Summary
In plants, exogenous transgene transcribing inverted-repeat (exo-IR) sequences produces double-stranded RNAs that are processed by DCL4. The generated 21-nt siRNAs function as mobile signals to trigger non-cell-autonomous silencing of target endogene in neighboring 10-15 cells. The potential involvement of nuclear silencing pathway components in signal spreading or sensing in target cells is not clear. Here, we demonstrate that the exo-IR silencer (exo-Pdsi) is negatively auto-regulated through methylation spreading, which acts in cis to reinforce self-silencing of the silencer. Mutations affecting nuclear proteins DRD1 and Pol V (NRPE1 or NRPD2) relieved exo-Pdsi self-silencing resulting in higher levels of Pdsi transcripts which increased non-cell-autonomous silencing of endo-PDS. Our results suggest that in an experimental silencing pathway, methylation spreading on a silencer transgene may not have a direct endogenous plant counterpart when protein-encoding gene is the target. DRD1-Pol V-dependent de novo methylation, by acting in cis to reinforce self-silencing of exo-IR may play a role in restraining inappropriate silencing of active protein-coding genes in plants.
Keywords: exo-IR, non-cell-autonomous silencing, PTGS, TGS, methylation spreading
Introduction
RNA silencing is a nucleotide sequence-specific process that includes RNA degradation, DNA methylation, heterochromatin formation and protein translation inhibition in eukaryotic genomes (for review see Baulcombe, 2004, 2005; Meister and Tuschl, 2004; Matzke and Birchler, 2005; Dunoyer and Voinnet, 2008; Brodersen et al. 2008; Voinnet, 2005, 2008; Chinnusamy and Zhu, 2009; Matzke et al. 2004, 2007, 2009; Heo and Kim, 2009). In plants, post-transcriptional gene silencing (PTGS) is a process that is induced by siRNAs and miRNAs, and it achieves specificity through RNA-RNA sequence recognition and base pairing. The proteins involved include an RNA-dependent RNA polymerase (RDR6), Dicer-like ribonucleases (DCL1,2,4) that generate siRNA and miRNA, and SGS3 which selectively binds 5′ overhang-containing dsRNA, an effector protein of the Argonaute family (AGO1,7) (Bartel, 2004; Gasciolli et al. 2005; Xie et al. 2005; Peragine et al. 2004; Dunoyer et al. 2005; Yoshikawa et al. 2005; Allen et al. 2005; Vazquez et al. 2004; Himber et al. 2003; Schwach et al. 2005; Mourrain et al. 2000; Elmayan et al. 2009; Fukunaga and Doudna 2009). In a different pathway that takes place in the nucleus siRNAs related to promoter sequences direct the silencing machinery to block transcription from the homologous promoters (transcriptional gene silencing, TGS).
Nuclear silencing has a great impact on the expression and constitution of the Arabidopsis genome (Zilberman and Henikoff, 2007, 2005). In plants, the nuclear silencing pathway comprises of RNA-directed DNA methylation (RdDM), which requires both 24-nt siRNAs and long noncoding RNA transcripts for de novo DNA methylation (Wierzbicki et al. 2008; Daxinger et al. 2008). The 24-nt siRNAs are generated in a pathway involving a plant-specific DNA-directed RNA polymerase (Pol IV, formally Pol IVa), RDR2 and DCL3 (Kanno et al. 2005; Herr et al. 2005; Onodera et al. 2005; Zhang et al. 2007; Xie et al. 2004; Matzke, 2005), whereas the noncoding RNAs are produced by another plant-specific DNA-directed RNA polymerase (Pol V, formally Pol IVb) that is structurally and functionally distinct from Pol IV (Wierzbicki et al. 2008; Haag et al. 2009; Pikaard et al. 2008; Chinnusamy and Zhu, 2009). The 24-nt siRNAs are loaded into AGO4 and/or AGO6 (Zilberman et al. 2004; Zheng et al. 2007; He et al. 2009) and then recruited to target genomic regions, by base-pairing with Pol V-dependent transcripts from the target loci (Wierzbicki et al. 2009). In this way, a 24-nt siRNAs/AGO4/Pol V-dependent RNA transcripts effector complex is formed that directs DNA methyltransferase DRM2-dependent de novo methylation of target genomic sequences (Cao and Jacobsen 2002; Li et al. 2006; Preuss et al. 2008; Mosher et al. 2008; Lahmy et al. 2009; Matzke et al. 2009). This process requires another plant-specific SNF2-like chromatin remodeling protein (DRD1) (Kanno et al. 2004, 2005; Chan et al. 2006; Huettel et al. 2006, 2007) and a hinge domain-containing protein (DMS3) (Kanno et al. 2008; Matzke et al. 2009). Recently, Pol II has been shown to play a central role in coordinating Pol IV and Pol V in siRNA-mediated DNA and histone methylation (Zheng et al. 2009). Pol II recruits AGO4 and Pol V to the silenced loci through both physical interactions with AGO4 and scaffold transcripts that are generated adjacent to the silenced loci. Pol II also acts in a feed-forward loop to promote siRNA accumulation from these loci by recruiting Pol IV to chromatin (Zheng et al. 2009). A new regulator of RdDM, RDM1, has also been shown to associate and co-localize with Pol II, AGO4 and DRM2 in the nucleus (Gao et al. 2010). RDM1 can bind single-stranded methylated DNA and is required for Pol IV- and Pol V-dependent 24-nt siRNA accumulation and RdDM (Gao et al. 2010).
A PTGS phenomenon termed silencing transitivity (Voinnet, 2008) requires RDR6 for 21-nt secondary siRNA biogenesis, transcription of the target gene and bidirectional spreading of methylation within transcribed regions (Vaistij et al. 2002; Van et al. 2003; Parizotto et al. 2004; Eamens et al. 2008). Daxinger et al. (2009) have also reported a stepwise pathway for biogenesis of 24-nt secondary siRNAs. They used a two-component transgene TGS silencing system (Kanno et al. 2008): a transgene target locus (T) and a transcribed inverted repeat (IR) silencer locus (S). They observed unidirectional spreading of DNA methylation downstream of the target during TGS. An absence of secondary siRNAs and a strong reduction DNA methylation spreading in the rdr2, nrpd1 (NRPD1 is the largest subunit of Pol IV) and dcl3 mutants revealed that TGS transitivity requires the Pol IV-RDR2-DCL3 pathway (Daxinger et al. 2009).
In addition to transitive silencing along the target DNA sequence, a non-cell autonomous process allows RNA silencing to spread from cell-to-cell and throughout the whole plant (Palauqui et al. 1997; Voinnet and Baulcombe, 1997; Voinnet et al. 2000; Klahre et al. 2002; Himber et al. 2003; Voinnet 2005; Dunoyer et al. 2005, 2007; Smith et al. 2007).
Double-stranded siRNAs were recently reported to be mobile silencing signals (Molnar et al. 2010; Dunoyer et al. 2010a; 2010b). Dunoyer et al. found that siRNAs produced by at least one of the endogenous IR loci are also functional as mobile silencing signals and mediate both PTGS and RdDM of the IR target. They suggested that endogenous IR loci are genetically indistinguishable from transgene constructs commonly used to study RNA silencing pathways in plants (Dunoyer et al. 2010a). However, it remains unclear whether this process is equivalent to the silencing of endogenous coding sequences. Why IR silencer transgenes expressed in plants are not always able to efficiently silence related endogenous coding genes? How do plants control or prevent unwanted silencing of active protein-coding genes? The biological significance of PTGS and RdDM acting on an exogenous IR silencer in induction/regulation of endogenous gene silencing is not known.
