Abstract
Vitamin A, retinol, circulates in blood bound to serum retinol binding protein (RBP) and is transported into cells by a membrane protein termed stimulated by retinoic acid 6 (STRA6). It was reported that serum levels of RBP are elevated in obese rodents and humans, and that increased level of RBP in blood causes insulin resistance. A molecular mechanism by which RBP can exert such an effect is suggested by the recent discovery that STRA6 is not only a vitamin A transporter but also functions as a surface signalling receptor. Binding of RBP-ROH to STRA6 induces the phosphorylation of a tyrosine residue in the receptor C-terminus, thereby activating a JAK/STAT signalling cascade. Consequently, in STRA6-expressing cells such as adipocytes, RBP-ROH induces the expression of STAT target genes, including SOCS3, which suppresses insulin signalling, and PPARγ, which enhances lipid accumulation. RBP-retinol thus joins the myriad of cytokines, growth factors and hormones which regulate gene transcription by activating cell surface receptors that signal through activation of Janus kinases and their associated transcription factors STATs.
1. Introduction
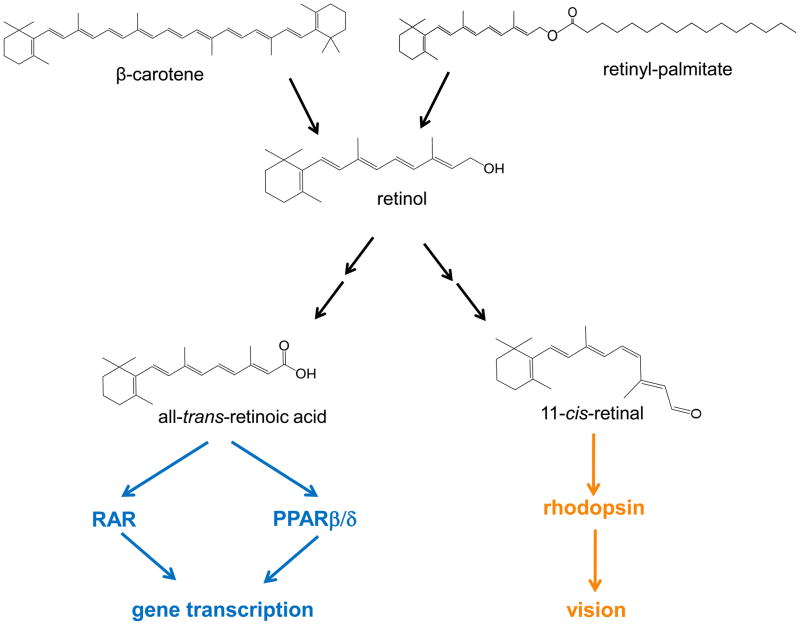
Vitamin A was recognized as an essential factor in foods about a century ago [1,2] and a substantial body of knowledge on the mechanisms that regulate its absorption and disposition in the body and on its biological functions has since accumulated [3]. The vitamin plays key roles in embryonic development, vision, immune function, and tissue remodeling and metabolism. It is usually believed that most of these functions are exerted not by the parental vitamin A molecule, retinol, but by active metabolites (Fig. 1). Hence,11-cis-retinal mediates phototransduction and is essential for vision, and all-trans-retinoic acid regulates gene transcription by activating the nuclear receptors retinoic acid receptors (RARs) and peroxisome proliferator-activated receptor β/δ (PPARβ/δ) [4,5,6,7]. Other retinoids, most notably 9-cis-retinoic acid, display transcriptional activities. However, while this isomer can efficiently activate the nuclear receptor retinoid X receptor (RXR), it has been difficult to establish whether it is in fact present in tissues that express RXR in vivo, other than the pancreas [8]. It thus remains unclear whether 9-cis-retinoic acid is a physiologically meaningful RXR ligand [9].
Figure 1. Chemical structures of vitamin A (retinol), its precursors, and two important active metabolites.
Retinol can be generated from β-carotene, present in plants, or from retinylesters, originating from animal sources. Retinol can then be metabolically transformed to active metabolites including all-trans-retinoic acid, which regulates gene transcription by activating the nuclear receptors RAR and PPARβ/δ, and 11-cis-retinal, which serves as a cofactor for the visual chromophore rhodopsin and is critical for vision.
Vitamin A is obtained from the diet either from animal sources, where it is present in the form of retinylesters, or from plants that contain carotenoids such as β-carotene (Fig. 1). In intestinal absorptive cells, retinol derived from either source is esterified to long chain fatty acids to form retinyesters. Retinylesters are then packaged in chylomicrons, secreted through the lymphatic system into blood and are taken up by the liver. The liver thus serves as the major storage for vitamin A in the body [10]. The mechanisms by which vitamin A needs are “sensed” by the liver and that trigger the release of retinol from its hepatic storage pool are unknown. However, when such a release is induced, retinol is mobilized from the liver bound to a protein called serum retinol binding protein (RBP). Other tissues, including adipose tissues, kidney, lung, heart, skeletal muscle, spleen, eye, and testis express RBP. However, corresponding to its function in vitamin A storage, the liver is the main site of synthesis and secretion of this protein. In blood, retinol-bound RBP is associated with a 55 KDa homotetrameric protein termed transthyretin (transporter of thyroxin and retinol, TTR), which, in addition to binding RBP, transports thyroxin (T4). The ternary retinol/RBP/TTR complex is the circulating vitamin A source for extrahepatic tissues. Uptake of retinol from blood into target cells is mediated by a protein called stimulated by retinoic acid 6 (STRA6), a cell surface transporter which binds RBP and facilitates the movement of retinol from the serum protein into cells [11,12]. In target cells, retinol can be stored in the form of retinylesters or it can be converted into the transcriptionally active metabolites retinoic acids. In retinal pigment epithelium in the eye, retinol can also be metabolized to 11-cis-retinal which is transported to photoreceptor cells where it serves to regenerate the visual pigment rhodopsin.
It is well documented that vitamin A is involved in lipid metabolism and insulin responses through its ability to activate the nuclear receptors termed retinoic acid receptors (RAR), and peroxisome proliferator-activated receptor β/δ (PPARβ/δ). Upon their activation, these receptors regulate the expression of proteins that control adipocyte differentiation, lipolysis, energy dissipation, fatty acid oxidation, and glucose transport [13,14,15,16,17,18]. Indeed, it has long been thought that the only function of RBP is to allow the hydrophobic vitamin A to circulate in blood, and that retinol participates in regulating energy homeostasis and insulin responsiveness solely through serving as a precursor for retinoic acid. However, more recently, it was reported that expression of RBP in adipose tissue and, correspondingly, serum levels of the protein, are markedly increased in obese mice and humans. It was further demonstrated that elevation in serum RBP levels causes insulin resistance [19]. By linking RBP to impairment of insulin responses in obese animals, these observations raise the intriguing possibility that the protein has biological activities other than to serve as the plasma carrier of vitamin A. In pursuing such a possibility, we discovered that association of retinol-bound RBP with the vitamin A transporter STRA6 triggers a signalling cascade mediated by the Janus kinase JAK2 and its associated transcription factors Signal Transducers and Activators of Transcription (STATs). The observations further revealed that activation of a JAK/STAT cascade by RBP-retinol results in upregulation of expression of STAT target genes including genes that inhibit insulin signalling and that control lipid homeostasis. Here, we review available information on the newly found signalling pathway initiated by retinol-bound RBP (holo-RBP). We summarize the observations that led to the surprising conclusions that the circulating RBP-retinol complex regulates gene transcription by a mechanism that is independent of the function of retinol as a precursor for retinoic acid, and that STRA6 functions as a signalling membrane receptor.
