Abstract
Healthcare information technology and genotyping technology are both advancing rapidly, creating new opportunities for medical and scientific discovery. The convergence of these two technologies is now facilitating genetic association studies of unprecedented size within the context of routine clinical care. As a result, the medical community will soon be presented with a number of novel opportunities to bring functional genomics to the bedside in the area of pharmacotherapy. By linking biological material to comprehensive medical records, large multi-institutional biobanks are now poised to advance the field of pharmacogenomics through three distinct mechanisms: (1) retrospective assessment of previously known findings in a clinical practice-based setting, (2) discovery of new associations in huge observational cohorts, and (3) prospective application in a setting capable of providing real-time decision support. The current review explores each of these translational mechanisms within an historical framework.
INTRODUCTION
Although the field of pharmacogenomics has the potential to transform the clinical practice of medicine, gene-based drug prescribing currently occurs rather infrequently. The Clinical Pharmacogenomics Implementation Consortium (CPIC) has been organized to move research findings into routine practice [http://PharmGKB.org ]. Personalized gene-based health care delivery may therefore soon shift the allocation of medical resources away from reactive treatment of disease, toward a more proactive approach based upon human variation (1,2). It is likely that electronic medical records (EMRs) will play a pivotal role in this transformation.
Prior to 2008, approximately 10% of all U.S. physicians had utilized a basic electronic health care record (3). Although the proportion of U.S. hospitals implementing a basic EMR system was similarly low (7.6%) prior to 2008 (4), application of EMRs is rapidly expanding in both the inpatient and outpatient settings (5). Nearly half of all large multispecialty group practices now utilize a comprehensive electronic record [http://www.computerworld.com].
Through the Patient Protection and Affordable Care Act of 2010, federal legislators have set an aggressive timeline encouraging the widespread implementation of EMRs (5). Providers that roll out EMRs by 2015, will realize additional incentives for the care of Medicare patients, but only if the application is determined to fulfill pre-specified “meaningful use” criteria. The meaningful use rules for 2011 and 2012 were released by the U.S. Department of Health and Human Services on July 13, 2010 [http://www.ama-assn.org]. A two-track approach has been established, containing criteria that are both mandatory (e.g., maintenance of active medication lists) and optional (e.g., decision support software capable of flagging drug-drug interactions).
To date, EMR deployment has not only improved patient care, but it has also established large practice-based longitudinal datasets ideal for the conduct of observational research (6,7). These datasets are rich in clinical information, available in formats that are both structured and unstructured. Structured data include diagnoses, clinical laboratory results, diagnostic imaging results, procedures ordered and performed, medications ordered and dispensed, and physician order entry. Structured medication data in particular have been used for a large variety of pharmacoepidemiology, pharmacoeconomic, and service-related health care investigations (8). When unstructured medication data are embedded in free text clinical notes, natural language processing (NLP) algorithms have proven useful in the accurate reconstruction of comprehensive drug exposure histories (9). NLP-based phenotyping approaches have been used successfully to identify genetic determinants of drug outcome in the context of toxicity and efficacy (10).
As a community, our ability to characterize the genetic architecture underlying treatment outcome also continues to improve due to advances in genotyping technology. Increases in throughput, and decreases in cost, are allowing investigators to move from candidate genes (pharmacogenetics), to genome-wide SNP scanning (pharmacogenomics) in cohorts of increasing size (11). It is likely that entire genomic sequences will soon be linked to individual EMRs (1,2). Because the convergence of these two rapidly expanding technologies (i.e., biomedical informatics and high-throughput genotyping) represents an unprecedented opportunity to bring functional genomics to the bedside, many large medical centers are constructing DNA biobanks in the context of routine clinical practice (12–14). These biobanks offer the advantages of scale, cost efficiency, and extremely dense longitudinal healthcare data.
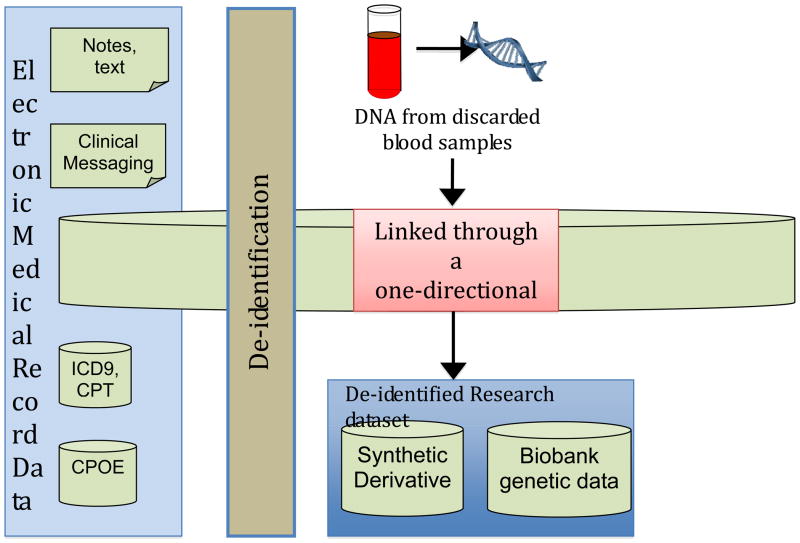
Many models have emerged for the construction of biobanks, including models based on recruitment and enrollment of subjects from a specific geographic region or practice community. Other approaches have employed novel informatics strategies to completely de-identify an EMR. The latter approach optimizes security, while allowing the de-identified samples to be linked to archived biological material in a cost effective manner (as depicted in Figure 1). Early results indicate that biobanks constructed from completely de-identified EMRs are robust in their ability to replicate genetic associations previously identified in disease-based cohorts (15).