Here, we used a previously reported transgenic chemical-inducible PDSi system to trigger the transcription of inverted repeat exo-IR dsRNA (exo-Pdsi) by a local chemical treatment to induce systemic silencing of endogenous PDS (endo-PDS) (Guo et al. 2003). We found that non-cell autonomous silencing involves the production of secondary siRNAs, and DNA methylation/spreading from the transcribed region to the promoter region occurred only in the exo-Pdsi silencer. Neither secondary siRNAs nor DNA methylation/spreading were found in the endo-PDS silencing target. By crossing PDSi plants with previously characterized mutants affected in PTGS and TGS, we found that the PTGS-related components, DCL4 and SGS3, but not RDR6, are required for exo-Pdsi to induce non-cell autonomous silencing. Among the TGS mutant progeny, unexpectively, three progenies from the crosses, drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi (mutations in the first and second largest subunits NRPE1 and NRPD2 of PolV), exhibited enhanced silencing on upper systemic leaves rather than impairing PDS silencing. The enhanced silencing in the drd1-pol v mutants was associated with an increase of 21-nt siRNAs but decreased methylation and relaxation of transcriptional silencing of the exo-Pdsi silencer. These results led us to conclude that in our PDSi inducible silencing system, DRD1-Pol V mediate spreading of DNA methylation along the transgenic exo-Pdsi sequence in a negative feedback loop to maintain exo-Pdsi self-silencing. As a consequence less 21-nt PDS-related siRNAs are produced which limit the spreading of PTGS of the endo-PDS gene.
Results
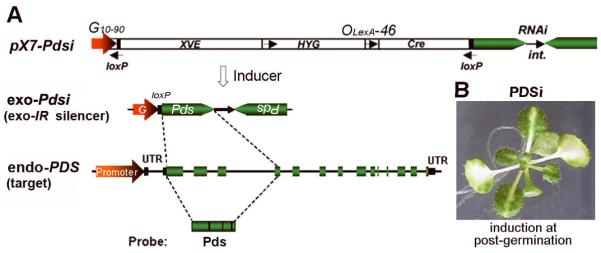
We have previously generated transgenic Arabidopsis thaliana PDSi plants, which carry a chemical-inducible Cre/loxP (CLX) recombination system. Chemical induction of transgenic plants can trigger the expression of an intron-containing inverted-repeat Pds transcript (Pdsi) leading to silencing of the endogenous PDS gene (endo-PDS). Silenced plants display a visual photobleaching phenotype in lower treated leaves, which can spread systemically to upper untreated leaves (Guo et al. 2003). Figure 1A shows structural features of the primary transgene (pX7-Pdsi) and the exo-IR (exo-Pdsi) silencer after CLX recombination, as well as the endogenous PDS genomic locus. Here, we investigated the effects of several PTGS and TGS mutants on triggering silencing of the endo-PDS (target) by the exo-Pdsi (silencer).
Figure 1. Schematic diagrams of the inducible RNAi construct and genomic structure of the PDS gene and phenotype of induced PDS silencing.
(A) A schematic diagram showing structural features of the inducible pX7-RNAi construct (see Guo et al. 2003 for detail), a chemical-inducible Cre/loxP (CLX) recombination system (Zuo and Chua 2000), to trigger the expression of an intron-containing inverted-repeat RNAi (IR). G10-90, a strong, synthetic, constitutive promoter._exo-Pdsi (exo-IR silencer): the reconstituted transcription unit derived from Cre/loxP-mediated DNA recombination after inducer treatment. endo-PDS (target): Genomic structure of the endogenous PDS gene. UTR, untranslated region of mRNA. Green boxes are exons of PDS separated by introns. Pds, fragment corresponding to the first 4 exons of PDS used for IR construction and probe for hybridization. (B) Phenotype of inducible PDSi at the post-germination stage. One-week-old seedlings were treated with inducer for one week and picture was taken at 10 days after removing inducer.
PDS silencing phenotypes in mutant backgrounds
A stable and homozygous A. thaliana inducible PDSi line (line 2) (Guo et al. 2003) showed a reproducible and uniform photobleaching phenotype in chemically-treated local leaves. However, upon induction at the post-germination stage the photobleaching was limited to areas near the veins in upper untreated leaves (Fig. 1B). This line was crossed with previously characterized homozygous mutants affected in RNA silencing, including PTGS pathway mutants rdr6-11, sgs3-11 and dcl4-2, and TGS pathway mutants drd1-6, nrpd2(drd2-4), nrpe1(drd3-7), nrpd1(nrpd1a-4)-, rdr2-1, dcl3-1, ago4-1 and drm1.2 (drm1/drm2 double mutant). The resulting F1 progeny were self-fertilized to generate a segregating F2 population. Homozygous F3 seeds were germinated on inductive medium. Except for dcl4/PDSi, which displayed very weak silencing, all of the other mutants showed a uniform photobleaching phenotype in cotyledons (similar to PDSi line-2 seedlings) at 14-day post-induction (dpi) (Supplemental Fig. S1). These results indicated that the chemical-inducible CLX recombination silencing system worked well upon induction in the crossed progeny. The faint silencing in dcl4/PDSi is consistent with the notion that DCL4 is the most important plant DCL for dsRNA cleavage to initiate PTGS (Gasciolli et al. 2005; Bouche et al. 2006; Deleris et al. 2006; Fusaro et al. 2006).
At the post-germination stage (see Material and Methods), induced silencing of the endo-PDS was not affected in rdr6/PDSi seedlings, consistent with the report that RDR6 is not required for IR-PTGS (Himber et al. 2003; Dunoyer et al. 2005). PDS silencing was reduced in sgs3/PDSi, which displayed a patched bleaching phenotype, and was greatly reduced in dcl4/PDSi seedlings (Fig. 2A). These results are consistent with the involvement of the PTGS pathway. Of the TGS mutants, nrpd1/PDSi and dcl3/PDSi displayed reduced, while ago4/PDSi, drm1.2/PDSi and rdr2/PDSi displayed unchanged bleaching phenotype compared to that of PDSi control seedlings (Fig. 2B). Surprisingly, three TGS mutant progeny from the crosses, drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi (nrpd2 and nrpe1 are mutations in the first and second largest subunits NRPE1 and NRPD2 of Pol V), exhibited enhanced silencing with extensive photobleaching on upper leaves (Fig. 2B)
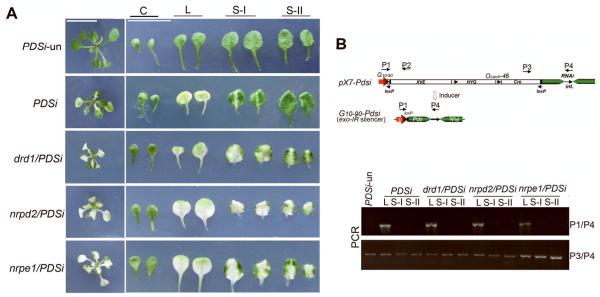
Figure 2. Phenotypes of PDS silencing induced at the post-germination stage in different silencing mutant genotypes.
(A) PDS silencing in PTGS mutant progeny from the crosses, dcl4/PDSi, rdr6/PDSi and sgs3/PDSi. (B) PDS silencing in TGS mutant progeny from the crosses, nrpd1/PDSi, dcl3-1/PDSi, ago4-1/PDSi, drm1.2/PDSi, rdr2-1/PDSi, drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi. Photographs were taken at two weeks after removing inducer. Scale bar = 1cm. Each genotype was labeled on top of picture. PDSi-un, without inducer treatment.
