Abstract
Background
Childhood germ cell tumors (cGCTs), believed to arise from transformed primordial germ cells by an unknown mechanism, provide a unique model system for investigating cell signaling, pluripotency and the microenvironment of neoplastic stem cells (NSCs) in vivo. This is the first report of proteomics of cGCTs.
Procedure
Four dysgerminomas (DYSs) and four childhood endodermal sinus tumors (cESTs), resembling self-renewing and differentiating NSCs, respectively, were selected. Proteomic studies were performed by 2-DE, SDS-PAGE and cLC/MS/MS with protein database searching.
Results
2-DE: 9 of 941spots were differentially regulated with greater than a 2-fold change in spot volume for at least 3 of 4 gels in each group. 2 of 9 spots had p-values for the t-test analysis of comparisons less than 0.001, while the remaining spots had p-values from 0.013 to 0.191. Top-ranked proteins were identified in 9 of 9 spots with 4.0 to 38% sequence coverage. APOA1, CRK and PDIA3 were up-regulated in cESTs. TFG, TYMP, VCP, RBBP, FKBP4 and BiP were up-regulated in DYSs. SDS-PAGE: Up-regulation of NF45 and FKBP4 was observed in 4 of 4 cESTs and DYSs, respectively. The fold-changes observed correspond with characteristic genetic changes.
Conclusion
Differential regulation of FKBP4 and NF45, combined with previous research on immunosuppressant binding, suggests that glucocorticoid receptor signaling merits further investigation in cGCTs and NSCs.
Keywords: proteomics, neoplastic stem cells, childhood germ cell tumor, glucocorticoid receptor signaling
INTRODUCTION
The premise for the cancer stem cell (CSC)1 hypothesis is that cancers originate from genetic mutations and epigenetic modifications of single neoplastic stem cells (NSCs) that perturb cell viability, proliferation and/or self-renewal and differentiation [1]. Thus, the distinction between NSCs and CSCs is that the latter can initiate only malignant tumors, while the former can also initiate benign tumors. CSCs have been isolated by fluorescence-activated cell sorting (FACS) of distinctive low abundance cell surface membrane proteins from: blood, colon, pancreatic, ovarian, brain, breast and lung cancers. If the CSC hypothesis is correct, then a thorough understanding of the proteome of NSCs will have a profound impact on cancer prevention, diagnosis and treatment [2].
Stem cells, including CSCs, embryonic stem cells (ESCs), embryonic germ cells and hematopoietic stem cells, are long-lived, self-renewing cells that can divide symmetrically or asymmetrically and differentiate into all other cell types. Because human stem cells can be differentiated and expanded in vitro, given the proper stimulus and microenvironment, they are the basis for many promising personalized/regenerative medicine and drug development strategies. For example, artificial up-regulation of a few proteins known as pluripotency factors (PFs) in non-pluripotent (somatic) cells can generate induced pluripotent stem cells [3]. Well-known PFs include low abundance transcription factors such as POU5F1 (aka Oct-4) and NANOG that are involved in cell signaling, self-renewal and tumorigenesis. Lesser known PFs include nuclear receptors that are also involved in steroidogenesis [4].
Childhood GCTs (cGCTs), believed to arise from pluripotent transformed primordial germ cells or gonocytes, by an unknown mechanism, provide a unique model system for investigating cell signaling, pluripotency and the microenvironment of NSCs. GCTs comprise approximately 3% of all malignant children’s tumors [5] and up to 60% of all malignancies in men between 20 and 40 years of age [6]. Pure GCTs include: germinomas [e.g., ovarian dysgerminomas (DYSs) and testicular seminomas], embryonal carcinomas (ECs), teratomas, choriocarcinomas, and endodermal sinus tumors (ESTs, aka yolk sac tumors). Akin to migration of hematopoietic stem cells from the bone marrow, migration of primordial germ cells and their transformation (thought to occur in utero), depends on cell signaling and the microenvironment. Akin to the pluripotency of ESCs, each subtype of GCT contains self-renewing and differentiating NSCs that may result in a variety of neoplastic structures resembling embryonic (endoderm, mesoderm, and ectoderm) and extra-embryonic (yolk sac, trophoblast) derivatives. Indeed, prior to the availability of true ESCs, GCTs were used as surrogates for research on stemness. Unlike ESCs and other cell culture models, tumors such as cGCTs more closely reflect real-world socioeconomic and environmental factors that contribute to the underlying biochemistry of NSCs in vivo, including ethnicity, age, nutrition, healthcare and chemical exposure.
Proteomics is the large-scale identification, characterization and quantification of proteins. The proteome is a product of the great molecular diversity encoded by atoms in nucleotides (genes and transcripts), amino acids and their post-translational modifications (PTMs), such as phosphorylation. Consequently, it is impossible to predict the relatively dynamic proteome from the genome. In humans, approximately 24,000 genes are translated into an estimated 2 million protein isoforms that may span up to 12 orders of magnitude in abundance. Likewise, there is a poor correlation between the transcriptome and proteome due to single nucleotide polymorphisms, alternative splicing, PTMs, limiting ribosomes available for translation, mRNA and protein stability and various unknown actors (e.g., metabolites and microRNA). For these reasons and others, the proteome is best understood by direct measurement of its protein components. However, the molecular heterogeneity of clinical specimens challenges the most advanced proteomics methods, which remain relatively immature compared to genomics and transcriptomics.
