aminoethanethiol hydrochloride in a mixture of dimethylformamide and deionized water to effect a UV-triggered thiol-ene “click” reaction.42-46 Dialysis of the reaction mixture followed by lyophilization removed all the small-molecule impurities and afforded the desired polymers as a fluffy powder. As expected, the thiol-ene reaction proceeded rapidly and completed in 10 min, yielding PAOBLG-AETs with nearly quantitative grafting efficiency (Figures S5 and S6).
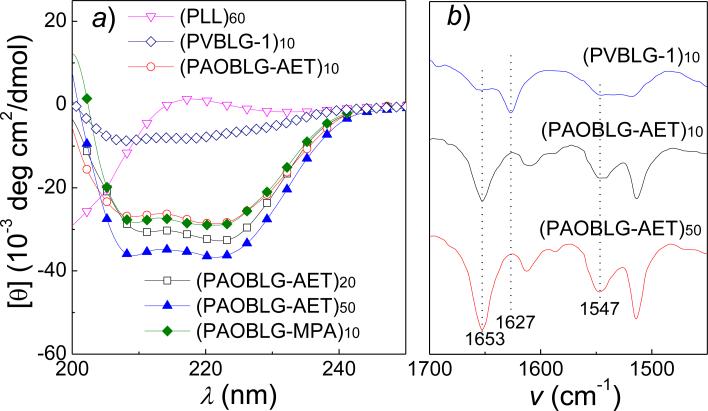
The PAOBLG-AETs are very soluble in water (> 20 mg·mL-1) because of the terminal ammonium groups on each of their side chains, in sharp contrast to PAOBLG which is insoluble in water. To determine whether the PAOBLG-AETs have the expected high helicity at low DP, we used circular dichroism (CD) spectroscopy to analyze the conformation of the PAOBLG-AETs at pH 2 at which all side-chain amines should be protonated and are charged. All three PAOBLG-AETs ((PAOBLG-AET)10, (PAOBLG-AET)20 and (PAOBLG-AET)50) showed the characteristic CD spectra of α-helix with two minima at 208 and 222 nm (Figure 2a). (PAOBLG-AET)10 (charge-backbone distance of 17 σ-bonds, Figure 1c) had a –[θ]222 value of 28.5 × 103 cm2·deg·dmol-1, which corresponds to a helicity of 81% (Figure 2a, Table 1)47, in sharp contrast to a 60-mer poly-L-lysine ((PLL)60, charge-backbone distance of 4 σ-bonds) that adopts a random coil conformation and (PVBLG-1)10 (charge-backbone distance of 11 σ-bonds, Figure 1b) that has a –[θ]222 value of 7.2 × 103 cm2·deg·dmol-1, which corresponds to a helicity of only 26% (Figure 1a and Table 1). The high helicity of (PAOBLG-AET)10 was further verified by FTIR (Figure 2b).48, 49 (PVBLG-1)10 has mixed conformations containing both α-helix (amide I band at 1653 and amide II band 1547 cm-1) and β-sheet (amide I band at 1627 cm-1) in solid state, while (PAOBLG-AET)10 has predominant α-helix (strong amide I band at 1653 and amide II band 1547 cm-1) and negligible β-sheet conformation. For PAOBLG-AETs with DP values of 20 and 50, the –[θ]222 values were 34.0 and 36.8 ×103 cm2·deg·dmol-1, corresponding to helicities of 94% and 100%, respectively (Figure 2a, and Table 1). (PAOBLG-AET)10 and (PAOBLG-AET)50 have nearly identical FTIR spectrum (Figure 2b), further validating the high helical content of (PAOBLG-AET)10.
Figure 2.
(a) CD spectra of various polypeptides bearing charged side chains ((PLL)60, (PVBLG-1)10 and (PAOBLG-AET)10, 20, 50 at in aqueous solution at pH 2 and (PAOBLG-MPA)10) in aqueous solution at pH 10, (b) Fourier-transform infrared spectra (FTIR) of (PVBLG-1)10 and (PAOBLG-AET)10 and (PAOBLG-AET)50.
Table 1.