Further analysis of enhanced silencing in drd1-pol v mutants
As the extensive bleaching phenotype in the drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi lines was unexpected, we examined the CLX recombination frequency of primary transgene pX7-Pdsi in these mutants. Fig 3A shows induced silencing of the endo-PDS at the post-germination stage and silencing phenotype of leaves of different age. Silenced leaves were collected from the drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi seedlings for DNA extraction and silenced leaves and leaves of similar age from PDSi, as well as from PDSi-un were used as controls. PCR amplification with designed primers (Fig. 3B) for specific detection of pX7-Pdsi CLX recombination was carried out. Consistent with the previous report (Guo et al., 2003), we found that CLX recombination occurred only in chemically-treated local leaves of PDSi. No recombination was detected in systemic silenced sample, un-silenced leaves and in PDSi-un (Fig. 3B). Together with the PCR amplification with primers P3/P4 (Fig. 3B) showing incomplete pX7-Pdsi recombination in local silenced bleaching leaves, our data support the view that the induction of PDSi silencing involves long-distance movement in addition to cell-to-cell movement of the silencing signal derived from local PDSi silencing (Guo et al., 2003). Similar PCR profiles comparable to those for PDSi local and systemic leaves were obtained for the three tested mutants (Fig. 3B), indicating that the recombination frequency was not affected by the drd1-pol v mutations. These result revealed that the enhanced silencing in these mutants must result from effects of the mutations on the spreading of silencing rather than increased recombination of the primary pX7-Pdsi transgene. This finding suggested that DRD1-Pol V-dependent processes negatively regulate spreading of endo-PDS silencing in WT Arabidopsis.
Figure 3. Analysis of DNA recombination of the primary transgene.
(A) PDS silencing phenotype in PDSi, drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi. Pairs of cotyledon (C), first true leaves (local silenced leaves, L) and second (S-I) and third true leaves (S-II) were shown. Photographs were taken at 10 days after removing inducer. Scale bar =1cm. (B) PCR analysis of of leaves samples of different age described in (A). P1-4 denotes primers used for PCR amplification.
High accumulation of the exo-Pdsi silencer dsRNA in local silenced leaves of drd1-pol v mutants
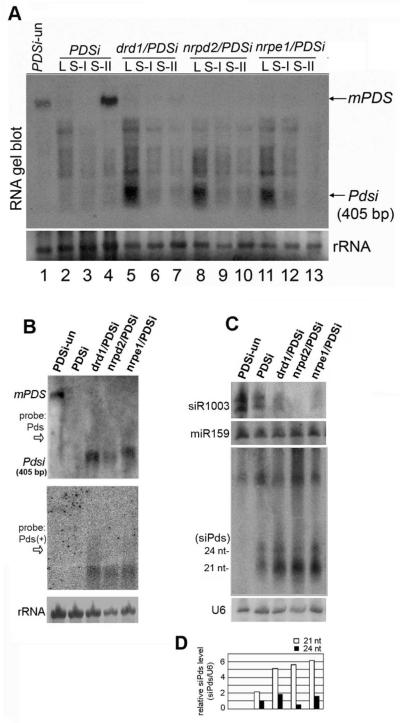
Next, we examined RNA levels of endo-PDS and exo-Pdsi. As expected, RNA gel blot analysis shows that the endo-PDS mRNA was silenced in local and systemic silenced samples of PDSi but was readily detected in systemic un-silenced leaves of PDSi and PDSi-un (Fig. 4A). Endo-PDS mRNA was not detected in any silenced leaf of the drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi seedlings with enhanced silencing genotype (Fig. 4A). Interestingly, exo-Pdsi RNA significantly accumulated in local silenced leaves of the 3 mutants but not in systemic silenced leaves of the same mutants nor in silenced PDSi samples (Fig. 4A). These results show that the recombination–generated exo-Pdsi RNA in local leaves was not self-silenced in these mutants affected in the DRD1-Pol V pathway (Fig. 3A, 4A). Accumulation of both plus and minus strands of exo-Pdsi dsRNA in the local leaves was further confirmed (Fig. 4B). These results agreed with the PCR data (Fig. 3B) and further supported that finding that pX7-Pdsi CLX recombination occurred only in chemically-treated local leaves and the enhancement of silencing spreading to systemic leaves in the drd1-pol v mutants. These results suggest that the exo-Pdsi silencer is normally self-silenced through a negative feedback loop that is dependent on the DRD1-Pol V pathway.
Figure 4. Analysis of endogenous PDS mRNA, Pdsi dsRNA and PDS-related siRNA accumulation.
(A) RNA gel blot analysis of leaves samples of different age with 32P-labeled Pds DNA probe shown in Figure 1A. Sample L, S-I, S-II were described in Figure 3. mPDS, endo-PDS mRNA. Pdsi, dsRNA of Pdsi transcript. Total RNAs loaded in each lane (15 μg) were visualized by methylene blue staining. (B) RNA gel blot analysis of local silenced leaves was carried out as described in (A). The membrane shown in the upper panel was striped and rehybridized with 32P-labeled sense Pds RNA probe for detection of antisense Pds transcript (bottom). (C) RNA gel blot analysis of Pds-derived siRNAs (siPds), and endogenous siR1003 and miR159 accumulation with 32P-labeled sense Pds RNA probe, or oligodeoxynucleotide probes specific for siR1003 and miR159. U6 RNA hybridization is shown as loading control. (D) Quantification of siPds relative to U6 RNA. The value of 24-nt siPds of PDSi was arbitrarily designated as 1. RNA extracted from each genotype was labeled on top. PDSi-un as described in Figure 2B.
Increased 21-nt siRNA levels in drd1-pol v mutants
We then analyzed the correlated small RNAs in local silenced leaves. Endogenous miR-159 was detected at a similar level in samples of all genotypes, and siR-1003 was absent or lower in the 3 tested mutant progeny from the crosses, further verifying the TGS mutant alleles (Fig. 4C). Using a Pdsi DNA fragment as a probe, we detected 21nt and 24nt siRNAs corresponding to the Pdsi region (designated as Pdsi-related, siPds) in all inducer-treated seedlings except in the control PDSi-un seedlings (Fig. 4C). The finding of two classes of siRNAs suggested that multiple DCL proteins can act in this silencing system. Notably, in seedlings of drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi showing enhanced endo-PDS silencing phenotype, the level of 21-nt siRNAs was much higher than in PDSi (Fig. 4C, D).
We used RNA transcripts corresponding to the middle and 5′UTR regions of the endo-PDS gene (sketch in Supplemental Fig. S2A, referred to as “pDs” and “pds-U”, respectively) to probe transitive secondary siRNAs related to endo-PDS RNA to compare with primary siPds (Fig. 4C and Supplemental Fig. S2B). Neither class of siRNAs could be detected by RNA gel blot analysis (Fig. S2C).
To analyze in detail the production of transitive siRNA, siRNA deep sequencing was performed with the silenced PDSi line and the three enhanced silencing lines. PDSi-un plants, without inducer treatment, were used as a control. Fig 4C shows that miR159 accumulated to a similar level in all the samples. Therefore, total siRNA reads obtained by deep sequencing from each genotype library were normalized to its own miR159 value, which was normalized to the PDSi-un miR159 counts (Fig. 5).