Proteomics of childhood GCTs (cGCTs) has never been investigated [7]2. Indeed, the scarcity and heterogeneity of cGCT specimens make their study a particularly challenging task undertaken by a handful of dedicated researchers. Reports of PFs as promising clinical biomarker candidates for GCTs [6,8–20] suggested that proteomic studies of cGCTs might be feasible. However, the cellular and molecular heterogeneity of even pure cGCT subtypes (e.g., teratomas) can challenge proteomics because low abundance proteins of interest may be masked by high abundance housekeeping proteins. Therefore, two relatively homogenous and unrelated cGCTs, DYSs and childhood ESTs (cESTs), were selected for our preliminary experiments because they each show a striking balance between self-renewal and differentiation and distinctly different patterns of genetic gain and loss (including the patterns seen in adult GCTs) and age of onset [8,9]. DYSs most closely resemble self-renewing ESCs histologically and in their gene expression pattern, and they demonstrate the characteristic genetic changes in adult GCTs. cESTs do not arise from differentiation of DYSs; however, they resemble differentiating ESCs showing more primitive endodermal differentiation and contain different genetic changes. Therefore, we analyzed self-renewing and differentiating cGCTs to gain insights on the proteome of NSCs. Gel-based (and gel-free) proteomics of cGCTs by capillary liquid chromatography-tandem mass spectrometry (cLC/MS/MS) with protein database searching, unlike antibody-based methods such as FACS, ELISA or Western blotting, enables mass-, sequence-, PTM-, and molecular formula-specific measurements (Supplemental Appendix II and Supplemental Figures 1–3).
METHODS
CGCT tissue specimens, snap frozen with liquid nitrogen and stored at −80°C, were obtained through the Cooperative Human Tissue Network (CHTN) with IRB approval. Frozen section analysis was performed to ensure >80% viable tumor cellularity, and no tumors were rejected for this reason. Four DYSs of children aged 8–18 and four cESTs of children 2 months to 3 years of age were examined. The ethnicity of these children is unknown, and no private identifying information was provided. Approximately 10 mg from each of 4 DYS and 4 cEST specimens was homogenized in a 1:100 cocktail of protease and phosphatase inhibitors (Pierce) and centrifuged at 21,000 x g for 10 minutes at 4°C. The supernatant was precipitated in TCA, re-suspended in 8M urea and centrifuged to separate the urea soluble proteins. Protein concentrations were determined by the EZ Quant kit (Invitrogen). Proteomics was performed by two-dimensional electrophoresis (2-DE) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with protein identification by unbiased and targeted cLC/MS/MS, respectively, with protein database searching (Supplemental Appendix I). Results were compared to the human genome and published work. Genes corresponding to differentially regulated proteins were mapped to human chromosomes with NCBI MapViewer (build 36.9, www.ncbi.nlm.nih.gov/mapview/). Biochemical pathway analysis was performed with Ingenuity Pathways Analysis (IngenuityR Systems, www.ingenuity.com).
RESULTS
2-DE
Nine of nine-hundred forty-one 2-DE spots (1%) were differentially regulated between cESTs and DYSs with greater than a two-fold change in spot volume for at least 3 of 4 gels (Figures 1 and 2). Two of nine spots had p-values for the t-test analysis of comparisons less than 0.001, while the remaining spots had p-values from 0.013 to 0.191 (Table 1). Top-ranked proteins were identified by unbiased cLC/MS/MS with protein database searching of in-gel digests for each of the 9 spots with 4.0 to 38% sequence coverage and at least two peptides per protein (routine criteria for protein identification): APOA1, CRK and PDIA3 were up-regulated in cESTs; TFG, TYMP, VCP, RBBP, FKBP4 (aka FKBP52) and BiP (aka HSPA5) were up-regulated in DYSs.
Figure 1.
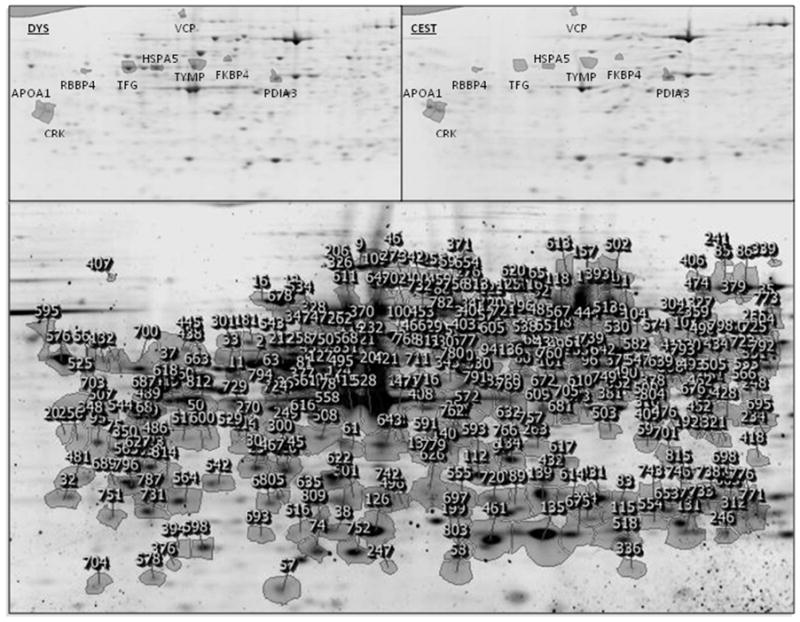
2-DE reference gel image for all cGCT specimens showing protein spot number assignments (bottom). Expanded views of protein spot boundaries for all cGCT specimens, labeled by their gene symbols as shown in Table 1, are shown for proteins that are up-regulated in DYSs vs. cESTs (top left panel) and CESTs vs. DYSs (top right panel). Top-ranked proteins were identified by unbiased cLC/MS/MS with protein database searching of in-gel digests for each of the 9 spots with 4.0 to 38% sequence coverage and at least two peptides per protein.