Conformation Analysis of Ionic Polypeptides
| Entry | Polypeptide | DP | -[θ]222 ×10-3 (cm2·deg·dmol-1)a | Helical content (%)b |
|---|---|---|---|---|
| 1 | (PLL)60 | 60 | - | 0 |
| 2 | (PVBLG-I)10 | 10 | 7.2 | 26 |
| 3 | (PAOBLG-AET)10 | 10 | 28.5 | 81 |
| 4 | (PAOBLG-AET)20 | 20 | 34.0 | 94 |
| 5 | (PAOBLG-AET)50 | 50 | 36.8 | 100 |
| 6 | (PAOBLG-MPA)10 | 10 | 29.6 | 84 |
The mean residue molar ellipticity [θ] was determined by following literature-reported formulas: Ellipticity ([θ]222 nm in cm2 deg dmol-1) = (millidegrees × mean residue weight)/(path length in millimetres × concentration of polypeptide in mg·ml-1)
The helical contents of the polypeptides were calculated using the following equation: percentage of α-helix = (-[θ]222 + 3000)/39,000.47
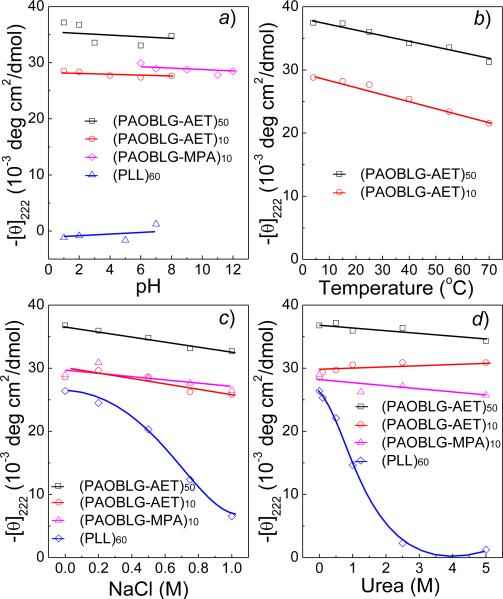
We next studied the helical stability of PAOBLG-AETs against changing environmental conditions, including changes in the pH and temperature and the presence of denaturing reagents. The -[θ]222 value of (PAOBLG-AET)10 remained unchanged when the solution's pH was increased from 1 to 8 (Figure 3a). At further increased pH values, (PAOBLG-AET)10 became less soluble because of de-protonation of some of its charged ammonium groups. (PAOBLG-AET)10 showed a lack of concentration dependence of its -[θ]222 value in helix-forming conditions, suggesting that it remained monomeric in aqueous solution (Figure S8). It displayed excellent helical stability against elevated temperature, with its -[θ]222 value decreasing 25% from 28,800 cm2·deg·dmol-1 at 4°C to 21,600 cm2·deg·dmol-1 at 70°C (Figure 3b), and against helix-destabilizing conditions such as high concentrations of NaCl (Figure 3c) and urea (Figure 3d). (PAOBLG-AET)10 showed unprecedented helical stability against any known α-peptides and amazingly maintained ~100% of its original helical content in 5M urea. (PAOBLGAET)50 showed very similar helical stability as (PAOBLG-AET)10 to those changing environmental conditions; the helical stabilities of both (PAOBLG-AET)10 and (PAOBLG-AET)50 were drastically different from that of PLL60 in high concentrations of NaCl (Figure 3c) and urea solutions (Figure 3d).
Figure 3.
(a) The pH dependence of residue molar ellipticity at 222 nm for (PAOBLG-AET)10, (PAOBLG-AET)50, (PAOBLG-MPA)10 and (PLL)60 at 0.05 mg·mL-1. (b) Temperature dependence of residue molar ellipticity at 222 nm for (PAOBLG-AET)10 and (PAOBLG-AET)50 at pH 2 and 0.05 mg·mL-1. (c) The salt-concentration dependence of residue ellipticity at 222 nm for (PAOBLG-AET)10 and (PAOBLG-AET)50 at pH 2 and (PAOBLG-MPA)10 and (PLL)60 at pH 10 (c = 0.05 mg/mL). (d) The helical stabilities of (PAOBLG-AET)10 and (PAOBLG-AET)50 at pH 2, and (PAOBLG-MPA)10 and (PLL)60 at pH 10 in the presence of urea (c = 0.05 mg/mL).
This novel strategy of distal charge placement on side chains to maintain both water solubility and high helicity in low MW polypeptide can also be extended to polypeptides bearing negatively charged side chains. (PAOBLG-MPA)10, a peptide with similar structure as (PAOBLG-AET)10 bearing carboxylate terminated side-chain with charge-backbone distance of 18 σ-bonds, was prepared via thiolene reaction of PAOBLG with 3-mercaptopropionic acid (Figures 1d and S7). (PAOBLG-MPA)10 had a helicity of 84% in aqueous solution at pH 9, when its carboxylate groups are completely deprotonated. The -[θ]222 value of (PAOBLG-MPA)10 remained unchanged when the solution's pH was decreased from 12 to 6 (Figure 3a). At further decreased pH values, (PAOBLG-MPA)10 became less soluble because of protonation of some of its charged carboxylate ions. (PAOBLG-MPA)10 showed very similar response as (PAOBLG-AET)10 against the helix-destabilizing conditions such as high concentrations of NaCl (Figure 3c) and urea (Figure 3d).