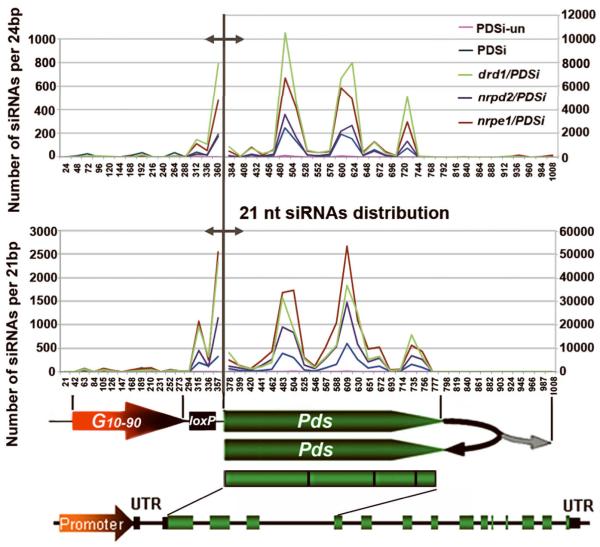
Figure 5. Distribution and comparison of 21- and 24-nt siPds obtained by deep sequencing in different genotypes.
Total miR159 reads obtained by deep sequencing from each genotype library was first normalized to PDSi-un’s miR159 counts to set the miR159 value for each genotype library. Total counts of siPds from each genotype were then normalized to its own miR159 value. Regions along the G10-90-loxP-Pdsi (1030 bp) with sequence identity to siPds was divided into 42 sections of 24 bp (upper panel) or 48 sections of 21 bp (lower panel) for counting number of siPds per 24bp or 21bp. Different Y axes represent number of siPds matched to the promoter region (left of the gray demarcation line) or the transcribed region (right of the gray demarcation line) related to position indicated in X axes. Structures of G10-90-loxP-Pdsi and the endogenous PDS gene are shown below to indicate the source of siRNA.
In all tested samples except PDSi-un, we found 24-nt siRNAs matching the exo-Pdsi sequences upstream of the Pdsi region, G10-90-loxP (Fig. 5). These results suggest that transitive silencing along the exo-Pdsi sequences indeed involved the Pol IV-dependent 24-nt synthesis nuclear pathway. Twenty one-nt siRNAs matching the G10-90-loxP region were also obtained in all samples except PDSi-un (Fig. 5) suggesting the involvement of PTGS in the degradation of Pol IV-related transcripts. The amounts of 21-nt siRNAs were correlated with the degree of the bleaching phenotype (Fig. 5 and Fig. 2).
In the transcribed Pdsi region, PDS-related siRNAs presumbaly include those derived from exo-Pdsi transcripts as well as those from endo-PDS RNA since both exo-Pdsi and endo-PDS RNA sequences are identical. Two types of siRNAs were obtained and their levels were higher in the 3 enhanced silencing mutants than in silenced PDSi (Fig. 5), consistent with their enhanced silencing phenotype and the siRNA hybridization analysis (Fig. 2 and 4).
Alignment analysis suggested that the 24-nt siRNAs were mainly derived from the exo-Pdsi silencer: when endo-PDS genomic DNA was aligned with the siRNAs, none of the siRNAs matched the PDS intronic regions. This result provides evidence that DRD1-Pol V-dependent processes do not target the endo-PDS DNA.
When endo-PDS RNA was aligned with PDS-related siRNAs, all the siRNAs were homologous to the Pdsi targeted region. This is consistent with the siRNAs hybridization analysis in which PDS-related siRNAs were not detected in regions beyond the Pdsi target region when pds-U or pDs were used as probes (supplemental Fig. S2), indicating that transitive silencing does not take place along the endo-PDS transcript. Taken together, our results indicate that the nuclear silencing DRD1-Pol V pathway presumably acts on the exo-Pdsi silencer DNA sequence but not on the endo-PDS DNA sequence.
Methylation status of the endogenous PDS DNA in wild type and the drd1-pol v mutants
The siRNA analysis results suggest that transitive silencing along the endo-PDS RNA did not occur upon silencing triggered by exo-Pdsi. To examine whether the induction of endo-PDS silencing required only PTGS for RNA degradation and did not involve DRD1-Pol V-mediated de novo DNA methylation, we investigated the methylation state of the endo-PDS target in local silenced leaves. Methylation-sensitive restriction-PCR (Chop-PCR) was first used to interrogate the endo-PDS DNA. PCR amplification corresponding to the 5′ termini of endo-PDS DNA (including the first two exons and the first intron) showed no obvious difference between PDSi and the 3 mutant genotypes upon inducer treatment. Almost no methylation was detected by digestion with either methylation-sensitive enzyme (AluI and HaeIII) (Supplemental Fig. S3A). Neither was there any difference between PDSi with and without (PDSi-un) inducer treatment (Supplemental Fig. S3A). These results are consistent with the comparable methylation state detected by sequencing of bisulphite-treated genomic DNA (Supplementary Fig. S3B).
DNA gel blot was used to further analyze endo-PDS genomic DNA including the promoter and the coding region. Digestion with methylation-sensitive enzyme confirmed the similar no/low methylation state of both the promoter and the coding region upon silencing induction in PDSi as well as the mutants (Supplemental Fig. S3C). Figure 6A shows results obtained from digestions using two of the methylation-sensitive enzymes. In the control PDSi-un sample, 3 specific bands (2823bp, 2229bp and 694bp) were detected (Fig. 6A, lane 1 and 5), indicating that a portion of DNA in CGATCG site in the promoter was methylated under native conditions. Upon induction, increased methylation at the CGATCG site in the promoter was observed in the silenced PDSi sample (Fig. 6A cf. lane 5 and 6). However, an equivalent pattern of hybridization bands was observed in each mutant genotype with or without induction of silencing (Fig. 6A). These results show that methylation changes at this site occurred in the mutants regardless of whether silencing was induced. Therefore, methylation at this site must result from properties of the various TGS mutant alleles. The trivial change of methylation in the PDS endogenous promoter in the mutant plants did not affect expression as the PDS mRNA was detected in the mutants at high levels similar to those in PDSi-un and the WT control (Fig. S3D). This result further suggests that increased methylation at the CGATCG site in PDSi after silencing induction may not be required for PDS silencing. Taken together, our results suggest that induction of endo-PDS silencing does not result in increased endo-PDS DNA methylation in the regions targeted by the exo-Pdsi nor does it significantly change the DNA methylation level of the native PDS promoter, even though the Pdsi target region is located at the 5′ terminus downstream of the promoter.
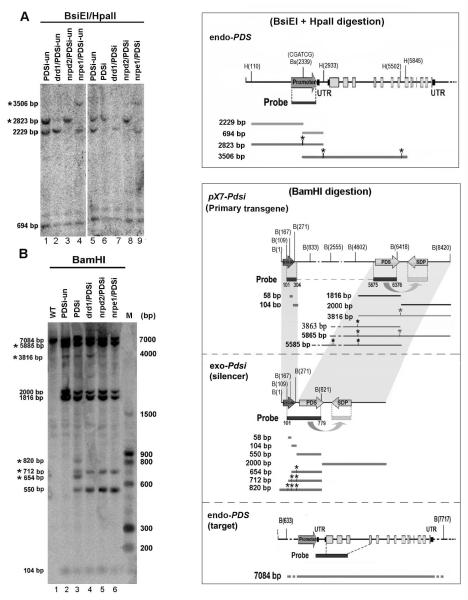
Figure 6. Analysis of DNA methylation of the endo PDS gene and the exo-Pdsi silencer.