Figure 2.
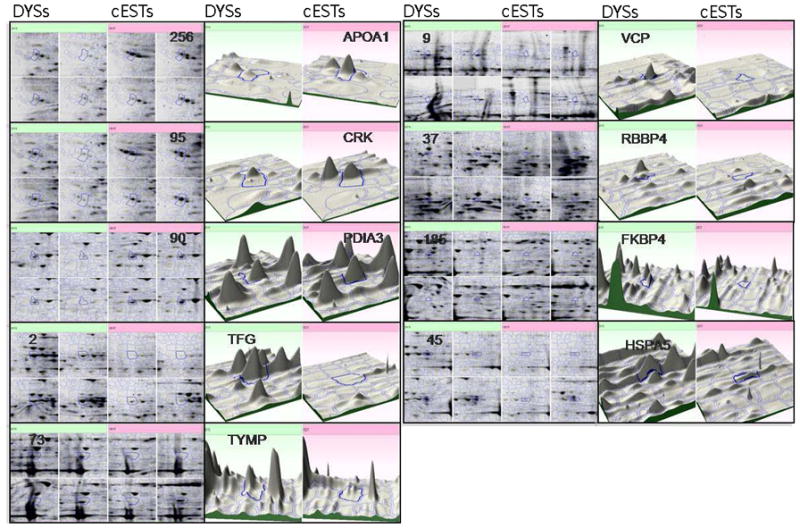
Expanded views of 2-DE gel images for individual cGCT specimens (4 DYSs and 4 cESTs), labeled by their gene symbols and protein spot numbers as shown in Table 1, are shown for proteins that are up-regulated in DYSs vs. cESTs (green, left quadrants) and CESTs vs. DYSs (pink, right quadrants). Three-dimensional plots illustrate the fold-change observed in representative pairs of specimens.
Table 1.
Proteins up-regulated in cESTs and DYSs.
| Protein (2-DE Spot No.) | Fold-Change | P-Value | Sequence Coverage (%) | Double-Charged Peptides (amino acid residues in protein) | Accession Number | Gene Symbol | Chromosomal Locus |
|---|---|---|---|---|---|---|---|
| Up-regulated in cESTs vs. DYSs | |||||||
| Pro-apolipoprotein (256) | 2.2 | 0.117 | 38 | 34–46; 95–100; 103–112; 140–146; 147–157; 160–166; 167–177; 184–194; 202–212; 213–221 |
AAA51747.1 GI:178775 |
APOA1 | 11q23-q24 |
| Proto-oncogene C-crk (95) | 2.0 | 0.013 | 6.0 | 21–31; 123–130 |
P46108.2 GI:15893932 |
CRK | 17p13.3 |
| Protein disulfide isomerase (90) | 2.1 | 0.030 | 23 | 63–73; 95–104; 108–119; 120–129; 131–140; 297–304; 336–344; 367–379; 426–433; 434–448; 483–496 |
CAA89996.1 GI:860986 |
PDIA3 | 15q15 |
| Up-regulated in DYSs vs. cESTs | |||||||
| TRKfused gene (2) | 5.7 | <0.001 | 4.0 | 48–57; 108–113 |
AAH41600.1 GI:11628401 |
TFG | 3q12.2 |
| Thymidine Phosphorylase (73) | 2.3 | 0.011 | 18 | 35–41; 49–55; 236–249; 266–279; 330–342; 346–22q13.33 358; 443–453; 454–464 | NP _001944.1 GI:4503445 |
TYMP | |
| Valosin containing protein (9) | 4.3 | 0.024 | 16 | 9–18; 46–53; 149–155; 218–225; 232–239; 240–251; 278–287; 366–377; 378–386; 600–614; 669–677; 701–708;701–709;754–766 |
NP_009057.1 GI:6005942 |
VCP | 9p13.3 |
| Retinoblastoma binding protein 4 (37) | 2.8 | <0.001 | 10 | 5–15; 16–22; 103–114; 252–258; 342–349 | NP _005601.1 GI:5032027 |
RBBP4 | 1p35.1 |
| FK506 binding protein 4 (185) | 3.2 | 0.191 | 32 | 8–28; 29–35; 40–52; 67–73; 67–74; 77–83; 139–152; 190–206; 235–244; 245–250; 345–354; 379–387; 409–417; 410–417; 446–459 | NP _002005.1 GI:4503729 |
FKBP4 | 12p13.33 |
| Binding imunoglobulin protein (45) | 2.7 | 0.012 | 13 | 51–61; 62–75; 83–97; 166–182; 354–368; 449–465 |
AAF13605.1 GI:6470150 |
BiP | 9q33-q34.1 |
For example, 2-DE spot number 185 was identified as FKBP4 with 32% sequence coverage. The theoretical monoisotopic mass of the unmodified form of a top-ranked tryptic peptide from FKBP4 spanning amino acid residues 139–152, GEDLTEEEDGGIIR, is 1531.70 Da. The observed monoisotopic mass for the precursor ion of this peptide of 1531.68 Da is in excellent agreement to the predicted value (< 11 ppm mass error). Likewise, the amino acid sequence assignment for its product ions, generated by gas-phase fragmentation, is in excellent agreement (< 353 ppm mass error). Lastly, confidence in our ion assignments by protein database searching is reflected by observation of a low expectation value of 2.3E-08 (probability of a random match in the protein database), 29 sequence-specific product ions, and multiply-charged ions.