Figure 1. One approach to the construction of a biobank for pharmacogenomic research.
Electronic medical records (EMRs) typically contain a combination of unstructured text reports and structured data. Structured data includes most laboratory values, vital signs, and such data as computerized provider order entry (CPOE) records. In addition, administrative billing codes (ICD9, CPT) form valuable components for electronic phenotyping. These data can then be de-identified using algorithms to remove personal health identifiers from text through a combination of statistical and pattern-matching techniques [ref 13]. Finally, de-identified medical records are linked to DNA samples using research unique identifiers, which can be generated using a one-way hash algorithm that prevents discovery of the input number (e.g., a medical record number).
The current review explores ways that existing EMRs may help facilitate the translation of pharmacogenomics into widespread clinical practice. Three distinct mechanisms are discussed: retrospective assessment of known findings in a clinical practice-based setting, discovery of novel associations in the context of gene-environment interaction, and prospective application in a setting capable of providing real-time decision support through bioinformatics and pharmacovigiliance. Each is presented in an historical context, with an emphasis on outlook.
I. Historical Overview
The clinical practice community began moving toward implementation of EMRs nearly half a century ago (16). While most early EMRs consisted primarily of billing codes, some contained diagnostic codes in site-specific lexicons. The majority of the early lexicons were rather cursory. Shortly thereafter, procedural codes and clinical laboratory data began to be archived in coded and easily extractable format (17). In most systems of care, clinical notes were entered as free text, in a manner that was diverse, complex, and site specific. Medication data were typically unstructured, only available within free text notes. More recently, computer-based prescribing has facilitated the entry of medication data in a format that is coded. National efforts are currently underway to harmonize both of these data formats (18).
Early in the last decade, lack of structured medication data presented an obstacle to the routine use of EMRs for studies addressing the genetic determinants underlying treatment outcome. As such, initial progress in the field of pharmacogenetics was limited primarily to cohorts that were enrolled in treatment trials (19). However, as clinical interest in this field grew, investigators began manually interrogating clinical practice-based datasets for drug-exposed cohorts of limited sample size (20). Within this setting, the process of case ascertainment was labor-intensive and expensive. Nonetheless, such studies were partly successful, particularly if they focused on predictors of toxicity (rather than efficacy) for drugs with clinically severe adverse drug reactions (ADRs) and a narrow therapeutic index (21–23).
More recently, the electronic reconstruction of comprehensive medication histories has allowed the field of pharmacogenomics to gain considerable momentum within datasets that are derived from routine clinical practice (9). Data derived from EMRs have proven highly accurate for quantifying disease phenotypes (onset and rate of progression) as well as treatment outcomes (efficacy and toxicity). Clinical diagnoses can be efficiently extracted from de-identified EMRs, through the application of algorithms integrating diagnostic codes, clinical laboratory data, and medication histories, and these traits are robust in their ability to replicate known associations previously characterized in disease-specific research cohorts (15). With the development of controlled vocabularies, their inclusion in centralized terminology systems such as the Unified Medical Language System (UMLS), and application of scalable information extraction strategies from the clinical narrative (24) [www.ohnlp.org] [www.i2b2.org], investigators are now able to extend this work to the characterization of treatment outcome using NLP software (25).
Standard evaluation metrics for the performance of NLP systems are shown in equations 1–4:
| (1) |
| (2) |
| (3) |
| (4) |
Detailed drug information can now be extracted from clinical text, including strength, route, and frequency. Jagannathan et al. (26) recently compared four commercial NLP engines, reporting a high F-measure of 93.2% for capturing drug names, but lower F-measures of 85.3%, 80.3%, and 48.3% for retrieving strength, route, and frequency. Newer approaches can increase F-measures to over 90% for strength, route, and frequency (9). A recent NLP challenge, in the Integrating Biology and the Bedside (i2b2) initiative, focuses specifically on the extraction of drug signature from clinical free text documents (https://www.i2b2.org/NLP/Medication/).
II. Current Efforts
Clearly, for pharmacogenetic and pharmacogenomic association studies to be successful within the context of EMRs, accurate drug-exposure histories need to be efficiently extracted and linked to treatment outcomes that are carefully defined. Methods used to quantify the toxicity of a given drug are typically quite different than methods used to quantify its clinical efficacy (27). As noted above, many of the early findings in this field were related to toxicity (21–23).
EMRs and Drug Toxicity
Data obtained from routine medical practice are uniquely suited for the investigation of genetic determinants underlying adverse drug reactions (ADRs), while - in comparison - data obtained from clinical trials may be less optimal for this task (28). This discrepancy is due, in part, to the fact that clinical trials often have a ‘run-in’ period where potential study subjects experiencing manifestations of drug intolerance are excluded prior to randomization. Patients with relevant co-morbidities may also be actively excluded from clinical trials. Such is not the case within practice-based datasets. As a result, the prevalence of ADRs within the community is often higher (and more variable) than that observed within randomized trials.
Consider the example of lipid lowering therapy. HMG-CoA reductase inhibitors (statins) are the most commonly prescribed class of medications in the U.S., and large multicenter trials have demonstrated unequivocally that these drugs reduce the risk of cardiovascular events in patients at risk. Although statin-related ADRs occur very infrequently in the context of monotherapy, the ADR event rate increases within the context of interacting medications (29,30). Many of these drug-drug interactions reflect alterations in pharmacokinetic processes.