2. The retinol-RBP-TTR complex
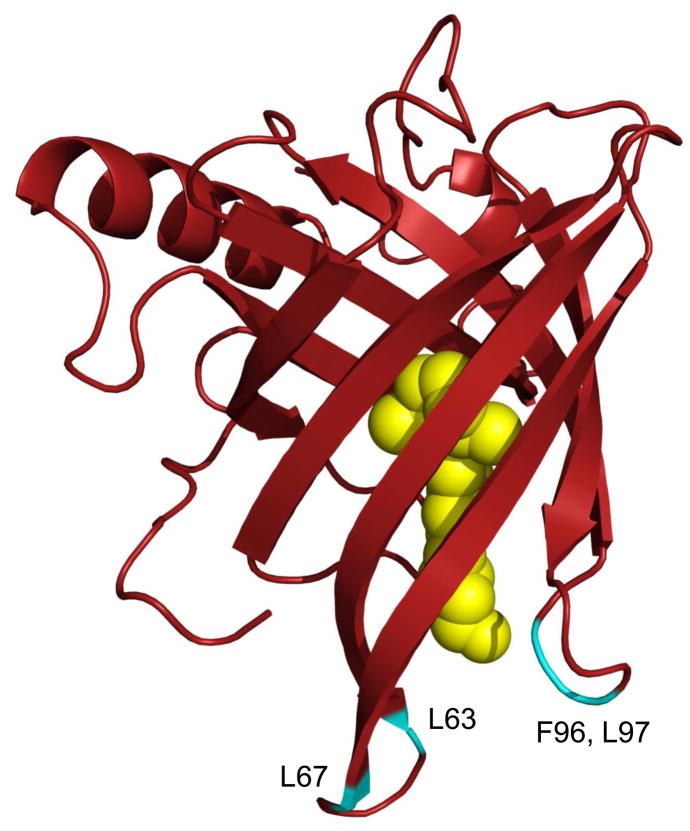
Retinol is secreted from the liver into blood bound to RBP, a member of the lipocalin family which includes small, mostly extracellular, proteins found in vertebrate and invertebrate animals, plants, and bacteria. Lipocalins have diverse functions but, like RBP, many of them serve as transporters for small hydrophobic molecules [20,21]. These proteins share a very low sequence homology but display a highly conserved overall fold. They are comprised of an eight-stranded antiparallel β-sheet that is folded over itself to form a β-barrel which constitutes the ligand binding pocket. The amino termini of lipocalins wrap around the back of the barrel, capping that side of the pocket. In contrast, the front of the β-barrel is open, providing a portal for the ligand which is flanked by a single loop scaffold. In RBP, retinol is encapsulated in the binding pocket with the β-ionone ring innermost and the hydroxyl head-group reaching to the protein surface where it is coordinated to a water molecule at the pocket entrance ([22,23], see Fig. 2). The association of retinol with RBP is stabilized mainly by hydrophobic interactions between the β-ionone ring and the isoprenoid chain with amino acid residues that line the interior of the pocket [23,24]. In addition to binding hydrophobic ligands, many lipocalins interact with accessory proteins. Indeed, holo-RBP is found in blood associated with the thyroxin transporter TTR. It is thought that complex formation between RBP with TTR serves to prevent loss of the low molecular weight RBP by glomerular filtration in the kidneys.
Figure 2. The three dimensional crystal structure of holo-retinol binding protein (RBP-ROH).
The human holo-RBP structure [23] (PDB ID 1BRP) was generated using Pymol (http://www.pymol.org/). The structure shows the eight stranded antiparallel β-sheet folded over itself to form a β-barrel. Retinol (yellow) is encapsulated by the barrel with the β-ionone ring buried in the binding pocket and the alcohol group is at the protein surface. Residues that stabilize the interactions of RBP with TTR are highlighted in blue. The location of these residues emphasize that interactions of RBP with TTR block the entrance to the RBP ligand-binding pocket.
The major sites of synthesis of TTR are the choroid plexus in the brain and the liver, and the protein is found in plasma and in cerebrospinal fluid [25]. Where RBP is assembled with TTR and how this process occurs are not fully understood but it has been suggested that the complete ternary retinol:RBP:TTR complex is formed in hepatocytes prior to secretion into blood [26]. In addition to transporting retinol and T4, TTR displays protease activities and participates in the biology of the nervous system [27]. Notably, TTR is one of the 30 human proteins known to be associated with amyloidoses disorders, i.e. pathologies characterized by aggregation of misfolded proteins which lead to the formation of extracellular deposits and impair organ function [28].
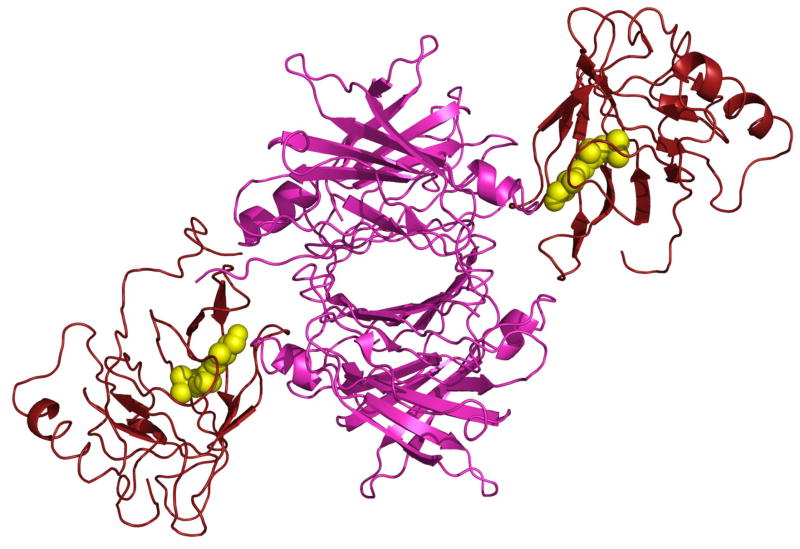
TTR is a tetrameric protein comprised of four identical subunits. In vitro, two RBP molecules can bind to the TTR tetramer, but, corresponding to the serum levels of the proteins, the retinol:RBP:TTR complex circulates in blood under normal circumstances at a 1:1:1 molar stoichiometry. The reported 3-dimensional crystal structure of the retinol:hRBP:hTTR complex [29] reveals that TTR tetramer is comprised of a dimer of dimers with the two RBPs bound to opposite dimers (Fig. 3). In the complex, the open end of the RBP β-barrel is positioned at the 2-fold dimer axes of TTR and the association is also stabilized by amino acid residues at the C-terminal of RBP (Fig. 3). Notably, association with TTR blocks the entrance to the ligand-binding pocket of RBP (Fig. 2 and Fig. 3). These observations raise the question of the mechanism that allows retinol to exit the protein prior to moving into target cells. The association of RBP with TTR displays an equilibrium dissociation constant (Kd) of 0.07 μM and critically requires the presence of the native ligand, retinol [30]. The higher stability of the RBP-TTR complex in the presence of retinol appears to emanate from participation of the hydroxyl group of retinol in the contacts with TTR [31], and from retinol-triggered conformational change in RBP that places a loop containing residues 34–37 in a position favorable for interaction with TTR [29]. Notably, RBP does not associate with TTR in the presence of either retinal or retinoic acid although these retinoids bind to RBP with affinities similar to that displayed by retinol [32]. It seems that the larger head groups of these retinoids sterically interfere with binding of RBP to its serum partner protein.