SDS-PAGE
Targeted cLC/MS/MS of tryptic digests of SDS-PAGE gel slices at ~ 50 kDa revealed up-regulation of NF45 in 4 of 4 cESTs (0 of 4 DYSs) and confirmed up-regulation of FKBP4 in 4 of 4 DYSs (0 of 4 cESTs) (Figure 3.). For example, the theoretical monoisotopic mass for the tryptic peptide selected for NF45 spanning amino acid residues 128–141, ILPTLEAVAALGNK, is 1408.83 Da. Likewise, the theoretical monoisotopic mass for the tryptic peptide for FKBP4 spanning amino acid residues 190–206, FEIGEGENLDLPYGLER, is 1949.94 Da. The observed monoisotopic masses of precursor and product ions are in excellent agreement (< 100 and 1000 ppm mass error, respectively) for each peptide observed in each sample. Akin to unbiased cLC/MS/MS of 2-DE protein spots, confidence in our ion assignments by protein database searching is reflected by observation of low expectation values, sequence-specific product ions, and multiply-charged ions. Additional confidence is obtained by observation of narrow cLC peaks with high signal-to-noise ratio in the extracted ion chromatograms for these peptides.
Figure 3.
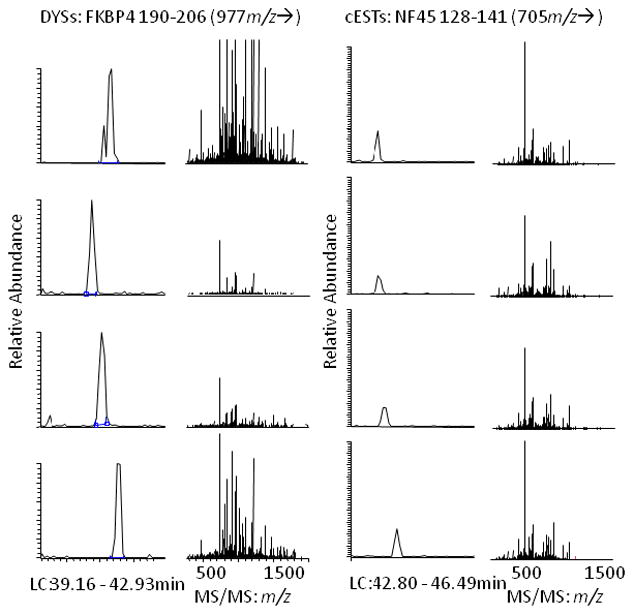
Extracted ion chromatograms (left) and amino acid sequence-specific MS/MS spectra (right) for targeted cLC/MS/MS of tryptic digests of SDS-PAGE gel slices at ~ 50 kDa revealed up-regulation of NF45 NF45 128–141 in 4 of 4 cESTs (0 of 4 DYSs) and confirmed up-regulation of FKBP4 190–206 in 4 of 4 DYSs (0 of 4 cESTs). Narrow cLC peaks with high signal-to-noise ratio in the extracted ion chromatograms for these peptides and protein database searching of their MS/MS spectra confirmed these results.
PTMs
Error tolerant protein database searching of MS/MS spectra revealed that no PTMs of FKBP4 or NF45 were unambiguously assigned.
DISCUSSION
This is the first report of proteomics of cGCTs2. The fold-changes observed for the proteins described below are in general agreement with characteristic genetic changes previously observed by cytogenetics. This was not entirely expected, given the poor correlation between mRNA and protein abundances in the literature and the intricate and unknown interactions of nucleotides, amino acids and their modifications.
The genes up-regulated in cESTs vs. DYSs are expected to be up-regulated during differentiation and/or down-regulated during self-renewal. Therefore, while important for cGCTs and NSCs, these genes are not expected to be PFs. APOA1, CRK and PDIA3 were revealed by 2-DE (Supplemental Appendix III), while Nuclear factor 45 (NF45, aka ILF2) was observed by SDS-PAGE. NF45 is a poorly understood phosphoprotein containing a rare DZF domain that, together with NF90, is involved in calcineurin (CN)-mediated T-cell activation and immunosuppressant (IS) binding [21–23].
The genes up-regulated in DYSs vs. cESTs are expected to be up-regulated during self-renewal and/or down-regulated during differentiation. Therefore, these genes are candidate PFs in cGCTs and NSCs. TFG, TYMP, VCP, RBBP4, FKBP4 and HSPA5 were revealed by 2-DE. The most significant of these, FKBP4, was up-regulated 3.2-fold at the protein level. FK506 binding protein 4 (FKBP4, aka FKBP52) is an immunophilin (IP), peptidyl-prolyl cis/trans isomerase, and phosphoprotein member of the FK506 binding protein family that, together with HSP90 and other proteins, is involved in CN-mediated T-cell activation, IS binding, and glucocorticoid receptor (GR, aka NR3C1) signaling [24–27]. Previously, transcriptomics of ESCs, EC cells, and seminomas revealed 330 PFs that were up-regulated at the mRNA level in all samples relative to somatic cell lines and normal testis [8]: a 2.2-, 8.3- and 2.6-fold up-regulation of FKBP4 mRNA was observed in ESCs, ECs, and seminomas, respectively. Likewise, previous proteomics work revealed 191 PFs, including a 3.3-fold up-regulation of FKBP4, that were up-regulated at the protein level in self-renewing ESCs [28]. Significantly, FKBP4 was also up-regulated in a proteomics study of invasive epithelial ovarian cancer [29]. The next most significant gene, retinoblastoma binding protein 4 (RBBP4), was up-regulated 2.8-fold at the protein level. RBBP4 is involved in tumorigenesis, histone acetylation and transcriptional silencing. Other retinoblastoma proteins are known to be involved in GR signaling. Previously, a 2.3-fold up-regulation of RBBP4 was observed in ECs [8]. Up-regulation of RBBP4 and FKBP4 at chromosomes 1p35.1 and 12p13.33, respectively, are in excellent agreement with cytogenetic studies where loss of chromosome 1p is often observed in cESTs [30–32] and gain of chromosome 12p is often observed in ESCs, ECs, seminomas and DYSs [8,28,33]. The remaining genes are involved in a variety of critical pathways (Supplemental Appendix III).