We have also observed that genetic variability in pharmacokinetic processes can contribute to the severity of statin-related ADRs (20). However, patient-to-patient variability in Phase I metabolism only represents a single component within any given patient’s capacity for drug disposition. The entire process is better understood when each component is considered within the context of absorption, distribution, metabolism, and elimination (ADME) (31). For example, many statins undergo additional modification through Phase II conjugation by enzymes within the UDP-glucuronosyltransferase (UGT) family (32). UGT1A1 and UGT1A3 are both capable of converting atorvastatin acid to a lactone derivative, and perturbations in atorvastatin kinetics previously attributed to UGT1A1*28 may in fact be due to genetic variability in UGT1A3 (UGT1A3 haplotypes are in allelic association with UGT1A1*28) (33). Membrane transporters also markedly influence statin disposition. The SEARCH Collaborative Group recently reported that simvastatin-induced muscle toxicity is associated with genetic variability in statin uptake (34). Other data suggest that variability in efflux may influence risk (35). Efforts are underway to replicate these findings using EMRs (30).
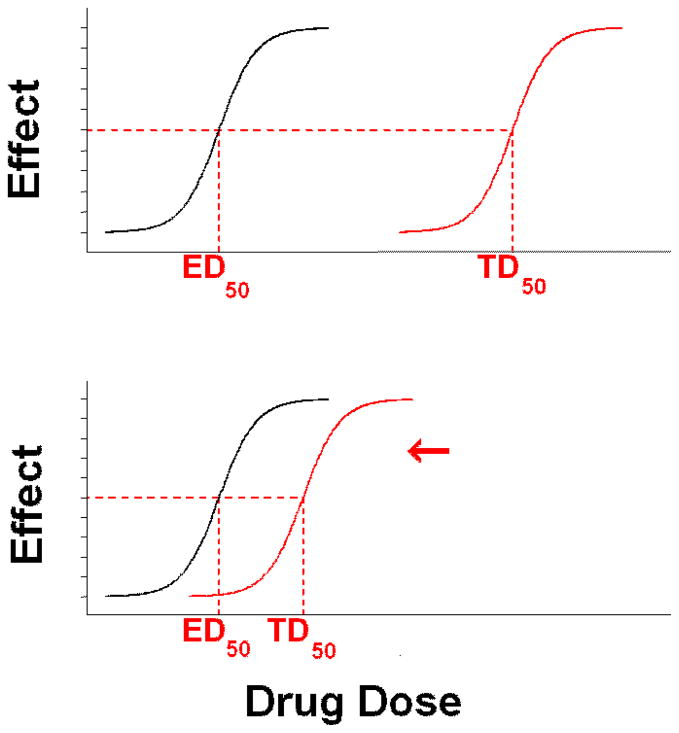
The ability to resolve genetic determinants of drug toxicity depends upon a number of factors (22). Unless the toxicity endpoint is rigorously defined, such studies are subject to misclassification bias. Two important properties must be considered: the clinical severity of the ADR, and the therapeutic index of the drug. Therapeutic index (TI) reflects the ratio of the dose of drug known to cause half maximal toxicity to the dose of the drug known to deliver half maximal efficacy (TD50/ED50) (31). Drugs with a wider TI (e.g., statins) tend to have ADRs that are less susceptible to genetic (or environmental) perturbations in kinetic processes. This may, in part, by why statin-related ADRs occur so rarely within the context of monotherapy (22). Conversely, ADRs tend to occur more frequently with drugs that have a narrower TI (e.g., anticoagulants, antineoplastics). This principle is illustrated in Figure 2.
Figure 2. Quantifying drug toxicity. Therapeutic Index (TI) = TD50/ED50.
ED50 = dose of a drug observed to yield half-maximal efficacy.
TD50 = dose of a drug observed to yield half-maximal toxicity.
To illustrate the importance of TI, consider the thromboembolic complications related to the use of tamoxifen. Tamoxifen is an antiestrogen used to treat breast cancer. World-wide, it is the most common endocrine therapy for estrogen receptor-positive breast cancer. Although aromatase inhibitors are also commonly prescribed for this indication, there are still ~1.5 million tamoxifen prescriptions written in the U.S. each year. While tamoxifen reduces the risk of breast cancer recurrence by ~50%, there is considerable variability in its efficacy and toxicity profile.
Deep venous thromboses (DVT) represent rare but serious side effects related to the use of tamoxifen. Because these events are rare, there may not be enough events in typical clinical trials to conduct adequately powered genotype-phenotype association studies. Therefore, EMRs have recently been used to identify sufficient numbers of breast cancer patients experiencing DVTs while on tamoxifen, to study this association. Genetic variants in the estrogen receptor were found to be associated with the occurrence of DVT, using banked DNA from ADR case patients, and frequency matched tamoxifen-exposed controls (36). Studies are now underway to confirm these results using additional EMRs linked to biobanks participating in the NIH-funded eMERGE network [https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page].