Figure 3. The three dimensional crystal structure of the retinol-RBP-TTR complex.
Human retinol-RBP-TTR [29] (PDB ID 1QAB) was generated using Pymol (http://www.pymol.org/). The TTR tetramer (magenta) is comprised of a dimer of dimers with two RBP molecules (red) bound to the opposite dimers. Interactions between RBP and TTR are mediated by residues at the entrance to the ligand binding pocket and span across the two TTR dimers.
3. STRA6
The tight interaction of retinol with RBP allows the poorly-soluble vitamin to circulate in plasma. However, target tissues for vitamin A do not take up the protein and, in order to reach the interior of cells, retinol must dissociate from RBP prior to uptake. It has long been postulated that there exists a receptor for RBP which functions to transport retinol from the protein into cells [33,34,35]. The identity of such a receptor has remained elusive until a recent report suggested that an integral plasma membrane protein, termed stimulated by retinoid acid gene 6 (STRA6), may function in this capacity [12]. It was demonstrated that STRA6 directly associates with RBP, that ectopic over-expression of STRA6 in cultured cells facilitates retinol uptake from the RBP-retinol complex, and that, conversely, reducing the expression level of STRA6 decreases retinol uptake. It was thus suggested that STRA6 is a retinol transporter that mediates the extraction of the vitamin from RBP and its transfer across plasma membranes and into target cells [12]. It was also proposed that STRA6 can function bi-directionally to both take up retinol from the circulation and to secrete the vitamin from cells [36]. Interestingly, it was reported that STRA6-mediated retinol uptake does not proceed in the absence of lecithin retinol acyl transferase (LRAT), an enzyme that metabolically traps retinol by converting it into retinylesters [36]. Hence, vitamin A uptake appears to be closely linked to its metabolism.
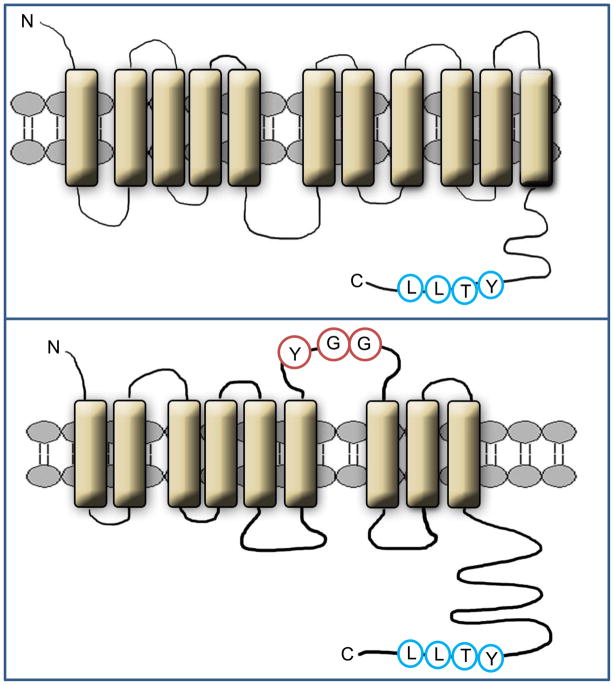
STRA6 lacks homology to any known protein. It is a largely hydrophobic protein which can be predicted by computer modeling to contain 11 trans-membrane helices, a number of loops, and a large cytosolic domain (Fig. 4, top). Alternatively, it was suggested, based on epitope tagging analysis, that the protein may be arranged in 9 trans-membrane helices [11] (Fig. 4, bottom). In the context of the latter model, it has been proposed that the interactions of STRA6 with RBP are stabilized by residues in an extracellular loop located between helix 6 and 7 [37] (Fig. 4, bottom). The details of the structure of STRA6 remain to be further elucidated. In the adult, STRA6 is expressed in blood-organ barriers, retinal pigment epithelial of the eye, brain, adipose tissue, spleen, kidney, testis, and female genital tract [38,39]. Interestingly, the expression level of STRA6 is elevated in colorectal, ovarian, and endometrium cancers, as well as in wilm’s kidney tumors and melanomas [38]. The functional significance of the increased expression of STRA6 in carcinoma cells is unknown.
Figure 4. Alternative models for the structure of STRA6.
Top: Computer model of STRA6 suggests that the protein is arranged in 11 transmembrane helices, a number of loops, and a large cytosolic domain. The phosphotyrosine motif in the cytosolic domain is highlighted. The model of STRA6 (GeneID 64220) was generated by the software http://bp.nuap.nagoya-u.ac.jp/sosui. Bottom: an alternative model in which STRA6 is arranged in 9 trans-membrane helices has been suggested [11]. It was proposed that highlighted residues in an extracellular loop located between helix 6 and 7 in this model stabilize the interactions of STRA6 with RBP [37].
Mutations in the STRA6 gene in humans lead to Matthew-Wood syndrome, a collection of defects in embryonic development resulting in malformations of multiple organ systems including severe microphthalmia, pulmonary agenesis, bilateral diaphragmatic eventration, duodenal stenosis, pancreatic malformations, and intrauterine growth retardation [39,40]. As RBP serves to deliver vitamin A to the embryo [41] and as the retinol metabolite retinoic acid plays key roles in embryonic development [42], developmental defects observed in the absence of STRA6 may reflect perturbation in retinoic acid homeostasis. It has been proposed in regard to this that such defects emanate from a failure to clear retinol from blood, resulting in nonspecific vitamin A excess in embryonic tissues [36]. Genetic analyses of families with Matthew-Wood syndrome revealed that disease-causing mutations can occur from insertion of a premature stop codon, from mutations within loops that connect the transmembrane helices, or from mutations in two residues at the C terminus of the protein [39]. Interestingly, one of the latter residues, T644, is located within a protein motif recognizable as a phosphotyrosine motif, a protein sequence often used by membrane signalling receptors to recruit downstream effectors (Fig. 4). The presence of such a motif in STRA6 and the apparent critical need for this sequence for proper function of the protein raise the intriguing possibility that STRA6 may be involved in cellular signalling, perhaps in response to RBP.
4. JAK/STAT pathways transduce extracellular signals to the nucleus
4.1 JAK/STAT signalling
In animals, from flies to humans, extracellular polypeptides such as cytokines, hormones, growth factors, and at least one adipokine, leptin, function by binding to cognate transmembrane receptors that, in turn, activate a signalling cascade mediated by the transcription factors termed Signal Transducers and Activators of Transcription (STAT) and their associated tyrosine kinases called Janus kinases (JAK). Activation of JAK/STAT pathways induced by extracellular signalling peptides and their receptors transduces extracellular signals to reprogram gene expression and thus to regulate multiple aspects of cellular behavior [43,44,45]. Members of the STAT family (STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6) harbor an SH2 domain which allows them to associate with phosphotyrosines in cell surface signalling receptors. In addition to STATs, cytokine receptors recruit JAKs (JAK1, JAK2, JAK3 and Tyk2). Binding of an extracellular ligand to its receptor results in phosphorylation and activation of a receptor-associated JAK. In turn, JAK phosphorylates a tyrosine residue in the cytosolic domain of the receptor, leading to recruitment of STAT. Subsequently, JAK catalyzes the phosphorylation of a conserved tyrosine residue near the STAT C-terminus. Activated STATs then form dimers that translocate to the nucleus, bind to DNA, and function as transcription factors. STAT dimers recognize a response element comprised of the sequence 5′-TT(N4-6)AA-3′ in regulatory region of target genes which, based on its original identification as a γ-interferon activation sequence, is usually referred to as a GAS element. STATs thus facilitate gene transcription in response to a myriad of cytokines, hormones, and growth factors (Fig. 4). STAT1 and STAT2 are closely involved in regulating immunity and inflammation and were reported to display tumor suppressive activities [46,47]. In contrast, STAT3, STAT5a, and STAT5b enhance cell cycle progression, angiogenesis, and survival, and they are considered to be oncogenes [48,49]. Target genes that mediate procarcinogenic activities of these STATS include the cell cycle regulators cyclin D1 and cyclin D3, the oncogene c-Myc, the growth factor VEGF, genes involved in migration and invasion such as MMP-2 and MMP-9, and anti-apoptotic genes including survivin, Mcl-1, and Bcl-XL [50]. In the context of the issues addressed here, STAT5 is of particular interest because it is recruited to cognate receptors by a consensus motif of the sequence YTXL [51,52], which corresponds to the YTLL sequence found at the C-terminus of STRA6. It is worth noting that, in addition to mediating cytokine signalling, STAT5 is an important component of signaling downstream of other receptors including some G-protein coupled receptors [53] and insulin and leptin receptors [54,55].