(A) Detailed analysis of methylation in endo-PDS promoter using two methylation-sensitive enzymes BsiEI and HpaII sites. DNA was isolated from leaf samples of each genotype with or without inducer treatment and digested with BsiEI and HpaII. DNA gel blots were analyzed with a 32P-labeled promoter DNA probe. PDSi, with inducer; PDSi-un, without inducer. BsiEI and HpaII sites along the endo-PDS genome are shown in the upper right rectangle. Sizes of DNA fragments resulting from complete and methylation-resistant digestion (labeled with *) that could be detected with the promoter probe are shown in the box on the right panel. These DNA fragments are indicated on the left of the gel. (B) Detailed analysis of methylation in primary pX7-Pdsi, exo-Pdsi and endo-PDS using methylation-sensitive enzymes BamHI sites. DNA samples and blotting were as described in (A), except digestion with BamHI. BamHI sites along each construct are shown in the box on the right. DNA probe covered both the G10-90 promoter and the Pds fragment is indicated under each construct. Sizes of DNA fragments resulting from complete and methylation-resistant digestion (*) that could be detected with the DNA probe are shown in the box. These DNA fragment are also marked on the left of gel. The positions of labeled marker DNA fragments are shown on the right.
DNA methylation status in the exo-Pdsi silencer in wild type and drd1-pol v mutants
Next, we examined the methylation state of the exo-Pdsi DNA, G10-90-loxP-Pdsi, which was generated from CLX recombination in local silenced leaves. Chop-PCR results provided suggestive evidence that exo-Pdsi DNA was generally methylated in silenced PDSi plants (Supplemental Fig. S3A). G10-90-loxP-Pdsi DNA of the enhanced silencing mutants drd1/PDSi, nrpd2/PDSi, nrpe1/PDSi showed little or no methylation (Supplemental Fig. S3A). DNA gel blot analysis showed that G10-90-loxP-Pdsi DNA was methylated in the silenced PDSi sample. BamHI-digestion resulted in four additional hybridization signals (*820bp, *712bp, *654bp and 550bp) (Fig. 6B, lane 3) compared to the BamHI-digested PDSi-un sample (Fig. 6B, lane 2). These results indicate that G10-90-loxP-Pdsi DNA (mostly in the promoter) was indeed methylated upon induction of its transcribed RNA silencing. Together with the detection of Pdsi upstream G10-90-loxP-derived transitive 24-nt siRNA (Fig. 5), and low level of 24-nt siRNA in the reduced silencing line nrpd1/PDSi (mutation in the first largest subunits NRPD1 of Pol IV) (Supplemental Fig. S2B), our results indicate that the induction of silencing by exo-Pdsi in PDSi involves a stepwise pathway for methylation spreading that presumably extends Pol IV-related nascent RNA synthesis from the methylated Pdsi region to the upstream G10-90-loxP region. Consequently, 24-nt siRNAs derived from Pol IV-related nascent RNAs mediate spreading of DNA methylation to regions upstream of Pdsi. By contrast, in the samples of drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi, the methylated bands of *820 and *654bp were not detected but the 550bp band increased in intensity, indicating low methylation in the G10-90-loxP promoter region of exo-Pdsi (Fig. 6B), although Pol IV-dependent G10-90-loxP-derived transitive 24-nt siRNAs were detected in these mutants (Fig. 5). This was consistent with the requirement of DRD1-Pol V in the RdDM process.
The DNA gel blot results also uncovered little or no methylation in the primary pX7-Pdsi transgene G10-90-loxP promoter or the far downstream Pdsi DNA regions because three BamHI-digested bands (2000bp, 1816bp, 104bp) and a weak methylated band (*3816bp) were detected in PDSi-un (Fig. 6B, lane 2). In silenced PDSi, except for the four bands (*820bp, *712bp, *654bp and 550bp) derived from the recombined exo-Pdsi, the digestion profiles of the primary pX7-Pdsi transgene DNA were comparable to those in PDSi-un (Fig. 6B). These observations suggest that DNA methylation did not target the primary pX7-Pdsi transgene DNA which was inactive in Pdsi transcription, in agreement with the transcription-dependence of transitive silencing (Vaistij et al. 2002). The DNA hybridization result also indicates the involvement of non-cell-autonomous silencing when endo-PDS silencing was induced post-germination. Consistent with the PCR result (Fig. 3B), in all inducer-treated samples, intensive hybridization signals from the primary transgene DNA (5585bp, 2000bp, 1816bp), and weaker signals derived from the CLX recombined DNA (Fig. 6B) were detected indicating that CLX recombination occurred only in a small portion of the cells upon strict inducer treatment. The extension of the bleached phenotype to systemic leaves in the 3 enhanced silencing lines suggests that non-cell-autonomous silencing was enhanced relative to the silenced PDSi sample, in which only vein-limited-bleaching was observed in systemic leaves (Fig. 3A).
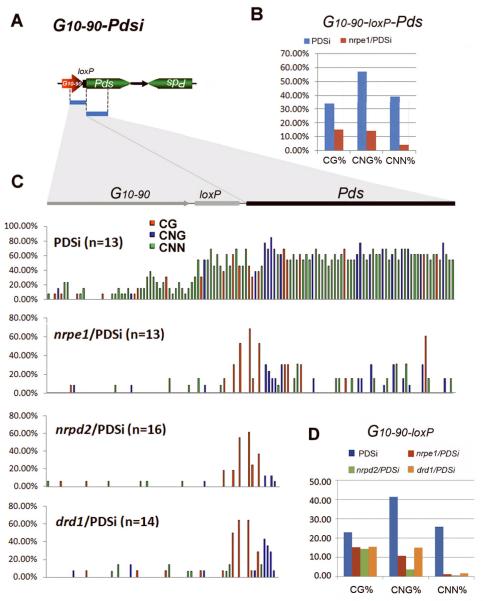
Detailed analysis by bisulfite sequencing confirmed the Chop-PCR and DNA gel blot results. In the PDSi-un sample, almost no methylation was detected in the primary pX7-Pdsi transgenic promoter and low methylation was detected in the Pdsi DNA region (Supplemental Fig. S4A). In the silenced PDSi sample, the exo-Pdsi DNA was highly methylated not only in the Pdsi but also the promoter G10-90 including the loxP region, with 33% CG, 57% CNG and 39% asymmetric CNN methylation (Fig. 7B, C and Supplemental Fig. S4B). Additionally, loxP was more intensively methylated adjacent to the transcribed region than at the distant promoter sequence (Fig. 7C and Supplemental Fig. S4B). Low levels of DNA methylation were detected in mutants with the enhanced silencing (Fig. 7B-D and Supplemental Fig. S4B).
Figure 7. Analysis of DNA methylation on the exo-Pdsi silencer by sequencing of bisulfite-treated DNA.
(A) Bisulfite sequencing was performed for the G10-90-loxP-Pds region in the exo-Pdsi silencer. (B, C, D) Percentage of CG, CNG and CNN methylation of G10-90-loxP-Pds or G10-90-loxP region in the indicated genotypes. The total number of clones analyzed is in parenthesis, and the original data are shown in Supplementary Figure S4B.
Taken together, our results indicate that the exo-Pdsi silencer DNA itself is a target for methylation during induced Pdsi silencing, and the methylation extends 5′ to its promoter in a process that requires DRD1 and Pol V (NRPD2 and NRPE1). No transitive silencing along the endo-PDS RNA occurred upon silencing triggered by exo-Pdsi; the induction of endo-PDS silencing required only PTGS for RNA degradation.