EMRs and Drug Efficacy
Tamoxifen is a moderately potent antiestrogen, however, it is metabolized into many metabolites that are known to be more potent than the parent compound. The most abundant of the high potency metabolites is endoxifen (4-OH-N-desmethyl-tamoxifen). This active metabolite is formed primarily by cytochrome P450 2D6 (CYP2D6). As expected, patients have reduced plasma concentrations of endoxifen if they have reduced CYP2D6 activity due to either genetic polymorphisms in CYP2D6 or the concurrent use of medications that inhibit CYP2D6 activity. Since some association studies have shown an association of CYP2D6 activity with tamoxifen outcome, recent efforts have used EMRs to determine if the concurrent use of CYP2D6 inhibiting drugs (e.g., antidepressants) by women taking tamoxifen is associated with increase breast cancer recurrence. These studies have included breast cancer health records from countries such as Denmark (37), the Netherlands (38), and Canada (39). Another study utilized records from the Medco, Inc., pharmacy benefits manager in the U.S. (40). These studies have shown mixed results. In some cases, CYP2D6 inhibitors were associated with breast cancer recurrence, but not in others. The discrepancy may be due to differences in the populations studied, sample sizes, and/or the methods used to mine the databases. This highlights the need for additional work focused on understanding how to optimally and consistently use EMRs.
Within the NIH-funded Pharmacogenomics Research Network (PGRN) [http://www.PharmGKB.org], there are several ongoing efforts to construct and standardize comprehensive treatment histories for patients exposed to tamoxifen within biobanks linked to EMRs. Our group has recently applied automated NLP-based extraction and information merging within the EMR to quantify each patient’s breast cancer treatment strategy within several PGRN nodes. Sensitivity and specificity were 90.27%–99.73% (positive and negative predictive values were 80.00%–99.93%), when compared to a manually abstracted dataset.
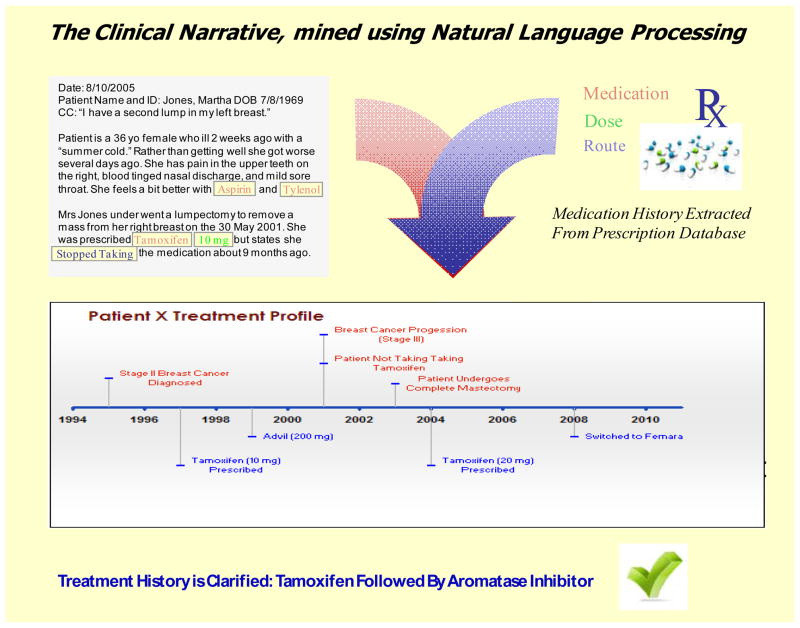
Figure 3 illustrates how the combination of prescription database querying, and NLP techniques applied to the clinical narrative, can be merged to assemble an automated treatment classification. Similar efforts are underway, in other systems of care (41), and for other drug-gene interactions (42). McAlpine and colleagues have effectively scanned EMR data to quantify drug exposure (including dose), as part of an investigation to determine the strength of association between genetic variability in CYP2D6 and therapeutic response to antidepressants.
Figure 3. Structured and unstructured data generate high-quality phenotypes.
Upper Left: Recent advances in Natural Language Processing (NLP) allow extremely accurate reconstruction of comprehensive medication histories. Upper Right: Structured medication data generated by computerized provider order entry software (e.g. name-value pairs, such as “medication = tamoxifen”) can be easier to collate and analyze. However, structured data must be normalized across diverse systems of care. The National Library of Medicine (NLM) has developed a terminology called RxNorm [http://www.nlm.nih.gov/research/umls/rxnorm/], linking drug names (dose, ingredient, and formulation) with drug vocabularies commonly used in pharmacy management systems (e.g., First Databank, Micromedex, MediSpan, MediSpan, Gold Standard Alchemy, Multum). Bottom: Structured and unstructured data can be merged to yield high-quality drug exposure phenotypes that facilitate pharmacogenomic studies using EMRs.
Thus, by altering drug levels in the circulation (or within a tissue microenvironment), many genetic predictors of toxicity (e.g., variable drug oxidation by cytochromes P450) are also known to alter drug efficacy. Genetic variability in Phase I drug metabolism of some statins not only influences the severity of ADRs (as noted above), it also impacts the degree to which these drugs decrease low density lipoprotein (LDL) cholesterol levels (43). Since the effect size appears to be relatively small (<10 mg/dl difference in LDL lowering per copy of the minor allele), the clinical significance of these findings remains uncertain (43).
Variability in pharmacodynamic candidate genes can have a similar effect. Variants in HMG CoA reductase clearly influence the lipid lowering efficacy of statins (44). Large, multi-center efforts are now underway to identify additional genes influencing statin ef cacy, primarily through the retrospective genotyping of archived biological materials obtained during prior randomized clinical trials (11). However, restricting these efforts to treatment trials would produce only very limited information because most efficacy data obtained during trials are limited to a single dose. Complete characterization of drug response requires a consideration of potency (ED50) as well as maximal efficacy. These properties can only be determined in patients exposed to multiple drug doses, and may best be derived from practice-based data.