Cytokine signalling mediated by JAK/STAT pathways is “switched off” by several types of negative regulators. The phosphotyrosine phosphatases SHPs, CD45, and PTP1B/TC-PTP downregulate cytokine signalling by dephosphorylating the activated cytokine receptors, JAK, and STAT [44]. Protein Inhibitor of Activated STAT (PIAS) inhibits the DNA binding and transcriptional activity of STATs both through direct interactions and through its intrinsic SUMO E3-ligase activity [56,57].
4.2 STAT target genes involved in regulating energy homeostasis and insulin responses
The activities of JAK/STAT cascades are also potently downregulated by proteins encoded by the direct STAT target genes called Suppressors of Cytokine Signaling (SOCS, including CIS, SOCS1-SOCS7). Following their upregulation by STAT, SOCS function as components of negative feedback loops that dampen cytokine signalling [44,58,59,60]. SOCS possess a central SH2 domain, a variable N-terminal domain, and a C-terminal 40-amino-acid module called the SOCS box. These proteins inhibit JAK/STAT signalling by competing with STATs for binding to phosphotyrosines in activated receptors and by blocking the catalytic activity of JAK. SOCS can also recruit ubiquitin ligases and, consequently, proteins with which they interact, such as JAK, become ubiquitinated and degraded by the proteasome. SOCS proteins have been implicated in inhibiting the activities of multiple extracellular signalling molecules, including interleukin-6 (IL-6), leukemia inhibitory factor (LIF), granulocyte colonystimulating factor (G-CSF), IL-10, growth hormone, and the interferons IFN-β and IFN-γ.
In the same vein, SOCS are potent inhibitors of the activities of two pathways that play central roles in regulating energy homeostasis and insulin responses. Specifically, upon binding of their respective ligands, the leptin receptor (LR) and the insulin receptor (IR) activate STATs, leading to upregulation of SOCS3 which, in turn, suppresses signalling [55,61,62,63,64]. In addition to inhibiting their own activities by the SOCS3-mediated negative feedback loop, insulin and leptin actions can be suppressed in response to induction of SOCS by other cytokines. For example, induction of SOCS3 by IL-6 leads to insulin resistance [65,66,67].
Leptin functions in hypothalamic neurons where it inhibits food intake by suppressing orexigenic neuropeptides and inducing the expression anorexigenic neuropeptides. The leptin receptor LRb is also expressed in peripheral tissues including skeletal muscle, liver, adipose tissue, and pancreatic β cells. In these, leptin is involved in the metabolism of glucose and lipids, cell proliferation and differentiation, and in cross-talk with other hormonal regulators, most notably, insulin [68,69]. For example, in muscle, leptin triggers lipid oxidation thereby enhancing insulin sensitivity [70]. Induction of SOCS3 upon activation of STAT in cells that respond to insulin and/or leptin would thus suppress signalling triggered by these cytokines and would lead to increased adiposity and impaired insulin responsiveness [71,72].
Another STAT-regulated gene closely involved in lipid metabolism and energy homeostasis is the nuclear receptor PPARγ, which was shown to be a direct target for STAT5 in circulating angiogenic cells and in adipocytes [73,74]. PPARγ is a master regulator of adipocyte biology. Its expression and activation during adipocyte differentiation induce the expression of multiple proteins that promote adipogenesis. In mature adipocytes, PPARγ regulates the expression of genes involved in hallmarks of adipocyte function such as triglyceride uptake and storage. Factors that increase the expression of PPARγ, e.g. STATs, would thus promote the formation of new adipocytes and enhance lipid accumulation in adipose tissue.
5. STRA6 transduces RBP-retinol signalling to trigger a JAK/STAT cascade that regulates insulin responses and lipid homeostasis
Previous studies revealed that, in obese and insulin resistant mice, synthesis of RBP in adipose tissue is enhanced and that the protein is secreted from this tissue into blood resulting in a marked elevation in its serum levels. It was further demonstrated that administration of RBP to lean mice leads to insulin resistance, and that mice lacking RBP are protected from insulin resistance induced by a high fat diet. These observations led to the surprising conclusion that RBP functions as an adipokine that contributes to obesity-induced insulin resistance [19]. In accordance, it was reported that treatment of mice with RBP impairs insulin signaling in muscle and in adipocytes and increases PEPCK expression and glucose production in the liver [19,75]. Both in rodents and humans, a strong correlation was found between elevated serum levels of RBP and obesity as well as various obesity-associated pathologies, including inflammation, fatty liver disease and insulin resistance [76,77,78,79]. It was therefore proposed that decreasing serum RBP may comprise a novel therapeutic approach for reversing insulin resistance [19,80,81,82]. One compound that was suggested to serve in this capacity is N-(4-hydroxyphenyl)retinamide (fenretinide) whose binding to RBP prevents its association with TTR, resulting in rapid loss of the small protein in the kidney. Fenretinide is currently being tested for treatment of insulin resistance in obese humans (Medical Research Foundation, University of California, San Diego (http://clinicaltrials.gov/ct2/show/study/NCT00546455). It is worth noting however that the efficacy of fenretinide as an insulin sensitizer may be mediated by mechanisms other than lowering serum RBP levels [82]. In addition, fenretinde inhibits the visual cycle and thus diminishes dark adaptation, i.e. it causes night blindness [83,84]. Such effects are however reversible upon cessation of drug intake. Whether RBP may be a target for treatment of insulin resistance remains to be established but the observations that the protein links between obesity and insulin resistance challenge the long-held notion that the only function of this protein is to transport vitamin A in blood. These observations raise important questions regarding the molecular mechanisms and the cellular components that mediate RBP-induced suppression of insulin responses.