Discussion
Here, we have shown that triggering silencing of the endogenous PDS gene (target) at post-germination by dsPds generated from the exo-Pdsi (silencer) involves both PTGS and TGS. The nuclear silencing pathway, which includes DRD1-Pol V-dependent RdDM, targets the exo-Pdsi DNA at the transcribed region and extends to the promoter region. By contrast, there is no effect on the genomic DNA of endo-PDS target. There were some methylation on GG and CNG in the Pds DNA sequence in primary transgene (pX7-Pdsi) before CLX recombination (Supplemental Fig. S4), compared to the same region of genomic DNA of endo-PDS target sequence (Supplemental Fig. S3B). The methylation changes in the primary transgene Pds DNA, probably caused by manipulation of the transgene construction process, may recruit DRD1-Pol V-dependent RdDM machinery to transcribe exo-Pdsi DNA and induce silencing. DRD1-Pol V-mediated methylation spreading along a transgenic DNA but not to its homologous endogenous coding sequence may represent a surveillance system in plants. Therefore, DRD1-Pol V-dependent RdDM required to reinforce self-silencing of exo-Pdsi may play a role to prevent inappropriate silencing of the endo-PDS transcript in WT plants.
Differential effects of nuclear silencing components on signal spreading and signal sensing
The RdDM-related mutations, drd1, nrpd2 and nrpe1 greatly reduced methylation at the exo-Pdsi transgene (Fig. 7 and supplemental Fig. S4B) resulting in relief of TGS of exo-Pdsi (Fig. 4B) that generates siRNA production (Fig. 4C and 5). These exo-Pdsi-related siRNAs function as mobile silencing signals (Molnar et al. 2010; Dunoyer et al. 2010a; 2010b). In drd1, nrpd2 and nrpe1 mutant backgrounds, there is increased levels and spreading of siRNAs between cells thereby targeting endo-PDS RNA for PTGS at a distance and resulting in enhanced silencing (Fig. 2B). Enhanced silencing was observed in mutations affecting proteins involved in the RdDM process (DRD1-Pol V (NRPE1 or NRPD2)). This suggests that RdDM proteins through binding siRNAs and cis-methylating the exo-Pdsi, establish a negative feedback loop to restrain signal spreading between cells and/or perturb signal sensing in recipient cells. Reducing methylation of exo-Pdsi DNA may not be the exclusive mechanism to relieve self-silencing of exo-Pdsi. Histone modifications that accompany DRD1-Pol V-dependent RdDM may also be involved in the exo-Pdsi self-silencing process. Further investigation of RdDM proteins involved in histone modifications will be helpful to address this issue.
Reducing photobleaching phenotype in nrpd1 and dcl3 mutants suggests that mutations affecting NRPD1 and DCL3 can reduce non-cell-autonomous silencing (Fig. 2B) without greatly affecting induction of local silencing (Fig. S1B). This observation suggests that NRPD1 and DCL3 may be individually necessary to detect the signal in recipient cells and trigger endo-PDS mRNA degradation. We noted that even when seeds were germinated on inductive medium (Fig. S1), only two cotyledons of the dcl3/PDSi mutant seedlings showed induced bleaching phenotype with some reduction compared to that of PDSi, whereas the first true leaves displayed splendid green phenotype as un-treated PDSi-un (Fig. S1). We confirmed incomplete recombination of the pX-Pdsi transgene in the silenced cotyledons of the PDSi and dcl3/PDSi plants (data not shown), indicating the involvement of non-cell-autonomous silencing in cotyledons even though silencing was induced at the seed-germination stage. This may further support the view that DCL3 is necessary to detect the signal in recipient cells and trigger endo-PDS silencing. There was no alteration of silencing of SUC:SUL (transgenic Arabidopsis carrying an exo-SULi silencer) in the dcl3 mutant (Dunoyer et al. 2007); however, the spread of silencing of in SUC:PDS (another similar exo-PDSi system) is enhanced by a deficiency of DCL3 (Smith et al. 2007). In the SUC:SUL system, there was no spreading of SUL silencing in nrpd1 or rdr2 mutants (Dunoyer et al. 2007). Both NRPD1 and RDR2 have been suggested to be necessary for physical trafficking of siRNA through plasmodesmata or for signal detection in recipient cells (Dunoyer at al., 2007). Note that in our inducible PDSi system, induction of silencing at the post-germination stage involved long-distance movement in addition to cell-to-cell movement of the silencing signal (Fig. 3 and 4). The induction of silencing at a distance may have different requirements for nucleolar-associated proteins. The observation of unchanged bleaching phenotype in the rdr2/PDSi similar to that of PDSi seedlings (Fig. 2B) may favor the view of weak non-cell-autonomous silencing in nrpd1 and dcl3 background. Pol IV-RDR2-DCL3 are the 3 nucleolar-associated proteins required to amplify the 24-nt siRNA trigger (Matzke et al. 2009), which mediates the spreading of methylation during TGS (Kanno et al. 2008; Daxinger et al. 2009). Pol IV may transcribe the methylated DNA template producing an aberrant RNA for RDR2 to generate dsRNA that is subsequently processed to 24-nt siRNAs by DCL3 (Matzke et al. 2009). Pol IV may also transcribe dsRNA in the amplification cycle (Vaucheret 2005; Matzke et al. 2009). This may explain no alteration of the non-cell-autonomous silencing phenotype in rdr2/PDSi compared to PDSi silencing induced at the post-germination stage. Pol IV may compensate for the RDR2 deficiency to amplify dsRNAs as substrates for DCL3 to generate 24-nt siRNAs. Therefore, the secondary siRNA-generating machinery mediated by Pol IV-RDR2-DCL3 may play roles in both translocation of the signal and signal detection in recipient cells. Future studies should test the effects of point mutants of nrpd1 and dcl3.
Endogenous PDS coding sequences not prone to methylation spreading
Previous work have reported that exo-IR derived siRNAs define target sequences for DRD1-Pol V-dependent RdDM and methylation spreading in a two-component transgene TGS silencing system, in which both the silencer and the target are exogenous transgenes (Kanno et al. 2008). This result shows that nuclear RdDM can also take place along a target gene when it is of transgenic origin. Recently, 24-nt siRNAs derived from an endogenous IR locus have been shown to be able to trigger both PTGS and RdDM of the endogenous IR target (Dunoyer et al. 2010a). The existence of highly abundant siRNAs for these endogenous IR loci in Arabidopsis indicates that these endogenous IR loci are substantially targeted by PTGS and RdDM (Dunoyer et al. 2010a). However, this RdDM acts on endogenous IR loci and targets genes of transgenic origin (Kanno et al. 2008) and exo-IR silencers (in this study) but has no significant effect on the endo-PDS coding sequence. Similarly, when the endogenous rbcS gene was silenced by virus-induced gene silencing (VIGS) no methylation of the rbcS DNA was detected (Jones et al. 1999). This important difference suggests that endogenous coding sequences, as opposed to IRs and transgenic DNA, are not prone to methylation and methylation spreading. We note that no PDS-related siRNAs matching the endo-PDS sequence beyond the exo-Pdsi targeted region were detected by small RNA deep sequencing nor RNA gel blot analysis (Supplemental Fig. S2C). This finding, along with the previous report of a lack of spreading of RDR6-dependent silencing using VIGS of endo-PDS (Vaistij et al. 2002), supports the view that spreading of PTGS along endogenous RNA target does not occur if the related gene does not support nuclear RdDM. It seems that the inability to relay silencing is not unique to the PDS gene. The endogenous Nia (nitrate reductase) and Nii (nitrite reductase) gene sequences per se can not be targeted for systemic silencing in grafting experiments (Palauqui et al. 1997). Induction of non-cell-autonomous silencing of these two genes was only observed when they were presented as transgenes.