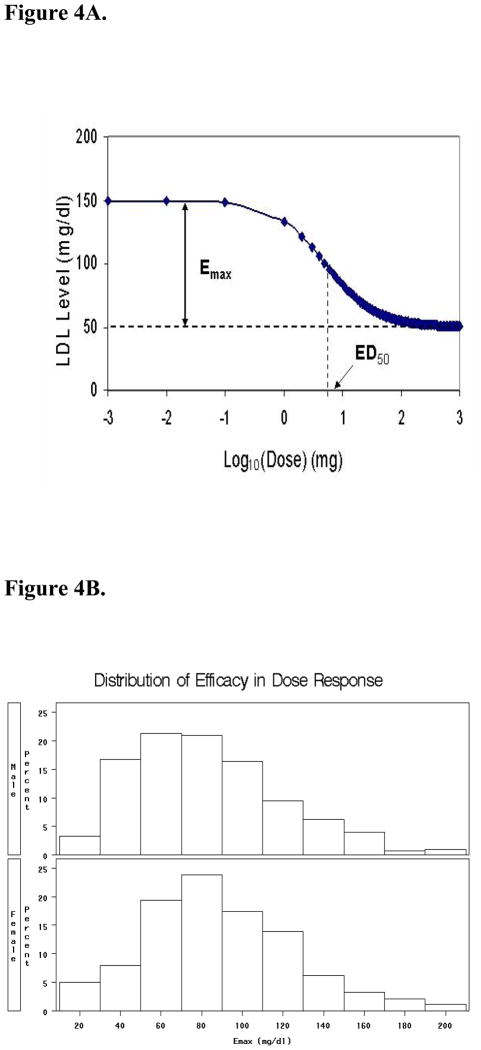
Without full characterization of efficacy in subjects exposed to drugs across a wide range of doses, clinical phenotypes cannot be fully characterized. To begin identifying genetic markers associated with the LDL cholesterol-lowering effect of atorvastatin across multiple doses, we previously interrogated EMR data contained within a large population-based biobank (27). NLP software was used to generate comprehensive retrospective drug exposure histories for the entire database (n = 20,000). For statin exposure, these algorithms were 100% sensitive and 96% specific, with an initial PPV of 87%. Through manual chart abstraction, these algorithms were optimized using programming that corrected for dosing discrepancies attributed to pill splitting; final PPV was 95%. Full dose response relationships were then constructed for all biobank participants exposed to atorvastatin. The result was a nested cohort of 3710 individuals for whom we subsequently derived rigorous phenotypic parameters for atorvastatin potency (ED50) and maximal atorvastatin efficacy (Emax) (27). The distribution of these traits is shown in Figure 4.
Figure 4. Quantifying drug efficacy within large populations, using EMRs.
4A. Dose-response for atorvastatin. LDL cholesterol plotted by dose.
4B. Gender-stratified distribution for Emax within an EMR biobank
The application of algorithms rigorously characterizing efficacy within EMR data is now being expanded to other classes of drugs across diverse systems of care. Through an initiative led by the PGRN (http://www.pharmgkb.org/contributors/pgrn/pat_profile.jsp), these approaches are being explored for the accurate characterization of drug outcome within multiple nodes of the HMO Research Network (HMORN). Network members include Harvard Pilgrim, and Fallon Healthcare (in the Northeastern US), Kaiser Permanente Georgia (in the Southeastern US), HealthPartners, Henry Ford, and Marshfield Clinic (in the Midwestern US), Kaiser Permanente Colorado, Kaiser Permanente Northwest, Group Health Cooperative, Lovelace Clinic, Kaiser Permanente Hawaii, Kaiser Permanente Southern California, and Kaiser Permanente Northern California (in the Western US). These institutions provide care to >10 million individuals (8).
Cross-network consortia are being established, and more than 250,000 DNA samples are already in hand. Kaiser Permanente Northern California, in particular, has recently embarked on one of the most ambitious biobanking efforts ever undertaken. As biological materials are linked to additional EMRs in these and other systems of care, the resulting data will advance the field of pharmacogenomics in two distinct ways: first, by assessing the generalizability of previously identified drug response genes within the community; and second, by facilitating discovery of previously unrecognized determinants of drug outcome in the context of clinical covariates.
III. Vision for the Future
The knowledge base relating variable outcomes in drug therapy to genomic variation is rapidly expanding. As noted above, the construction and integration of biobanks linked to EMRs can follow a variety of designs. Biobanks enrolling subjects through the process of informed consent must comply with laws regarding privacy, at the local, state and federal level (10,12). Clinical data and biological materials that are de-identified in accordance with provisions of 45 CFR 46 may be used for ‘non-human subjects’ research (15). As these and other EMR-based data sources are merged for large scale pharmacogenomic studies, all efforts must have ongoing oversight by Institutional Review Boards, internal and external ethics committees, community advisory boards, legal departments, and the Federal Office of Human Research Protection.
Data security and privacy are paramount. The Safe Harbor provision of the HIPAA Privacy Rule requires the removal of 18 personal identifiers (including demographic data) prior to any form of public disclosure. Additional layers of security are being developed and tested as the clinical and scientific communities manage increasingly complex genomic datasets (45).
Decision Support
Further, as efforts to merge biobanks grow, there is increasing difficulty in envisioning how pharmacogenomic knowledge can be brought to the bedside without advanced informatics tools that would include examination of the levels of evidence, as well as advice on how to manage individual subjects. The scientific and clinical communities have only begun to leverage the wide variety of phenotypic data available in EMRs to optimize studies of drug outcome. Medication information from the structured (e.g., electronic prescribing systems) and free-text components of each EMR (e.g., from clinical notes) will need to be merged, as shown in Figure 3. Additional data sources will allow assessment of adherence. Pharmacy claim data have been standardized for many years, and are widely used in health services research, especially for Pharmacy Benefit Management programs, Medicaid, and, recently, Part D of Medicare.