Differential regulation of FKBP4 and NF45, combined with previous research on IS binding in lymphoid cells and nuclear receptors in stem cells, suggests that GR signaling merits further investigation in cGCTs and NSCs. Treatment of cGCTS does not generally involve ISs. Rather, combination chemotherapy protocols with cisplatin or carboplatin, etoposide and bleomycin have been very successful. However, previous work on IS resistance and GR signaling in lymphoid cells is illuminating. The IS drugs known as glucocorticoids, such as prednis(ol)one and dexamethasone, and IP-/cyclophilin-binding drugs such as FK506 (aka tacrolimus), cyclosporin and rapamycin (aka sirolimus), are among the most frequently prescribed cancer, immunosuppressive and anti-inflammatory drugs in the world [34]. In lymphoblasts, ISs are believed to induce apoptosis by binding to the GR and GR complexes containing FKBPs and other proteins [e.g., the rapamycin-FKBP12 complex that binds to the mammalian target of rapamycin (mTOR)]. Competition for the GR by FKBP4 (aka FKBP52) and FKBP5 (aka FKBP51) may determine whether lymphoblasts are IS-sensitive or –resistant, respectively, in childhood acute lymphoblastic leukemia [35–43]. Presumably, this is due to the increased association of FKBP4, relative to FKBP5, to the cytoskeletal protein dynein, resulting in increased transport of IS-GR complexes to the nucleus and binding to GC response elements that activate or repress transcription of various genes involved in IS-induced apoptosis [44]. Limiting increased expression of the GR and binding of IS-GR complexes directly to transcription factors such as NF-κB may also be responsible for IS resistance. In lymphocytes, ISs inhibit CN-mediated dephosphorylation and nuclear translocation of NF45, reducing binding of NF45 to antigen receptor response elements/nuclear factor of activated T-cell (NFAT) DNA target sequences and down-regulating transcription of interleukin-2 (IL-2) (i.e., T-cell activation) [21,45]. Likewise, phosphorylation of FKBP4 by casein kinase II, known to compete for substrates with CN, has been shown to prevent association with HSP90 in GC-GR complexes while not affecting FK506 binding [26]. Moreover, the LxVP CN binding site for IS-IP complexes is shared by NFATs containing RHD domains [46], and a natural ligand for IPs, FK506 binding protein 48, has also been identified [47] that up-regulates transcription of IL-2 in the presence of CN inhibitors [48]. The GR signaling mechanisms presented above are controversial [49] and other mechanisms (Supplemental Figure IV), including those involved in apoptosis, have also been shown to be important [50]. In stem cells, nuclear receptors other than the GR are known to be involved in both maintaining the pluripotent state and in steroidogenesis. For example, steroidogenic factor 1 and liver receptor homolog 1 regulate Oct-4 expression in the NCCIT adult GCT cell line [4]. Therefore, it is not completely unexpected that GR signaling, where the GR is the major receptor for endogenous glucocorticoids (e.g., cortisol or hydroxyxortisone) and ISs, may be important to cGCTs and NSCs.
Observation of differential phosphorylation of proteins in the GR signaling pathway is required to prove active GR signaling in cGCTs and NSCs. However, no PTMs, particularly phosphorylation, were unambiguously assigned to specific amino acid residues by their characteristic mass shifts (e.g., −80 Da or −98 Da for HPO3 or H3PO4, respectively, at Ser, Thr, or Tyr residues) despite observation of unmodified tryptic peptides spanning many of the amino acid residues known to have PTMs (i.e., phosphorylation of FKBP4 at Thr143, Tyr411, Ser451 and Ser453). This is not at all surprising given the following: masking of unknown (and presumably low) abundance PTMs relative to unmodified amino acid residues (aka low site occupancy); the low amount of total protein that can be loaded onto gels; the low copy number of nuclear and membrane proteins known to be important to stem cells; and the scarcity and heterogeneity of cGCT specimens. Masking of PTMs with low site occupancy can be overcome through greater fractionation of proteins or peptides combined with immuno-enrichment/-depletion with antibodies, and milligrams of total protein can be loaded onto gel-free preparative devices to improve the characterization of proteins with low copy number. While antibody-based methods such as FACS, ELISA and Western blotting suffer less masking effects and often require less total protein, these techniques are also less specific than gel-based (or gel-free) proteomics, often cross-react, often have uncharacterized binding epitopes, and may not be commercially available for the PTM-modified amino acid residue of interest. Regardless of the analytical strategy, the omission of protease and phosphatase inhibitors during cGCT specimen collection, combined with the scarcity and heterogeneity of these specimens, remain serious stumbling blocks for proving active GR signaling in cGCTs and NSCs.
Through gel-based proteomics, we provide evidence that self-renewing cGCTs (DYSs) up-regulate FKBP4 while differentiating cGCTs (cESTs) up-regulate NF45. While these results are preliminary, FKBP4 and NF45 are promising biomarker candidates towards more accurate diagnosis of cGCT subtypes than current combined cytohistological and molecular [e.g., placental alkaline phosphatase, beta-human chorionic gonadotropin and alpha-fetoprotein] methods. Differential regulation of FKBP4 and NF45, combined with previous research, suggests that GR signaling may be very important to pluripotency and the microenvironment of NSCs in vivo. However, observation of differential phosphorylation of proteins in the GR signaling pathway, required to prove active GR signaling in cGCTs and NSCs, remains to be shown.