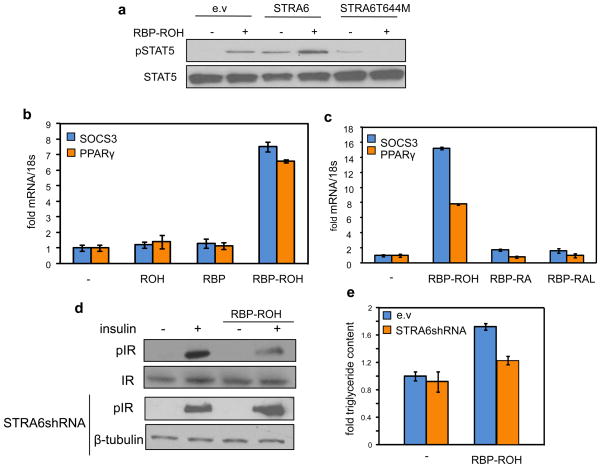
RBP is known to associate with two proteins, its binding partner in serum TTR and the retinol transporter STRA6. In considering possible mechanisms by which RBP may affect insulin signalling, it was noted that the cytosolic domain of STRA6 contains a stretch of residues that conform to a consensus phosphotyrosine motif [39,74] (Fig. 4). Phosphotyrosines are often found in surface receptors that transduce extracellular signals by activating JAK/STAT cascades. The presence of such a motif in STRA6 suggests the intriguing possibility that, in addition to serving as a vitamin A transporter, STRA6 may function as a signalling receptor which is activated by RBP. Recent studies indeed established that retinol-bound RBP (RBP-ROH) serves as an extracellular ligand that activates STRA6 which, in turn, modulates cellular responses by triggering JAK/STAT signalling [74]. In support of this notion, it was demonstrated that treatment of STRA6-expressing cells with RBP-ROH triggers phosphorylation in the phosphotyrosine motif at the cytosolic domain of STRA6, induces recruitment of JAK2 and STAT5 to STRA6, and leads to phosphorylation of STAT5 ([74], Fig. 6a). It was further shown that RBP-ROH-induced activation of STAT results in upregulation of the expression of STAT target genes (Fig. 5b). As this activity did not require de novo protein synthesis, the data indicated that it is a direct response [74]. Importantly, neither RBP nor retinol triggered JAK/STAT signalling when administered alone, and retinoic acid had no effect on this cascade either alone or when complexed with RBP (Fig. 6b, 6c). These observations establish that the RBP-ROH complex functions like classical cytokines and like another adipokine, leptin, to activate a STRA6/JAK2/STAT5 pathway. Hence, RBP-ROH regulates gene transcription in a manner that does not involve the known transcriptionally active vitamin A metabolite retinoic acid or its associated nuclear receptors. It is worth noting that ectopic expression of STRA6 variants that lack a functional SH2-binding motif, including a STRA6-T644M mutant found in Matthew-Wood patients, inhibits the ability of RBP-ROH to activate STAT ([74], Fig. 6a). These observations raise the possibility that impairment of this pathway may contribute to the development of Matthew-Wood-associated pathologies.
Figure 6. RBP-ROH activates STAT5 and induces STAT target genes to inhibit insulin signalling and enhance lipid accumulation.
(a) Treatment of HepG2 cells with RBP-ROH triggers STAT5 phosphorylation. The effect is enhanced upon ectopic expression of STRA6 and abolished in the presence of a STRA6 lacking the SH2 binding motif. (b) RBP-ROH, but neither RBP nor ROH alone, induces the expression of the STAT target genes SOCS3 and PPARγ. (c) The ability of RBP-ROH to induce STAT target genes is not recapitulated by RBP-bound retinoic acid (RA) or retinal (RAL). (d) In cultured adipocytes, RBP-ROH suppresses the ability of insulin to trigger the phosphorylation of the insulin receptor, and does so in a STRA6-dependent fashion. (e) RBP-ROH enhances lipid accumulation in cultured adipocytes in a STRA6-depndent fashion. For details see [74].
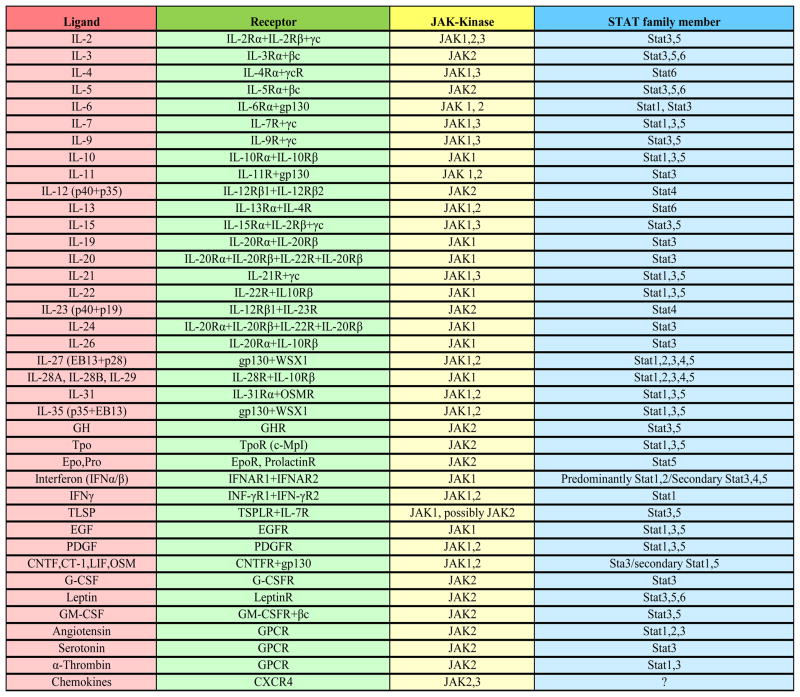
Figure 5. Extracellular signalling molecules that utilize JAK/STAT pathways.
The Table depicts known cytokines, hormones and growth factors that signal through cognate cell surface receptors to activate JAK/STAT signalling (adapted from Cell signaling, http://www.cellsignal.com/).
At least two genes whose expression is directly controlled by STATs are known to be involved in regulation of insulin responses and lipid homeostasis. One of these, SOCS3, is a potent inhibitor of signalling by cytokine receptors, including the insulin and leptin receptors [61,62,85]. The other is PPARγ, a key regulator of adipocyte differentiation and adipose lipid storage [86,87]. Activation of STAT5 by RBP-ROH in STRA6-expressing cells induces the expression of both of these genes ([74], Fig. 6b). In accordance with upregulation of SOCS3, RBP-ROH was found to suppress the activation of the insulin receptor and its ability to signal to downstream effectors in cultured adipocytes and an in vivo mouse model, and to do so in a STRA6-dependent fashion ([74], Fig. 6d). Upregulation of PPARγ upon treatment of adipocytes with RBP-ROH is accompanied by a STRA6-depndent increase in triglyceride accumulation ([74], Fig. 6e).
Taken together, these observations demonstrate that STRA6 functions as a signalling surface receptor which, upon its activation by extracellular RBP-ROH, triggers a JAK/STAT cascade to induce the expression of STAT target genes. RBP-ROH thus joins the more than 30 extracellular cytokines, hormones, and growth factors that signal through surface receptors associated with JAKs and STATs (Fig. 5). The model that emerges from these observations (Fig. 7) also suggests a mechanism through which the RBP-ROH complex is involved in regulating insulin responses and lipid homeostasis.