The stepwise pathway for RdDM spreading acts in a negative feedback loop
The nuclear silencing pathway acts in cis on the exo-IR transgene to greatly influence silencing of the endogenous target gene, even when the latter sequence is not prone to DNA methylation and subjected to PTGS. In spite of its requirement for different components, the involvement of nucleolar-associated proteins in the nuclear silencing pathway to trigger non-cell autonomous silencing of endogenous gene is firmly established. These proteins perturb either the movement of the silencing signal or its perception by recipient cells. Results from induced endo-PDS silencing at the post-germination stage revealed that the stepwise pathway for methylation spreading is required to reinforce self-silencing of exo-IR in a negative feedback loop. Consequently, triggering silencing of endogenous coding RNA by mere siRNA-guided degradation without recruiting the secondary siRNA-generating machinery to relay PTGS amplification would result in a gradual attenuation and eventual cessation of non-cell autonomous silencing. This explains the vein-limited photobleached phenotype in PDSi. Therefore, the stepwise pathway for RdDM spreading acting in cis to reinforce self-silencing of an exo-IR silencer clearly plays an important role in preventing undesired silencing of native protein-coding genes. Our finding may explain a general phenomenon of low and diverse efficiencies in inducing silencing of endogenous coding genes even when using inverted repeat transgenes as a silencer.
Materials and methods
Plant materials and growth conditions
The inducible PDSi line 2 in the Columbia (Col-0) ecotype background was described previously (Guo et al. 2003). rdr6-11, sgs3-11 and dcl4-2 were obtained from R. Scott Poethig. drd1-6, drd2-4 (nrpd2), drd3-7(nrpe1) mutants were a gift from Marjori Matzke. ago4-1 and drm1.2 were kindly provided by Steven E Jacobsen and Xiaofeng Cao, respectively. nrpd1a-4 (SALK_083051), dcl3-1 (SALK_005512) and rdr2-1(SAIL_1277) mutants were obtained from T-DNA insertion collection (Alonso et al. 2003. http://signal.salk.edu/tdna_protocols.html. Sessions et al. 2002. http://www.Arabidopsis.org/abrc/pCSA110.pdf). Arabidopsis plants were grown on MS medium containing 3% (W/V) sucrose and 0.8% (W/V) agar or grown in soil at 22 _C with a 16/8 h light/dark cycle.
Genetic crossing , screening of homologous lines and phenotype assessment
rdr6-11, sgs3-11 and dcl4-2, drd1-6, nrpd2(drd2-4), nrpe1(drd3-7), nrpd1(nrpd1a-4)-, rdr2-1, dcl3-1, ago4-1 and drm1.2 lines were crossed to the PDSi line 2 to generate mutant progeny expressing an pX7-Pdsi transgene. F2 seeds were sowed on MS medium containing 20 mg/L hygromycin B. Two-week-old hygromycin-resistant seedlings were transferred to soil and leaves were collected for genotyping to screen for homologous mutant lines. The primers used for genotyping mutants are shown in Supplemental Table S1. Seeds of homologous mutant lines were used for induction of PDS silencing. To induce silencing at the germination-stage, seeds were sowed in inductive medium containing 2umol/L 17β-estradiol. To induce silencing at the post-germination stage, one-week seedlings were transferred to inductive medium for one week and then returned to normal MS medium.
RNA and siRNA analysis
Total RNA was extracted from 4-week-old seedlings of each indicated genotype with or without inducer treatment using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Fifteen ug of total RNA was loaded for high-molecular-weight RNA blots which were probed with [α-32P]dCTP-labeled Pds cDNA using the the Rediprime™ II Random primer Labeling system (Amersham, Buckinghamshire, UK). For low molecular-weight RNA blots, 50ug of total RNA was loaded. [α-32P]UTP labeled transcript or [α-32P] ATP-labeled specific oligonucleotide sequences were used as probes.
Large scale Arabidopsis small RNA sequencing and analysis
Large-scale siRNA sequencing was performed by Beijing Genomics Institute (BGI, Beijing, China) using total RNAs of PDSi-un, PDSi, drd1/PDSi, nrpd2/PDSi and nrpe1/PDSi. Twenty ug of total RNA sample was used for small RNA deep sequencing. After removing adaptor/acceptor sequences from the raw reads obtained from deep sequencing, the remaining small RNA sequences were mapped to the endo-PDS and G10-90-Pdsi genome using Perl scripts. More than 1.0 million reads were obtained for each sample. Data analysis was performed using Microsoft Office Excel 2007.
DNA methylation assay
For methylation-sensitive restriction-PCR (Chop-PCR), genomic DNA (500 ng) was digested with the methylation-sensitive restriction enzyme HaeIII or AluI for 4 hours, before being used for PCR amplification. For gene-specific primers, see Supplemental Table S1. PCR conditions were 5 min at 94°C followed by 35 cycles of 30 sec at 94°C, 30 sec at 60°C, and 30 sec at 72°C. For the DNA gel blot hybridization assay, 5 ug of genomic DNA was digested with the methylation-sensitive enzymes as indicated, separated on 1.0% agarose gel and then transferred onto Hybond Nt (Amersham, Buckinghamshire, UK) membrane. 32P dCTP-labeled probes were used for DNA gel blot hybridization to determine their DNA methylation status. For bisulfite sequencing, 2 ug of genomic DNA was used for bisulfite treatment with the EpiTect Bisulfite Kit (QIAGEN N.V. Netherlands). The collected DNA was used for amplification. The PCR product was cloned into the pGM-T vector (Tiangen, China), and at least 10 individual clones were sequenced for each sample. Images representing methylation status of individual sequences were generated by CyMATE (Hetzl et al. 2007).