In summary, polypeptides with long side chain bearing positive/negative charge groups were synthesized by controlled ROP of AOB-l-Glu-NCA and subsequent thiol-ene reaction. Because of their elongated hydrophobic side chains and distally situated charges, these polypeptides are highly water-soluble and have very high helicity even with a DP value as low as 10. Furthermore, the helical structures of these low MW polypeptide electrolytes were stable to changes in pH, temperature, NaCl, and urea. To our knowledge, PAOBLG-AET(MPA) is the shortest, charged peptide to show such high helicity, remarkable helical stability and water solubility. Our study demonstates that elongating the hydrophobic side chain bearing a terminal charge group can serve as a general strategy for the design of water-soluble polypeptide with high helicity and high helical stability.
Supplementary Material
Acknowledgment
J.C. acknowledges the supports from the NSF (CHE-0809420), the NIH (Director's New Innovator Award 1DP2OD007246-01 and 1R21EB009486 A), and the Center for Nanoscale Science and Technology. We thank Professor Martin Gruebele and Dr. Apratim Dhar for allowing us to use their CD spectrometer.
Footnotes
Supporting Information Available: Experimental details and the spectroscopy and analytical data for the synthesis and characterization of PAOBLG-AET and PAOBLG-MPA. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Engler AC, Lee HI, Hammond PT. Angew. Chem., Int. Ed. 2009;48:9334–9338. doi: 10.1002/anie.200904070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rao JY, Luo ZF, Ge ZS, Liu H, Liu SY. Biomacromolecules. 2007;8:3871–3878. doi: 10.1021/bm700830b. [DOI] [PubMed] [Google Scholar]
- 3.Guo JS, Huang YB, Jing XB, Chen XS. Polymer. 2009;50:2847–2855. [Google Scholar]
- 4.Robertson DE, Farid RS, Moser CC, Urbauer JL, Mulholland SE, Pidikiti R, Lear JD, Wand AJ, Degrado WF, Dutton PL. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 5.Hill RB, Raleigh DP, Lombardi A, Degrado WF. Acc. Chem. Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryson JW, Betz SF, Lu HS, Suich DJ, Zhou HXX, Oneil KT, Degrado WF. Science. 1995;270:935–941. doi: 10.1126/science.270.5238.935. [DOI] [PubMed] [Google Scholar]
- 7.Li ZB, Deming TJ. Soft Matter. 2010;6:2546–2551. [Google Scholar]
- 8.Nowak AP, Breedveld V, Pakstis L, Ozbas B, Pine DJ, Pochan D, Deming TJ. Nature. 2002;417:424–428. doi: 10.1038/417424a. [DOI] [PubMed] [Google Scholar]
- 9.Shim MS, Kwon YJ. Biomaterials. 2010;31:3404–3413. doi: 10.1016/j.biomaterials.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 10.Schlaad H, Smarsly B, Losik M. Macromolecules. 2004;37:2210–2214. [Google Scholar]
- 11.Klok HA, Lecommandoux S. Peptide Hybrid Polymers. Vol. 202. Springer-Verlag Berlin; Berlin: 2006. Solid-state structure, organization and properties of peptide - Synthetic hybrid block copolymers. pp. 75–111. [Google Scholar]
- 12.Hadjichristidis N, Iatrou H, Pitsikalis M, Sakellariou G. Chem. Rev. 2009;109:5528–5578. doi: 10.1021/cr900049t. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Yoshitomi H, Morikawa M, Ando S, Takiguchi H, Inoue T, Sugihara G. Biopolymers. 1995;36:391–398. doi: 10.1002/bip.360360312. [DOI] [PubMed] [Google Scholar]
- 14.Munoz V, Serrano L. Nat. Struct. Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 15.Ma MT, Hoang HN, Scully CCG, Appleton TG, Fairlie DP. J. Am. Chem. Soc. 2009;131:4505–4512. doi: 10.1021/ja900047w. [DOI] [PubMed] [Google Scholar]
- 16.Graff DK, PastranaRios B, Venyaminov SY, Prendergast FG. J. Am. Chem. Soc. 1997;119:11282–11294. [Google Scholar]
- 17.Chen YX, Mant CT, Farmer SW, Hancock REW, Vasil ML, Hodges RS. J. Biol. Chem. 2005;280:12316–12329. doi: 10.1074/jbc.M413406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafmeister CE, Po J, Verdine GL. J. Am. Chem. Soc. 2000;122:5891–5892. [Google Scholar]