Supplementary Material
Supplemental Figure 1. Illustration of A) cLC/MS and B) cLC/MS/MS for mass, amino acid sequence, post-translational modification (PTM), and molecular formula assignment for the theoretical peptide MIDNIGHT. The isotopic envelope for the precursor ion and expected m/z values for b- and y-type product ions are shown.
Supplemental Figure 2. MS/MS spectrum and sequence assignment for tryptic peptide GEFGGFGSVSGK that spans amino acid residues 103–114 in human retinoblastoma binding protein 4 (RBBP4). Sequence-specific product ions, where charge is retained at the N- or C-terminus following peptide backbone bond cleavage, are labeled as b- and y-type ions, respectively. RBBP4 was identified with 10% sequence coverage from a 2-DE spot digest by cLC/MS/MS as shown by the underlined tryptic peptides in the amino acid sequence.
Supplemental Figure 3. A comparison of peptide fragmentation by collision-induced dissociation (CID) and electron transfer dissociation (ETD) via cLC/MS/MS of the human serum albumin peptide FKDLGEENFK with a linear ion trap tandem mass spectrometer.
Supplemental Figure 4. The glucocorticoid receptor (GR) signaling pathway (Ingenuity Pathway Analysis), including proteins observed in this work: fk506 binding proteins 4 and 5 [aka FKBP4 (FKBP52) and FKBP5 (FKBP51, respectively], heat shock proteins 70 and 90 (HSP70 and HSP90, respectively), calreticulin and dynein.
Acknowledgments
This work was supported by a Pediatric LRP grant to W.E.H. (mentored by E.J.P.) from the National Institutes of Health (NIH). We thank the RCMI Proteomics & Protein Biomarkers Cores at UTSA (NIH G12 RR013646) for assistance with experiment design, sample preparation, data collection, results interpretation and manuscript preparation. We thank the Computational Biology Initiative (UTSA/UTHSCSA) for providing access and training to the analysis software. We express our gratitude to: Kevin Hakala and Dr. Susan Weintraub for assistance with sample preparation and 2-DE; Dr. Sharon Murphy and Dr. Vivienne Rebel for insightful discussion (UTHSCSA); and Dr. Jaclyn Y. Hung (UTHSCSA) and Dr. Christopher Navara (UTSA) for critically reviewing this manuscript. Lastly, we acknowledge the support of the Cancer Therapy and Research Center (CTRC) at UTHSCSA, an NCI-designated Cancer Center (NIH P30CA54174).
Footnotes
key abbreviations: cEST, childhood endodermal sinus tumor; cGCT, childhood germ cell tumor; CN, calcineurin; CSC, cancer stem cell; DYS, dysgerminoma; EC, embryonal carcinoma; ESC, embryonic stem cell; EST, endodermal sinus tumor; FKBP4, FK506 binding protein 4; GR, glucocorticoid receptor; IP, immunophilin; IS, immunosuppressant; NF45, nuclear factor 45; NSC, neoplastic stem cell; PF, pluripotency factor
Gel-based proteomics of adult seminomas showed down-regulation of glutathione S-transferase M3 relative to non-neoplastic germ cell tissue.
The authors have declared no conflicts of interest.
References
- 1.Reya T, Morrison SJ, Clarke MF, et al. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 2.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112(13):4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 3.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448(7151):313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 4.Mullen EM, Gu P, Cooney AJ. Nuclear Receptors in Regulation of Mouse ES Cell Pluripotency and Differentiation. PPAR Res. 2007;2007:61563. doi: 10.1155/2007/61563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bussey KJ, Lawce HJ, Olson SB, et al. Chromosome abnormalities of eighty-one pediatric germ cell tumors: sex-, age-, site-, and histopathology-related differences--a Children’s Cancer Group study. Genes Chromosomes Cancer. 1999;25(2):134–146. [PubMed] [Google Scholar]
- 6.Juric D, Sale S, Hromas RA, et al. Gene expression profiling differentiates germ cell tumors from other cancers and defines subtype-specific signatures. Proc Natl Acad Sci U S A. 2005;102(49):17763–17768. doi: 10.1073/pnas.0509082102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann U, Junker H, Kramer F, et al. Comparative proteomic analysis of neoplastic and non-neoplastic germ cell tissue. Biol Chem. 2006;387(4):437–440. doi: 10.1515/BC.2006.058. [DOI] [PubMed] [Google Scholar]
- 8.Sperger JM, Chen X, Draper JS, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100(23):13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Looijenga LH, Stoop H, de Leeuw HP, et al. POU5F1 (OCT3/4) identifies cells with pluripotent potential in human germ cell tumors. Cancer Res. 2003;63(9):2244–2250. [PubMed] [Google Scholar]
- 10.Ezeh UI, Turek PJ, Reijo RA, et al. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104(10):2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 11.Hart AH, Hartley L, Parker K, et al. The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer. 2005;104(10):2092–2098. doi: 10.1002/cncr.21435. [DOI] [PubMed] [Google Scholar]
- 12.Honecker F, Stoop H, Mayer F, et al. Germ cell lineage differentiation in non-seminomatous germ cell tumours. J Pathol. 2006;208(3):395–400. doi: 10.1002/path.1872. [DOI] [PubMed] [Google Scholar]