Supplementary Material
Acknowledgments
We thank R. Scott Poethig for rdr6-11, sgs3-11 and dcl4-2 mutant seeds, Marjori Matzke for drd1-6, drd2-4 (nrpd2), drd3-7(nrpe1) and nrpd1a-4(nrpd1) mutant seeds, Steven E Jacobsen and Xiaofeng Cao for ago4-1 and drm1.2 seeds, and the ABRC fordcl3-1 (SALK_005512) and rdr2-1(SAIL_1277) mutants. This research was supported by grants from the National Science Foundation of China (Grants 90919010 and 31030009) to H-S. G. N-H. C. was supported by NIH grant GM44640.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing. Trends Biochem Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Bouche N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Cao X, Jacobsen SE. Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Zhang X, Shah G, Chien JS, Jacobsen SE. RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PLoS Genet. 2006;2:e83. doi: 10.1371/journal.pgen.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu JK. RNA-directed DNA methylation and demethylation in plants. Sci China C Life Sci. 2009;52:331–343. doi: 10.1007/s11427-009-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Kanno T, Bucher E, van WJ, Naumann U, Matzke AJ, Matzke M. A stepwise pathway for biogenesis of 24-nt secondary siRNAs and spreading of DNA methylation. EMBO J. 2009;28:48–57. doi: 10.1038/emboj.2008.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Kanno T, Matzke M. Pol V transcribes to silence. Cell. 2008;135:592–594. doi: 10.1016/j.cell.2008.10.027. [DOI] [PubMed] [Google Scholar]
- Deleris A, Gallego-Bartolome J, Bao J, Kasschau KD, Carrington JC, Voinnet O. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010a;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Small RNA Duplexes Function as Mobile Silencing Signals Between Plant Cells. Science. 2010b;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- Dunoyer P, Voinnet O. Mixing and matching: the essence of plant systemic silencing? Trends Genet. 2008;24:151–154. doi: 10.1016/j.tig.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Eamens A, Vaistij FE, Jones L. NRPD1a and NRPD1b are required to maintain post-transcriptional RNA silencing and RNA-directed DNA methylation in Arabidopsis. Plant J. 2008;55:596–606. doi: 10.1111/j.1365-313X.2008.03525.x. [DOI] [PubMed] [Google Scholar]
- Elmayan T, Adenot X, Gissot L, Lauressergues D, Gy I, Vaucheret H. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276:835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro AF, Matthew L, Smith NA, Curtin SJ, Dedic-Hagan J, Ellacott GA, Watson JM, Wang MB, Brosnan C, Carroll BJ, Waterhouse PM. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Liu HL, Daxinger L, Pontes O, He X, Qian W, Lin H, Xie M, Lorkovic ZJ, Zhang S, Miki D, Zhan X, Pontier D, Lagrange T, Jin H, Matzke AJ, Matzke M, Pikaard CS, Zhu JK. An RNA polymerase II- and AGO4-associated protein acts in RNA-directed DNA methylation. Nature. 2010;465:106–109. doi: 10.1038/nature09025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Guo HS, Fei JF, Xie Q, Chua NH. A chemical-regulated inducible RNAi system in plants. Plant J. 2003;34:383–392. doi: 10.1046/j.1365-313x.2003.01723.x. [DOI] [PubMed] [Google Scholar]
- Haag JR, Pontes O, Pikaard CS. Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS One. 2009;4:e4110. doi: 10.1371/journal.pone.0004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He XJ, Hsu YF, Pontes O, Zhu J, Lu J, Bressan RA, Pikaard C, Wang CS, Zhu JK. NRPD4, a protein related to the RPB4 subunit of RNA polymerase II, is a component of RNA polymerases IV and V and is required for RNA-directed DNA methylation. Genes Dev. 2009;23:318–330. doi: 10.1101/gad.1765209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hetzl J, Foerster AM, Raidl G, Mittelsten SO. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJ, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huettel B, Kanno T, Daxinger L, Bucher E, van WJ, Matzke AJ, Matzke M. RNA-directed DNA methylation mediated by DRD1 and Pol IVb: a versatile pathway for transcriptional gene silencing in plants. Biochim Biophys Acta. 2007;1769:358–374. doi: 10.1016/j.bbaexp.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Jones L, Hamilton AJ, Voinnet O, Thomas CL, Maule AJ, Baulcombe DC. RNA-DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell. 1999;11:2291–2301. doi: 10.1105/tpc.11.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Aufsatz W, Jaligot E, Mette MF, Matzke M, Matzke AJ. A SNF2-like protein facilitates dynamic control of DNA methylation. EMBO Rep. 2005;6:649–655. doi: 10.1038/sj.embor.7400446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno T, Bucher E, Daxinger L, Huettel B, Bohmdorfer G, Gregor W, Kreil DP, Matzke M, Matzke AJ. A structural-maintenance-of-chromosomes hinge domain-containing protein is required for RNA-directed DNA methylation. Nat Genet. 2008;40:670–675. doi: 10.1038/ng.119. [DOI] [PubMed] [Google Scholar]
- Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJ. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Klahre U, Crete P, Leuenberger SA, Iglesias VA, Meins FJ. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc Natl Acad Sci U S A. 2002;99:11981–11986. doi: 10.1073/pnas.182204199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmy S, Pontier D, Cavel E, Vega D, El-Shami M, Kanno T, Lagrange T. PolV(PolIVb) function in RNA-directed DNA methylation requires the conserved active site and an additional plant-specific subunit. Proc Natl Acad Sci U S A. 2009;106:941–946. doi: 10.1073/pnas.0810310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE. An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell. 2006;126:93–106. doi: 10.1016/j.cell.2006.05.032. [DOI] [PubMed] [Google Scholar]
- Lister R, O MR, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Rev Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ. Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta. 2004;1677:129–141. doi: 10.1016/j.bbaexp.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–376. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJ. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small Silencing RNAs in Plants Are Mobile and Direct Epigenetic Modification in Recipient Cells. Science. 2010 doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proc Natl Acad Sci U S A. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Ream T, Wierzbicki AT. Roles of RNA polymerase IV in gene silencing. Trends Plant Sci. 2008;13:390–397. doi: 10.1016/j.tplants.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Costa-Nunes P, Vithayathil P, Pikaard CS. RNA Polymerase V Functions in Arabidopsis Interphase Heterochromatin Organization Independently of the 24-nt siRNA-Directed DNA Methylation Pathway. Mol Plant. 2009;2:700–710. doi: 10.1093/mp/ssp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss SB, Costa-Nunes P, Tucker S, Pontes O, Lawrence RJ, Mosher R, Kasschau KD, Carrington JC, Baulcombe DC, Viegas W, Pikaard CS. Multimegabase silencing in nucleolar dominance involves siRNA-directed DNA methylation and specific methylcytosine-binding proteins. Mol Cell. 2008;32:673–684. doi: 10.1016/j.molcel.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle IR, Pontes O, Melnyk CW, Smith LM, Baulcombe DC. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010;24:986–991. doi: 10.1101/gad.579910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, Clarke JD, Cotton D, Bullis D, Snell J, Miguel T, Hutchison D, Kimmerly B, Mitzel T, Katagiri F, Glazebrook J, Law M, Goff SA. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Baulcombe DC. Dissection of silencing signal movement in Arabidopsis. Plant Signal Behav. 2007;2:501–502. doi: 10.4161/psb.2.6.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van HH, Bleys A, Depicker A. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 2003;131:245–253. doi: 10.1104/pp.009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. RNA polymerase IV and transcriptional silencing. Nat Genet. 2005;37:659–660. doi: 10.1038/ng0705-659. [DOI] [PubMed] [Google Scholar]
- Vaughn MW, Tanurdzic M, Lippman Z, Jiang H, Carrasquillo R, Rabinowicz PD, Dedhia N, McCombie WR, Agier N, Bulski A, Colot V, Doerge RW, Martienssen RA. Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 2007;5:e174. doi: 10.1371/journal.pbio.0050174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Non-cell autonomous RNA silencing. FEBS Lett. 2005;579:5858–5871. doi: 10.1016/j.febslet.2005.09.039. [DOI] [PubMed] [Google Scholar]
- Voinnet O. Post-transcriptional RNA silencing in plant-microbe interactions: a touch of robustness and versatility. Curr Opin Plant Biol. 2008;11:464–470. doi: 10.1016/j.pbi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Lederer C, Baulcombe DC. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard CS. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, Pikaard CS. RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet. 2009;41:630–634. doi: 10.1038/ng.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Wang Z, Li S, Yu B, Liu JY, Chen X. Intergenic transcription by RNA Polymerase II coordinates Pol IV and Pol V in siRNA-directed transcriptional gene silencing in Arabidopsis. Genes Dev. 2009 doi: 10.1101/gad.1868009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Zhu J, Kapoor A, Zhu JK. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 2007;26:1691–1701. doi: 10.1038/sj.emboj.7601603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Epigenetic inheritance in Arabidopsis: selective silence. Curr Opin Genet Dev. 2005;15:557–562. doi: 10.1016/j.gde.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development. 2007;134:3959–3965. doi: 10.1242/dev.001131. [DOI] [PubMed] [Google Scholar]
- Zuo J, Chua NH. Chemical-inducible systems for regulated expression of plant genes. Curr Opin Biotechnol. 2000;11:146–151. doi: 10.1016/s0958-1669(00)00073-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.