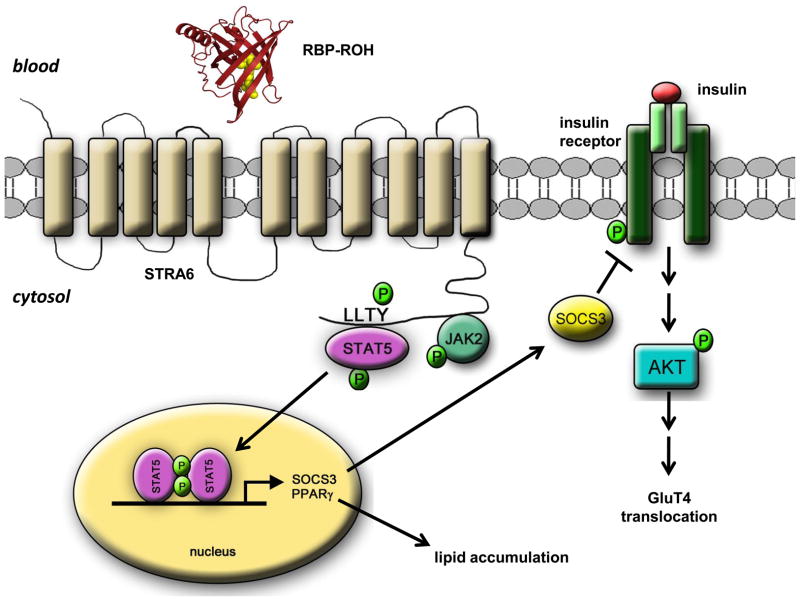
Figure 7. Model of the RBP-ROH/STRA6/JAK/STAT pathway.
Binding of RBP-ROH to the extracellular moiety of STRA6 triggers tyrosine phosphorylation in the receptor’s cytosolic domain. Phosphorylated STRA6 recruits and activates JAK2 which, in turn, phosphorylates STAT5. Activated STAT5 translocates to the nucleus to upregulate the expression of target genes, including SOCS3, which inhibits insulin signalling, and PPARγ, which enhances lipid accumulation.
6. Open Questions
The identification of the novel signalling cascade mediated by RBP-ROH, STRA6, JAK2, and STAT5 establish that STRA6 is not only a vitamin A transporter but also a surface signalling receptor. An important question that remains open is whether the two functions of the receptor are inter-related. Does signalling by STRA6 modulate STRA6-mediated retinol uptake? Conversely, is the uptake necessary for signalling?
Cytokine receptors often communicate with more than one signalling cascades. While it has been demonstrated that STRA6 activates a STAT/JAK pathway, it is possible that the receptor also functions through other cascades. Whether STRA6 transduces RBP-ROH signalling through multiple pathways remain to be clarified.
Available information demonstrates that RBP-ROH and STRA6 regulate the expression of genes involved in insulin responses and lipid homeostasis. However, the pathway must also control the expression of other genes, most likely in a tissue- and cell-specific manner. The involvement of RBP-ROH and STRA6 in other biological functions remains to be investigated. Notably in regard to this, mutation in the SH2 binding motif of STRA6 is associated with embryonic defects classified within the Matthew-Wood syndrome. It would be of great interest to understand whether and how signalling by STRA6 is involved in development.
STAT3, STAT5a, and STAT5b promote cell cycle progression, angiogenesis, and survival. The observations that the expression of STRA6 is upregulated in a number of cancers and that RBP-ROH-induced signalling by this receptor activates STAT5, suggest that the newly found cascade may be involved in cancer development. Whether this notion is correct and the exact roles that STRA6 plays in tumor initiation and growth remain to be clarified.
It has been reported that administration of RBP to mice results in upregulation of expression of hepatic PEPCK. As the liver does not express STRA6, this activity cannot be attributed to direct RBP-ROH/STRA6 signalling. Possibly, the response reflects a secondary, indirect effect resulting from systemic induction of insulin resistance by RBP. The mechanism by which RBP affects gene expression in the liver remains to be elucidated.
Finally, the structural features of STRA6 that allow this unique protein to associate with its accessory proteins and to facilitate vitamin A uptake as well as trigger signalling await additional investigations. Importantly in regard to this query, the observations that, in the circulating retinol-RBP-TTR complex, the entrance to the ligand-binding pocket of RBP is blocked by TTR raise the question of the mechanism that allows retinol to exit the protein prior to moving into target cells. Presumably, STRA6 is involved in dissociating TTR from RBP but the details of the process through which this is accomplished are unknown.
Highlights.
Holo-RBP, which transports vitamin A in blood, is a signalling molecule
STRA6 functions both as a vitamin A transporter and as a surface signalling receptor activated by holo-RBP
Activation of STRA6 by RBP-ROH triggers a JAK/STAT cascade, thereby inducing gene trascription
Some genes induced by RBP-ROH/STRA6/JAK/STAT signalling are involved in regulating insulin responses and lipid metabolism
Acknowledgments
We thank Hui Jin for important contributions to this work. Work from the authors laboratory was supported by NIH grants DK060684 and DK088669 to N.N. D.C.B. was partially supported by NIH grant DK073195T32.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCollum EV, Davis M. The necessity of certain lipins in the diet during growth. J Biol Chem. 1913;15:167–175. [Google Scholar]
- 2.Osborne TB, Mendel LB. The vitamins in green foods. J Biol Chem. 1919;37:187–200. [Google Scholar]
- 3.Noy N. Vitamin A. In: Stipanuk MH, editor. Biochemical, physiological, & molecular aspects of human nutrition. 2. Saunders Elsevier; St. Louis: 2006. [Google Scholar]
- 4.Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–954. [PubMed] [Google Scholar]
- 5.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LX. Retinoic acid receptors. Pharmacol Rev. 2006;58:712–725. doi: 10.1124/pr.58.4.4. [DOI] [PubMed] [Google Scholar]
- 6.Shaw N, Elholm M, Noy N. Retinoic acid is a high affinity selective ligand for the peroxisome proliferator-activated receptor beta/delta. J Biol Chem. 2003;278:41589–41592. doi: 10.1074/jbc.C300368200. [DOI] [PubMed] [Google Scholar]
- 7.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kane MA, Folias AE, Pingitore A, Perri M, Obrochta KM, Krois CR, Cione E, Ryu JY, Napoli JL. Identification of 9-cis-retinoic acid as a pancreas-specific autacoid that attenuates glucose-stimulated insulin secretion. Proc Natl Acad Sci U S A. 2010;107:21884–21889. doi: 10.1073/pnas.1008859107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. International Union of Pharmacology. LXIII. Retinoid X receptors. Pharmacol Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 10.Harrison EH. Mechanisms of digestion and absorption of dietary vitamin A. Annu Rev Nutr. 2005;25:87–103. doi: 10.1146/annurev.nutr.25.050304.092614. [DOI] [PubMed] [Google Scholar]
- 11.Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008;47:5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 13.Berry DC, Noy N. All-trans-retinoic acid represses obesity and insulin resistance by activating both peroxisome proliferation-activated receptor beta/delta and retinoic acid receptor. Mol Cell Biol. 2009;29:3286–3296. doi: 10.1128/MCB.01742-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry DC, Soltanian H, Noy N. Repression of cellular retinoic acid-binding protein II during adipocyte differentiation. J Biol Chem. 2010;285:15324–15332. doi: 10.1074/jbc.M110.110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YX, Lee CH, Tiep S, Yu RT, Ham J, Kang H, Evans RM. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell. 2003;113:159–170. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 17.Schwarz EJ, Reginato MJ, Shao D, Krakow SL, Lazar MA. Retinoic acid blocks adipogenesis by inhibiting C/EBPbeta-mediated transcription. Mol Cell Biol. 1997;17:1552–1561. doi: 10.1128/mcb.17.3.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neele DM, de Wit EC, Princen HM. Inhibition of apolipoprotein(a) synthesis in cynomolgus monkey hepatocytes by retinoids via involvement of the retinoic acid receptor. Biochem Pharmacol. 1999;58:263–271. doi: 10.1016/s0006-2952(99)00105-7. [DOI] [PubMed] [Google Scholar]