Data normalization strategies (using locally-adopted formats and terminologies) need to be shared across large healthcare networks. The mapping of clinically-relevant terms to community- vetted and -adopted ontologies will ensure semantic inter-operability and ease of study replication. Extending the example developed in Figure 3, variations in terms representing exposure to tamoxifen can be mapped to the same RxNORM code (10324), so that investigators avoid the creation of exhaustive lists of terms representing the same phenotypic data element.
Normalization procedures such as these will enable the establishment of large patient registries, with unique biological and clinical characteristics, across-institutions. Supported by NIH funding, many medical centers are already applying NLP techniques for information extraction from the clinical narrative to investigate rare medication side effects prospectively. Although the need for NLP analysis can be reduced by increased structured data entry in EMRs, novel approaches to the characterization of free text will continue to be essential. For example, while structured applications in medical oncology now automatically encode chemotherapy regimens ordered (and medications actually delivered to the patient), free text interrogation is still needed to understand why divergence occurs between some orders and the actual delivery.
The rapid advancements in electronic phenotyping approaches introduced above have occurred in parallel with equally robust progress in the area of genotyping technologies. Genome-wide SNP scanning arrays have become increasingly dense (e.g., now containing >1 million SNPS), and increasingly cost-effective, over the past decade. As a result, more genetic information is available at lower cost. Exome scanning (i.e., a focused approach that sequences all exons for all genes) is being conducted in cohorts of increasing sample size, and it seems inevitable that entire genomes will soon be included in each individual patient’s EMR (1,2).
Ultimately, the clinical utility of this phenotypic and genotypic information will depend upon biomedical informatics application platforms that provide efficient real-time decision support at the point of care. Furthermore, the expansion of knowledge generated by ongoing genotype-phenotype association studies will make it imperative that decision support platforms are flexible enough to incorporate future knowledge. Decision support software must allow reinterpretation of clinical genetic data based upon discoveries not yet made (46). There is also a considerable need to educate and train healthcare professionals in the use of these data. A recent survey carried out by the American Medical Association (AMA), in collaboration with Medco health solutions Inc., found that although 98% of provider participants agreed about the utility of genetic testing in drug therapy, only 10% actually felt that they had been adequately informed about the process. Only 26% had received some form of formalized training (https://www.medcoresearch.com/community/pharmacogenomics/physicansurvey). This stands in strong contrast to interview data which clearly indicate that most patients expect their healthcare professional to explain the clinical utility of pharmacogenetic tests (47).
Clinical Implementation
In order to ease the transition of this information into routine clinical practice, Gardner and colleagues have proposed the addition of genetic information to an existing drug interaction database, to further enhance clinical decision support and optimize patient safety (48). This approach is clinically appealing. Relevant genetic data can be formatted as an Extensible Markup Language (XML) document, and the additional information can be added using XML tags, and implemented through existing software. Such software is typically already in place at many large medical centers. The real challenge will be when and how to use this information. Potential application paradigms include (A) gene-based drug selection, (B) gene-based drug dosing, (C) medication reconciliation, (D) “push” phenotyping, and (E) pharmacovigilance.
Clearly an individual’s maintenance dose of warfarin is strongly influenced by variability in pharmacodynamic and pharmacokinetic candidate genes, and this relationship can be utilized to inform the process of gene-based drug dosing. Although similar strategies are being developed for managing the impact of pharmacokinetic candidate genes on clopidogrel efficacy, gene-based drug selection (the prescription of an alternate thienopyridine, for example) represents a suitable alternative. It is important to note, however, that these strategies need not be limited to the period of drug initiation. Both can be applied, in the context of a variety of drugs, during the process of medication reconciliation, particularly when patients move from a system of care without an EMR to a system of care with an EMR (49).
Furthermore, even in the absence of genetic information, EMRs can facilitate gene-based drug dosing by flagging patients in real time who appear to be developing an ADR, and prompting providers to consider genotyping such patients in an effort to optimize risk assessment. EMRs can also be used, at the population level, to identify trends in the development of new ADRs, pharmacovigilence, especially during the postmarketing period. Compelling data now support the claim that cardiovascular ADRs related to the use of highly potent COX-2 inhibitors may have been identified earlier using such an EMR-based approach (50).
In 2007, the US Congress also passed the Food and Drug Administration Amendments Act, directing FDA to increase the post market monitoring of drug safety. To accomplish this, FDA established the “Sentinel Initiative” with goals of detecting drug safety signals earlier and more accurately. This is now being accomplished through the real-time monitoring of EMRs and medical claims databases. The monitoring of multiple databases throughout the health care system requires the sophisticated integration of diverse EMR databases. While many cross-institutional networks have already been constructed to facilitate this process (e.g., the PGRN, the eMERGE network, and the HMO Research Network), it is likely that new funding initiatives and patient advocacy groups will drive the implementation of even larger consortia. Such efforts will generate the data needed to define the role of EMRs in pharmacogenomics and drug safety.
Outlook
Pharmacogenetic associations of various effect sizes are streaming into the literature at an increasing rate. It is not possible for individual practitioners to keep track of these relationships without assistance from information technology systems. However, with electronic decision support, clinical practice now has the potential to utilize this information. Genetic data will likely soon be deposited preemptively into each patient’s EMR, and robust biomedical informatics platforms will be positioned to interrogate this information during the process of clinical decision making, in real time. It appears that EMRs have brought personalized medicine to our doorstep.