- 19.Stephens OM, Kim S, Welch BD, Hodsdon ME, Kay MS, Schepartz A. J. Am. Chem. Soc. 2005;127:13126–13127. doi: 10.1021/ja053444+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohl CA, Chakrabartty A, Baldwin RL. Protein Sci. 1996;5:2623–2637. doi: 10.1002/pro.5560051225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobson CM, Sali A, Karplus M. Angew. Chem., Int. Ed. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 22.Dobson CM. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 23.Marqusee S, Robbins VH, Baldwin RL. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dill KA. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 25.Albert JS, Hamilton AD. Biochemistry. 1995;34:984–990. doi: 10.1021/bi00003a033. [DOI] [PubMed] [Google Scholar]
- 26.Seebach D, Abele S, Gademann K, Guichard G, Hintermann T, Jaun B, Matthews JL, Schreiber JV. Helv. Chim. Acta. 1998;81:932–982. [Google Scholar]
- 27.Marqusee S, Baldwin RL. Proc. Natl. Acad. Sci. U. S. A. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng RP, DeGrado WF. J. Am. Chem. Soc. 2001;123:5162–5163. doi: 10.1021/ja010438e. [DOI] [PubMed] [Google Scholar]
- 29.Bierzynski A, Kim PS, Baldwin RL. Proc. Natl. Acad. Sci. U. S. A. 1982;79:2470–2474. doi: 10.1073/pnas.79.8.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blackwell HE, Grubbs RH. Angew. Chem., Int. Ed. 1998;37:3281–3284. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 31.Madden MM, Vera CIR, Song WJ, Lin Q. Chem. Commun. 2009:5588–5590. doi: 10.1039/b912094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M, Nowak AP, Deming TJ, Pochan DJ. J. Am. Chem. Soc. 1999;121:12210–12211. [Google Scholar]
- 33.Lotan N, Yaron A, Berger A. Biopolymers. 1966;4:365–368. [Google Scholar]
- 34.Chen C, Wang Z, Li Z. Biomacromolecules. 2011 doi: 10.1021/bm200849m. DOI: 10.1021/bm200849m. [DOI] [PubMed] [Google Scholar]
- 35.Lu H, Wang J, Bai Y, Lang JW, Liu S, Lin Y, Cheng J. Nat. Commun. 2011;2:206. doi: 10.1038/ncomms1209. [DOI] [PubMed] [Google Scholar]
- 36.Lu H, Cheng JJ. J. Am. Chem. Soc. 2007;129:14114–14115. doi: 10.1021/ja074961q. [DOI] [PubMed] [Google Scholar]
- 37.Lu H, Cheng JJ. J. Am. Chem. Soc. 2008;130:12562–12563. doi: 10.1021/ja803304x. [DOI] [PubMed] [Google Scholar]
- 38.Lu H, Wang J, Lin Y, Cheng JJ. J. Am. Chem. Soc. 2009;131:13582–13583. doi: 10.1021/ja903425x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon L, Goodman JM. J. Org. Chem. 2007;72:9656–9662. doi: 10.1021/jo702088c. [DOI] [PubMed] [Google Scholar]
- 40.Kember MR, Buchard A, Williams CK. Chem. Commun. 2011;47:141–163. doi: 10.1039/c0cc02207a. [DOI] [PubMed] [Google Scholar]
- 41.Lu H, Bai Y, Wang J, Gabrielson NP, Wang F, Lin Y, Cheng J. Macromolecules. 2011 doi: 10.1021/ma201164n. 10.1021/ma201164n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoyle CE, Bowman CN. Angew. Chem., Int. Ed. 2010;49:1540–1573. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 43.Dondoni A. Angew. Chem., Int. Ed. 2008;47:8995–8997. doi: 10.1002/anie.200802516. [DOI] [PubMed] [Google Scholar]
- 44.Campos LM, Meinel I, Guino RG, Schierhorn M, Gupta N, Stucky GD, Hawker CJ. Adv. Mater. 2008;20:3728–3733. [Google Scholar]
- 45.Killops KL, Campos LM, Hawker CJ. J. Am. Chem. Soc. 2008;130:5062–5064. doi: 10.1021/ja8006325. [DOI] [PubMed] [Google Scholar]
- 46.Lowe AB. Polym Chem. 2010;1:17–36. [Google Scholar]
- 47.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 48.Elliott A, Bradbury EM. J. Mol. Biol. 1962;5:574–576. doi: 10.1016/s0022-2836(62)80121-1. [DOI] [PubMed] [Google Scholar]
- 49.Miyazawa T, Blout ER. J. Am. Chem. Soc. 1961;83:712–719. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.