- 19.Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- 20.Flower DR, North AC, Sansom CE. The lipocalin protein family: structural and sequence overview. Biochim Biophys Acta. 2000;1482:9–24. doi: 10.1016/s0167-4838(00)00148-5. [DOI] [PubMed] [Google Scholar]
- 21.Salier JP, Akerstrom B, Borregaard N, Flower DR. Lipocalins in bioscience: the first family gathering. Bioessays. 2004;26:456–458. doi: 10.1002/bies.20013. [DOI] [PubMed] [Google Scholar]
- 22.Zanotti G, Berni R, Monaco HL. Crystal structure of liganded and unliganded forms of bovine plasma retinol-binding protein. J Biol Chem. 1993;268:10728–10738. [PubMed] [Google Scholar]
- 23.Newcomer ME, Jones TA, Aqvist J, Sundelin J, Eriksson U, Rask L, Peterson PA. The three-dimensional structure of retinol-binding protein. Embo J. 1984;3:1451–1454. doi: 10.1002/j.1460-2075.1984.tb01995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noy N, Xu ZJ. Thermodynamic parameters of the binding of retinol to binding proteins and to membranes. Biochemistry. 1990;29:3888–3892. doi: 10.1021/bi00468a014. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber G. The evolutionary and integrative roles of transthyretin in thyroid hormone homeostasis. J Endocrinol. 2002;175:61–73. doi: 10.1677/joe.0.1750061. [DOI] [PubMed] [Google Scholar]
- 26.Selvaraj SR, Bhatia V, Tatu U. Oxidative folding and assembly with transthyretin are sequential events in the biogenesis of retinol binding protein in the endoplasmic reticulum. Mol Biol Cell. 2008;19:5579–5592. doi: 10.1091/mbc.E08-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liz MA, Mar FM, Franquinho F, Sousa MM. Aboard transthyretin: From transport to cleavage. IUBMB Life. 2010;62:429–435. doi: 10.1002/iub.340. [DOI] [PubMed] [Google Scholar]
- 28.Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cell Mol Life Sci. 2009;66:3095–3101. doi: 10.1007/s00018-009-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naylor HM, Newcomer ME. The structure of human retinol-binding protein (RBP) with its carrier protein transthyretin reveals an interaction with the carboxy terminus of RBP. Biochemistry. 1999;38:2647–2653. doi: 10.1021/bi982291i. [DOI] [PubMed] [Google Scholar]
- 30.Noy N, Xu ZJ. Interactions of retinol with binding proteins: implications for the mechanism of uptake by cells. Biochemistry. 1990;29:3878–3883. doi: 10.1021/bi00468a012. [DOI] [PubMed] [Google Scholar]
- 31.Monaco HL, Rizzi M, Coda A. Structure of a complex of two plasma proteins: transthyretin and retinol-binding protein. Science. 1995;268:1039–1041. doi: 10.1126/science.7754382. [DOI] [PubMed] [Google Scholar]
- 32.Noy N, Slosberg E, Scarlata S. Interactions of retinol with binding proteins: studies with retinol-binding protein and with transthyretin. Biochemistry. 1992;31:11118–11124. doi: 10.1021/bi00160a023. [DOI] [PubMed] [Google Scholar]
- 33.Sivaprasadarao A, Boudjelal M, Findlay JB. Solubilization and purification of the retinol-binding protein receptor from human placental membranes. Biochem J. 1994;302:245–251. doi: 10.1042/bj3020245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundaram M, Sivaprasadarao A, DeSousa MM, Findlay JB. The transfer of retinol from serum retinol-binding protein to cellular retinol-binding protein is mediated by a membrane receptor. J Biol Chem. 1998;273:3336–3342. doi: 10.1074/jbc.273.6.3336. [DOI] [PubMed] [Google Scholar]
- 35.Flannery JG, O’Day W, Pfeffer BA, Horowitz J, Bok D. Uptake, processing and release of retinoids by cultured human retinal pigment epithelium. Exp Eye Res. 1990;51:717–728. doi: 10.1016/0014-4835(90)90057-2. [DOI] [PubMed] [Google Scholar]
- 36.Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J Biol Chem. 2008;283:15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szeto W, Jiang W, Tice DA, Rubinfeld B, Hollingshead PG, Fong SE, Dugger DL, Pham T, Yansura DG, Wong TA, Grimaldi JC, Corpuz RT, Singh JS, Frantz GD, Devaux B, Crowley CW, Schwall RH, Eberhard DA, Rastelli L, Polakis P, Pennica D. Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 2001;61:4197–4205. [PubMed] [Google Scholar]
- 39.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quadro L, Hamberger L, Gottesman ME, Wang F, Colantuoni V, Blaner WS, Mendelsohn CL. Pathways of vitamin A delivery to the embryo: insights from a new tunable model of embryonic vitamin A deficiency. Endocrinology. 2005;146:4479–4490. doi: 10.1210/en.2005-0158. [DOI] [PubMed] [Google Scholar]
- 42.Mark M, Ghyselinck NB, Chambon P. Function of retinoic acid receptors during embryonic development. Nucl Recept Signal. 2009;7:e002. doi: 10.1621/nrs.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aaronson DS, Horvath CM. A road map for those who don’t know JAK-STAT. Science. 2002;296:1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 44.O’Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002;109(Suppl):S121–131. doi: 10.1016/s0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- 45.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 46.Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D. Gene modulation and immunoregulatory roles of interferon gamma. Cytokine. 2010;50:1–14. doi: 10.1016/j.cyto.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 47.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr Med Chem. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 48.Quesnelle KM, Boehm AL, Grandis JR. STAT-mediated EGFR signaling in cancer. J Cell Biochem. 2007;102:311–319. doi: 10.1002/jcb.21475. [DOI] [PubMed] [Google Scholar]
- 49.He G, Karin M. NF-kappaB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–241. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- 51.Klingmuller U, Bergelson S, Hsiao JG, Lodish HF. Multiple tyrosine residues in the cytosolic domain of the erythropoietin receptor promote activation of STAT5. Proc Natl Acad Sci U S A. 1996;93:8324–8328. doi: 10.1073/pnas.93.16.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McWhinney CD, Dostal D, Baker K. Angiotensin II activates Stat5 through Jak2 kinase in cardiac myocytes. J Mol Cell Cardiol. 1998;30:751–761. doi: 10.1006/jmcc.1998.0639. [DOI] [PubMed] [Google Scholar]
- 53.Guillet-Deniau I, Burnol AF, Girard J. Identification and localization of a skeletal muscle secrotonin 5-HT2A receptor coupled to the Jak/STAT pathway. J Biol Chem. 1997;272:14825–14829. doi: 10.1074/jbc.272.23.14825. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Sadowski HB, Kohanski RA, Wang LH. Stat5 is a physiological substrate of the insulin receptor. Proc Natl Acad Sci U S A. 1997;94:2295–2300. doi: 10.1073/pnas.94.6.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong F, Ren J. Fitness or fatness--the debate continues for the role of leptin in obesity-associated heart dysfunction. Curr Diabetes Rev. 2007;3:159–164. doi: 10.2174/157339907781368959. [DOI] [PubMed] [Google Scholar]