Acknowledgments
R01DK080007, U01HL069757, U01HG004608, U01 HL65962, RC2GM092618, R01CA139246, NCI DCCPS supplement to U01 HG 04599, UO1GM61388, Breast SPORE CA 116201, U54LM008748, R01GM088076, U01GM061373.
Footnotes
WEBLINKS
[https://www.i2b2.org ] June 6, 2010
[https://www.ohnlp.org] June 13, 2010
[http://informatics.mayo.edu ] June 6, 2010
[http://www.computerworld.com ] March 2, 2010
[http://www.ama-assn.org ] August 20, 2010
[http://www.nlm.nih.gov/research/umls/rxnorm/] June 6, 2010
[http://www.nlm.nih.gov/research/umls/] June 13, 2010
[http://PharmGKB.org ] August 20, 2010
[http://www.gwas.net ] March 18, 2010
References
- 1.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–35. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lifton RP. Individual genomes on the horizon. N Engl J Med. 2010;362:1235–6. doi: 10.1056/NEJMe1001090. [DOI] [PubMed] [Google Scholar]
- 3.DesRoches CM, et al. Electronic health records in ambulatory care--a national survey of physicians. N Engl J Med. 2008;359:50–60. doi: 10.1056/NEJMsa0802005. [DOI] [PubMed] [Google Scholar]
- 4.Jha AK, et al. Use of electronic health records in U.S. hospitals. N Engl J Med. 2009;360:1628–1638. doi: 10.1056/NEJMsa0900592. [DOI] [PubMed] [Google Scholar]
- 5.Shea S, Hripcsak G. Accelerating the use of electronic health records in physician practices. N Engl J Med. 2010;362:192–195. doi: 10.1056/NEJMp0910140. [DOI] [PubMed] [Google Scholar]
- 6.Pakhomov S, Bjornsen S, Hanson P, Smith S. Quality performance measurement using the text of electronic medical records. Med Decis Making. 2008;28:462–70. doi: 10.1177/0272989X08315253. [DOI] [PubMed] [Google Scholar]
- 7.Denny JC, Miller RA, Waitman LR, Arrieta MA, Peterson JF. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform. 2009;78 (Suppl 1):S34–42. doi: 10.1016/j.ijmedinf.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KA, et al. The HMO Research Network. In: Strom BL, editor. Pharmacoepidemiology. 4. West Sussex, England: John Wiley & Sons Ltd; pp. 261–270. [Google Scholar]
- 9.Xu H, Stenner S, Doan S, Johnson K, Waitman L, Denny JC. MedEx – a medication information exaction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty CA, Wilke RA. Biobanking and pharmacogenetics. Pharmacogenom. 2010;11:637–641. doi: 10.2217/pgs.10.13. [DOI] [PubMed] [Google Scholar]
- 11.Barber MJ, et al. Genome-wide association of lipid-lowering response to statins in combined study populations. PLoS One. 2010;5(3):e9763. doi: 10.1371/journal.pone.0009763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarty CA, et al. Marsh eld Clinic Personalized Medicine Research Project (PMRP): design, methods and recruitment for a large population-based biobank. Personalized Med. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 13.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ormond KE, Cirino AL, Helenowski IB, Chisholm RL, Wolf WA. Assessing the understanding of biobank participants. Am J Med Genet A. 2009;149A(2):188–98. doi: 10.1002/ajmg.a.32635. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–72. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald CJ. Protocol-based computer reminders, the quality of care and the non-perfectability of man. N Engl J Med. 1976;295:1351–5. doi: 10.1056/NEJM197612092952405. [DOI] [PubMed] [Google Scholar]
- 17.McDonald CJ, Murray R, Jeris D, Bhargava B, Seeger J, Blevins L. A computer-based record and clinical monitoring system for ambulatory care. Am J Public Health. 1977;67:240–5. doi: 10.2105/ajph.67.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meystre SM, et al. Extracting Information from Textual Documents in the Electronic Health Record: A Review of Recent Research. Methods Inf Med. 2008;47(Suppl 1):128–144. [PubMed] [Google Scholar]
- 19.Evans WE, et al. Conventional compared with individualized chemotherapy for childhood acute lymphoblastic leukemia. N Engl J Med. 1998;338:499–505. doi: 10.1056/NEJM199802193380803. [DOI] [PubMed] [Google Scholar]
- 20.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–91. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 22.Wilke RA, et al. Identifying genetic risk factors for serious adverse drug reactions – current progress and challenges. Nature Rev Drug Discov. 2007;6:904–916. doi: 10.1038/nrd2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein TE, et al. for the International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman C. A broad-coverage natural language processing system. Proc AMIA Symp. 2000:270–4. [PMC free article] [PubMed] [Google Scholar]
- 25.Chhieng D, et al. Use of natural language programming to extract medication from unstructured electronic medical records. AMIA. 2007:908. [PubMed] [Google Scholar]
- 26.Jagannathan V, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform. 2009;78:284–91. doi: 10.1016/j.ijmedinf.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Wilke RA, Berg RL, Linneman JG, Zhao CF, McCarty CA, Krauss RM. Characterization of LDL-cholesterol lowering efficacy for atorvastatin in a population-based DNA Biorepository. Basic Clin Pharmacol Toxicol. 2008;103:354–359. doi: 10.1111/j.1742-7843.2008.00291.x. [DOI] [PubMed] [Google Scholar]