- 13.Hoei-Hansen CE, Sehested A, Juhler M, et al. New evidence for the origin of intracranial germ cell tumours from primordial germ cells: expression of pluripotency and cell differentiation markers. J Pathol. 2006;209(1):25–33. doi: 10.1002/path.1948. [DOI] [PubMed] [Google Scholar]
- 14.Rajpert-De Meyts E, Hanstein R, Jorgensen N, et al. Developmental expression of POU5F1 (OCT-3/4) in normal and dysgenetic human gonads. Hum Reprod. 2004;19(6):1338–1344. doi: 10.1093/humrep/deh265. [DOI] [PubMed] [Google Scholar]
- 15.Palumbo C, van Roozendaal K, Gillis AJ, et al. Expression of the PDGF alpha-receptor 1.5 kb transcript, OCT-4, and c-KIT in human normal and malignant tissues. Implications for the early diagnosis of testicular germ cell tumours and for our understanding of regulatory mechanisms. J Pathol. 2002;196(4):467–477. doi: 10.1002/path.1064. [DOI] [PubMed] [Google Scholar]
- 16.Almstrup K, Hoei-Hansen CE, Wirkner U, et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res. 2004;64(14):4736–4743. doi: 10.1158/0008-5472.CAN-04-0679. [DOI] [PubMed] [Google Scholar]
- 17.Almstrup K, Hoei-Hansen CE, Nielsen JE, et al. Genome-wide gene expression profiling of testicular carcinoma in situ progression into overt tumours. Br J Cancer. 2005;92(10):1934–1941. doi: 10.1038/sj.bjc.6602560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Jong J, Looijenga LH. Stem cell marker OCT3/4 in tumor biology and germ cell tumor diagnostics: history and future. Crit Rev Oncog. 2006;12(3–4):171–203. doi: 10.1615/critrevoncog.v12.i3-4.10. [DOI] [PubMed] [Google Scholar]
- 19.Stoop H, Honecker F, van de Geijn GJ, et al. Stem cell factor as a novel diagnostic marker for early malignant germ cells. J Pathol. 2008;216(1):43–54. doi: 10.1002/path.2378. [DOI] [PubMed] [Google Scholar]
- 20.Cheng L, Sung MT, Cossu-Rocca P, et al. OCT4: biological functions and clinical applications as a marker of germ cell neoplasia. J Pathol. 2007;211(1):1–9. doi: 10.1002/path.2105. [DOI] [PubMed] [Google Scholar]
- 21.Kao PN, Chen L, Brock G, et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269(32):20691–20699. [PubMed] [Google Scholar]
- 22.Doerks T, Copley RR, Schultz J, et al. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12(1):47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem. 1994;269(32):20682–20690. [PubMed] [Google Scholar]
- 24.Cox MB, Riggs DL, Hessling M, et al. FK506-binding protein 52 phosphorylation: a potential mechanism for regulating steroid hormone receptor activity. Mol Endocrinol. 2007;21(12):2956–2967. doi: 10.1210/me.2006-0547. [DOI] [PubMed] [Google Scholar]
- 25.Siekierka JJ, Hung SH, Poe M, et al. A cytosolic binding protein for the immunosuppressant FK506 has peptidyl-prolyl isomerase activity but is distinct from cyclophilin. Nature. 1989;341(6244):755–757. doi: 10.1038/341755a0. [DOI] [PubMed] [Google Scholar]
- 26.Miyata Y, Chambraud B, Radanyi C, et al. Phosphorylation of the immunosuppressant FK506-binding protein FKBP52 by casein kinase II: regulation of HSP90-binding activity of FKBP52. Proc Natl Acad Sci U S A. 1997;94(26):14500–14505. doi: 10.1073/pnas.94.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Clark AF, Yorio T. FK506-binding protein 51 regulates nuclear transport of the glucocorticoid receptor beta and glucocorticoid responsiveness. Invest Ophthalmol Vis Sci. 2008;49(3):1037–1047. doi: 10.1167/iovs.07-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Hoof D, Passier R, Ward-Van Oostwaard D, et al. A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics. 2006;5(7):1261–1273. doi: 10.1074/mcp.M500405-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Jones MB, Krutzsch H, Shu H, et al. Proteomic analysis and identification of new biomarkers and therapeutic targets for invasive ovarian cancer. Proteomics. 2002;2(1):76–84. [PubMed] [Google Scholar]
- 30.Perlman EJ, Valentine MB, Griffin CA, et al. Deletion of 1p36 in childhood endodermal sinus tumors by two-color fluorescence in situ hybridization: a pediatric oncology group study. Genes Chromosomes Cancer. 1996;16(1):15–20. doi: 10.1002/(SICI)1098-2264(199605)16:1<15::AID-GCC2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Schuster AE, Fritsch MK, et al. Deletion mapping of 6q21–26 and frequency of 1p36 deletion in childhood endodermal sinus tumors by microsatellite analysis. Oncogene. 2001;20(55):8042–8044. doi: 10.1038/sj.onc.1204961. [DOI] [PubMed] [Google Scholar]
- 32.Palmer RD, Foster NA, Vowler SL, et al. Malignant germ cell tumours of childhood: new associations of genomic imbalance. Br J Cancer. 2007;96(4):667–676. doi: 10.1038/sj.bjc.6603602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer RD, Barbosa-Morais NL, Gooding EL, et al. Pediatric malignant germ cell tumors show characteristic transcriptome profiles. Cancer Res. 2008;68(11):4239–4247. doi: 10.1158/0008-5472.CAN-07-5560. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt S, Rainer J, Ploner C, et al. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11 (Suppl 1):S45–55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 35.Bachmann PS, Gorman R, Papa RA, et al. Divergent mechanisms of glucocorticoid resistance in experimental models of pediatric acute lymphoblastic leukemia. Cancer Res. 2007;67(9):4482–4490. doi: 10.1158/0008-5472.CAN-06-4244. [DOI] [PubMed] [Google Scholar]