- 56.Liu B, Yang Y, Chernishof V, Loo RR, Jang H, Tahk S, Yang R, Mink S, Shultz D, Bellone CJ, Loo JA, Shuai K. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 2007;129:903–914. doi: 10.1016/j.cell.2007.03.056. [DOI] [PubMed] [Google Scholar]
- 57.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169–1176. doi: 10.1038/ni1012. [DOI] [PubMed] [Google Scholar]
- 59.Starr R, Willson TA, Viney EM, Murray LJ, Rayner JR, Jenkins BJ, Gonda TJ, Alexander WS, Metcalf D, Nicola NA, Hilton DJ. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 60.Croker BA, Kiu H, Nicholson SE. SOCS regulation of the JAK/STAT signalling pathway. Semin Cell Dev Biol. 2008;19:414–422. doi: 10.1016/j.semcdb.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emanuelli B, Peraldi P, Filloux C, Sawka-Verhelle D, Hilton D, Van Obberghen E. SOCS-3 is an insulin-induced negative regulator of insulin signaling. J Biol Chem. 2000;275:15985–15991. doi: 10.1074/jbc.275.21.15985. [DOI] [PubMed] [Google Scholar]
- 62.Emanuelli B, Peraldi P, Filloux C, Chavey C, Freidinger K, Hilton DJ, Hotamisligil GS, Van Obberghen E. SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-alpha in the adipose tissue of obese mice. J Biol Chem. 2001;276:47944–47949. doi: 10.1074/jbc.M104602200. [DOI] [PubMed] [Google Scholar]
- 63.Krebs DL, Hilton DJ. A new role for SOCS in insulin action. Suppressor of cytokine signaling. Sci STKE 2003. 2003:PE6. doi: 10.1126/stke.2003.169.pe6. [DOI] [PubMed] [Google Scholar]
- 64.Pirola L, Johnston AM, Van Obberghen E. Modulation of insulin action. Diabetologia. 2004;47:170–184. doi: 10.1007/s00125-003-1313-3. [DOI] [PubMed] [Google Scholar]
- 65.Senn JJ, Klover PJ, Nowak IA, Zimmers TA, Koniaris LG, Furlanetto RW, Mooney RA. Suppressor of cytokine signaling-3 (SOCS-3), a potential mediator of interleukin-6-dependent insulin resistance in hepatocytes. J Biol Chem. 2003;278:13740–13746. doi: 10.1074/jbc.M210689200. [DOI] [PubMed] [Google Scholar]
- 66.Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G. Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab. 2001;280:E745–751. doi: 10.1152/ajpendo.2001.280.5.E745. [DOI] [PubMed] [Google Scholar]
- 67.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 68.Friedman JM. The function of leptin in nutrition, weight, and physiology. Nutr Rev. 2002;60:S1-14. doi: 10.1301/002966402320634878. discussion S68-84, 85–17. [DOI] [PubMed] [Google Scholar]
- 69.Sweeney G. Leptin signalling. Cell Signal. 2002;14:655–663. doi: 10.1016/s0898-6568(02)00006-2. [DOI] [PubMed] [Google Scholar]
- 70.Muoio DM, Dohm GL, Fiedorek FT, Jr, Tapscott EB, Coleman RA. Leptin directly alters lipid partitioning in skeletal muscle. Diabetes. 1997;46:1360–1363. doi: 10.2337/diab.46.8.1360. [DOI] [PubMed] [Google Scholar]
- 71.Shimabukuro M, Koyama K, Chen G, Wang MY, Trieu F, Lee Y, Newgard CB, Unger RH. Direct antidiabetic effect of leptin through triglyceride depletion of tissues. Proc Natl Acad Sci U S A. 1997;94:4637–4641. doi: 10.1073/pnas.94.9.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Unger RH. Leptin physiology: a second look. Regul Pept. 2000;92:87–95. doi: 10.1016/s0167-0115(00)00154-3. [DOI] [PubMed] [Google Scholar]
- 73.Dentelli P, Trombetta A, Togliatto G, Zeoli A, Rosso A, Uberti B, Orso F, Taverna D, Pegoraro L, Brizzi MF. Formation of STAT5/PPARgamma transcriptional complex modulates angiogenic cell bioavailability in diabetes. Arterioscler Thromb Vasc Biol. 2009;29:114–120. doi: 10.1161/ATVBAHA.108.172247. [DOI] [PubMed] [Google Scholar]
- 74.Berry DC, Jin H, Majumdar A, Noy N. Signalling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci U S A. 2011;108:4340–4345. doi: 10.1073/pnas.1011115108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ost A, Danielsson A, Liden M, Eriksson U, Nystrom FH, Stralfors P. Retinol-binding protein-4 attenuates insulin-induced phosphorylation of IRS1 and ERK1/2 in primary human adipocytes. FASEB J. 2007;21:3696–3704. doi: 10.1096/fj.07-8173com. [DOI] [PubMed] [Google Scholar]
- 76.Kovacs P, Geyer M, Berndt J, Kloting N, Graham TE, Bottcher Y, Enigk B, Tonjes A, Schleinitz D, Schon MR, Kahn BB, Bluher M, Stumvoll M. Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes. 2007;56:3095–3100. doi: 10.2337/db06-1647. [DOI] [PubMed] [Google Scholar]
- 77.Graham TE, Yang Q, Bluher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, Kahn BB. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med. 2006;354:2552–2563. doi: 10.1056/NEJMoa054862. [DOI] [PubMed] [Google Scholar]
- 78.Tonjes A, Bluher M, Stumvoll M. Retinol-binding protein 4 and new adipocytokines in nonalcoholic fatty liver disease. Curr Pharm Des. 2010;16:1921–1928. doi: 10.2174/138161210791208938. [DOI] [PubMed] [Google Scholar]
- 79.Galic S, Oakhill JS, Steinberg GR. Adipose tissue as an endocrine organ. Mol Cell Endocrinol. 2010;316:129–139. doi: 10.1016/j.mce.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 80.Motani A, Wang Z, Conn M, Siegler K, Zhang Y, Liu Q, Johnstone S, Xu H, Thibault S, Wang Y, Fan P, Connors R, Le H, Xu G, Walker N, Shan B, Coward P. Identification and characterization of a non-retinoid ligand for retinol-binding protein 4 which lowers serum retinol-binding protein 4 levels in vivo. J Biol Chem. 2009;284:7673–7680. doi: 10.1074/jbc.M809654200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johansson H, Gandini S, Guerrieri-Gonzaga A, Iodice S, Ruscica M, Bonanni B, Gulisano M, Magni P, Formelli F, Decensi A. Effect of fenretinide and low-dose tamoxifen on insulin sensitivity in premenopausal women at high risk for breast cancer. Cancer Res. 2008;68:9512–9518. doi: 10.1158/0008-5472.CAN-08-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Preitner F, Mody N, Graham TE, Peroni OD, Kahn BB. Long-term Fenretinide treatment prevents high-fat diet-induced obesity, insulin resistance, and hepatic steatosis. Am J Physiol Endocrinol Metab. 2009;297:E1420–1429. doi: 10.1152/ajpendo.00362.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baglietto L, Torrisi R, Arena G, Tosetti F, Gonzaga AG, Pasquetti W, Robertson C, Decensi A. Ocular effects of fenretinide, a vitamin A analog, in a chemoprevention trial of bladder cancer. Cancer Detect Prev. 2000;24:369–375. [PubMed] [Google Scholar]
- 84.Maeda A, Maeda T, Golczak M, Imanishi Y, Leahy P, Kubota R, Palczewski K. Effects of potent inhibitors of the retinoid cycle on visual function and photoreceptor protection from light damage in mice. Mol Pharmacol. 2006;70:1220–1229. doi: 10.1124/mol.106.026823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tups A. Physiological models of leptin resistance. J Neuroendocrinol. 2009;21:961–971. doi: 10.1111/j.1365-2826.2009.01916.x. [DOI] [PubMed] [Google Scholar]
- 86.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 87.Lefterova MI, Lazar MA. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20:107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]