- 28.Vandenbroucke JP, Psaty BM. Benefits and risks of drug treatments: how to combine the best evidence on benefits with the best data about adverse effects. JAMA. 2008;300:2417–9. doi: 10.1001/jama.2008.723. [DOI] [PubMed] [Google Scholar]
- 29.McClure DL, Valuck RJ, Glanz M, Murphy JR, Hokanson JE. Statin and statin-fibrate use was significantly associated with increased myositis risk in a managed care population. J Clin Epidemiol. 2007;60:812–818. doi: 10.1016/j.jclinepi.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Mareedu RK, et al. Use of an electronic medical record to characterize cases of intermediate statin-induced muscle toxicity. Prev Cardiol. 2009;12:88–94. doi: 10.1111/j.1751-7141.2009.00028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilke RA, Reif DG, Moore JH. Combinatorial pharmacogenetics. Nature Rev Drug Discov. 2005;4:911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- 32.Prueksaritanont T, et al. Effects of fibrates on metabolism of statins in human hepatocytes. Drug Metab Dispos. 2002;30:1280–1287. doi: 10.1124/dmd.30.11.1280. [DOI] [PubMed] [Google Scholar]
- 33.Riedmaier S, et al. UDP-glucuronosyltransferase (UGT) polymorphisms affect atorvastatin lactonization in vitro and in vivo. Clin Pharmacol Ther. 2010;87:65–73. doi: 10.1038/clpt.2009.181. [DOI] [PubMed] [Google Scholar]
- 34.Link E, et al. The SEARCH Collaborative Group. SLCO1B1 variants and statin-induced myopathy-a genomewide study. N Engl J Med. 2008;359:789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 35.Keskitalo JE, Zolk O, Fromm MF, Kurkinen KJ, Neuvonen PJ, Niemi M. ABCG2 polymorphism markedly affects the pharmacokinetics of atorvastatin and rosuvastatin. Clin Pharmacol Ther. 2009;86:197–203. doi: 10.1038/clpt.2009.79. [DOI] [PubMed] [Google Scholar]
- 36.Onitilo AA, et al. Estrogen receptor genotype is associated with risk of venous thromboembolism during tamoxifen therapy. Breast Cancer Res Treat. 2009;115:643–50. doi: 10.1007/s10549-008-0264-2. [DOI] [PubMed] [Google Scholar]
- 37.Lash TL, et al. Breast cancer recurrence risk related to concurrent use of SSRI antidepressants and tamoxifen. Acta Oncol. 2010;49:305–12. doi: 10.3109/02841860903575273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dezentjé VO, et al. Effect of concomitant CYP2D6 inhibitor use and tamoxifen adherence on breast cancer recurrence in early-stage breast cancer. J Clin Oncol. 2010;28:2423–9. doi: 10.1200/JCO.2009.25.0894. [DOI] [PubMed] [Google Scholar]
- 39.Kelly CM, et al. Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ. 2010;340:c693. doi: 10.1136/bmj.c693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aubert RE, et al. Risk of breast cancer recurrence in women initiating tamoxifen with CYP2D6 inhibitors. J Clin Oncol. 2009;27:9s abstr CRA508. [Google Scholar]
- 41.Liao K, et al. Electronic Medical Records for Discovery Research in Rheumatoid Arthritis. Arthritis Care and Research. 2010 August;62(8) doi: 10.1002/acr.20184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McAlpine DE, O’Kane DJ, Black JL, Mrazek DA. Cytochrome P450 2D6 genotype variation and venlafaxine dosage. Mayo Clin Proc. 2007;82:1065–8. doi: 10.4065/82.9.1065. [DOI] [PubMed] [Google Scholar]
- 43.Kivistö KT, et al. Lipid-lowering response to statins is affected by CYP3A5 polymorphism. Pharmacogenet. 2004;14:523–5. doi: 10.1097/01.fpc.0000114762.78957.a5. [DOI] [PubMed] [Google Scholar]
- 44.Krauss RM, et al. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circ. 2008;117:1537–44. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 45.Loukides G, et al. Anonymization of electronic medical records for validating genome-wide association studies. Proc Natl Acad Sci U S A. 2010;107:7898–903. doi: 10.1073/pnas.0911686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell DR, Mitchell JA. Status of clinical gene sequencing data reporting and associated risks for information loss. J Biomed Inform. 2007;40:47–54. doi: 10.1016/j.jbi.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 47.Fargher EA, Eddy C, Newman W, Qasim F, Tricker K, Elliott RA, Payne K. Patients’ and healthcare professionals’ views on pharmacogenetic testing and its future delivery in the NHS. Pharmacogenom. 2007;8:1511–9. doi: 10.2217/14622416.8.11.1511. [DOI] [PubMed] [Google Scholar]
- 48.Gardner D. Using genomics to help predict drug interactions. J Biomed Inform. 2004;37:139–46. doi: 10.1016/j.jbi.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 49.Bassi J, Lau F, Bardal S. Use of information technology in medication reconciliation: a scoping review. Ann Pharmacother. 2010;44:885–97. doi: 10.1345/aph.1M699. [DOI] [PubMed] [Google Scholar]
- 50.Brownstein JS, Sordo M, Kohane IS, Mandl KD. The Tell-Tale Heart: Population-Based Surveillance Reveals an Association of Rofecoxib and Celecoxib with Myocardial Infarction. PLoS One. 2007;2(9):e840. doi: 10.1371/journal.pone.0000840. [DOI] [PMC free article] [PubMed] [Google Scholar]