- 36.Gu L, Gao J, Li Q, et al. Rapamycin reverses NPM-ALK-induced glucocorticoid resistance in lymphoid tumor cells by inhibiting mTOR signaling pathway, enhancing G1 cell cycle arrest and apoptosis. Leukemia. 2008;22(11):2091–2096. doi: 10.1038/leu.2008.204. [DOI] [PubMed] [Google Scholar]
- 37.Kaspers GJ, Pieters R, Klumper E, et al. Glucocorticoid resistance in childhood leukemia. Leuk Lymphoma. 1994;13(3–4):187–201. doi: 10.3109/10428199409056282. [DOI] [PubMed] [Google Scholar]
- 38.Ploner C, Schmidt S, Presul E, et al. Glucocorticoid-induced apoptosis and glucocorticoid resistance in acute lymphoblastic leukemia. J Steroid Biochem Mol Biol. 2005;93(2–5):153–160. doi: 10.1016/j.jsbmb.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 39.Pottier N, Yang W, Assem M, et al. The SWI/SNF chromatin-remodeling complex and glucocorticoid resistance in acute lymphoblastic leukemia. J Natl Cancer Inst. 2008;100(24):1792–1803. doi: 10.1093/jnci/djn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renner K, Ausserlechner MJ, Kofler R. A conceptual view on glucocorticoid-lnduced apoptosis, cell cycle arrest and glucocorticoid resistance in lymphoblastic leukemia. Curr Mol Med. 2003;3(8):707–717. doi: 10.2174/1566524033479357. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt S, Irving JA, Minto L, et al. Glucocorticoid resistance in two key models of acute lymphoblastic leukemia occurs at the level of the glucocorticoid receptor. FASEB J. 2006;20(14):2600–2602. doi: 10.1096/fj.06-6214fje. [DOI] [PubMed] [Google Scholar]
- 42.Tissing WJ, Lauten M, Meijerink JP, et al. Expression of the glucocorticoid receptor and its isoforms in relation to glucocorticoid resistance in childhood acute lymphocytic leukemia. Haematologica. 2005;90(9):1279–1281. [PubMed] [Google Scholar]
- 43.Schmidt S, Rainer J, Riml S, et al. Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood. 2006;107(5):2061–2069. doi: 10.1182/blood-2005-07-2853. [DOI] [PubMed] [Google Scholar]
- 44.Wochnik GM, Ruegg J, Abel GA, et al. FK506-binding proteins 51 and 52 differentially regulate dynein interaction and nuclear translocation of the glucocorticoid receptor in mammalian cells. J Biol Chem. 2005;280(6):4609–4616. doi: 10.1074/jbc.M407498200. [DOI] [PubMed] [Google Scholar]
- 45.Zhao G, Shi L, Qiu D, et al. NF45/ILF2 tissue expression, promoter analysis, and interleukin–2 transactivating function. Exp Cell Res. 2005;305(2):312–323. doi: 10.1016/j.yexcr.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez A, Roy J, Martinez-Martinez S, et al. A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol Cell. 2009;33(5):616–626. doi: 10.1016/j.molcel.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chambraud B, Radanyi C, Camonis JH, et al. FAP48, a new protein that forms specific complexes with both immunophilins FKBP59 and FKBP12. Prevention by the immunosuppressant drugs FK506 and rapamycin. J Biol Chem. 1996;271(51):32923–32929. doi: 10.1074/jbc.271.51.32923. [DOI] [PubMed] [Google Scholar]
- 48.Krummrei U, Baulieu EE, Chambraud B. The FKBP-associated protein FAP48 is an antiproliferative molecule and a player in T cell activation that increases IL2 synthesis. Proc Natl Acad Sci U S A. 2003;100(5):2444–2449. doi: 10.1073/pnas.0438007100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tissing WJ, Meijerink JP, den Boer ML, et al. mRNA expression levels of (co)chaperone molecules of the glucocorticoid receptor are not involved in glucocorticoid resistance in pediatric ALL. Leukemia. 2005;19(5):727–733. doi: 10.1038/sj.leu.2403681. [DOI] [PubMed] [Google Scholar]
- 50.Meijsing SH, Pufall MA, So AY, et al. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Illustration of A) cLC/MS and B) cLC/MS/MS for mass, amino acid sequence, post-translational modification (PTM), and molecular formula assignment for the theoretical peptide MIDNIGHT. The isotopic envelope for the precursor ion and expected m/z values for b- and y-type product ions are shown.
Supplemental Figure 2. MS/MS spectrum and sequence assignment for tryptic peptide GEFGGFGSVSGK that spans amino acid residues 103–114 in human retinoblastoma binding protein 4 (RBBP4). Sequence-specific product ions, where charge is retained at the N- or C-terminus following peptide backbone bond cleavage, are labeled as b- and y-type ions, respectively. RBBP4 was identified with 10% sequence coverage from a 2-DE spot digest by cLC/MS/MS as shown by the underlined tryptic peptides in the amino acid sequence.
Supplemental Figure 3. A comparison of peptide fragmentation by collision-induced dissociation (CID) and electron transfer dissociation (ETD) via cLC/MS/MS of the human serum albumin peptide FKDLGEENFK with a linear ion trap tandem mass spectrometer.
Supplemental Figure 4. The glucocorticoid receptor (GR) signaling pathway (Ingenuity Pathway Analysis), including proteins observed in this work: fk506 binding proteins 4 and 5 [aka FKBP4 (FKBP52) and FKBP5 (FKBP51, respectively], heat shock proteins 70 and 90 (HSP70 and HSP90, respectively), calreticulin and dynein.


