Abstract
MicroRNAs (miRNAs) are small non-coding RNAs, which regulate gene expression by inhibiting translation or promoting degradation of specific target messenger RNAs (mRNAs). Alteration of the levels of a number of miRNAs is common in solid and hematological tumors. We have shown previously that miR-214 regulates Ezh2 in skeletal muscle and embryonic stem cells. The current study was aimed at examining the role of miR-214 in breast cancer where miR-214 levels are reduced but whether this phenomenon bears a functional relevance is unknown. MiR-214 expression was inversely correlated with Ezh2 mRNA and protein levels in breast cancer cell lines and at least one copy of the miR-214 alleles was found to be deleted in 24% (6/25) of primary breast tumors. Experimental increase of miR-214 in breast cancer cell lines correlated with reduction of Ezh2 protein levels, a known marker of invasion and aggressive breast cancer behavior. Supporting a direct targeting mechanism, miR-214 decreased luciferase activity from a construct containing the Ezh2 3′ untranslated region. Expression of miR-214 specifically reduced cell proliferation of breast cancer cells and inhibited the invasive potential of a highly metastatic breast cancer cell line. These findings indicate that reduced miR-214 levels may contribute to breast tumorigenesis by allowing abnormally elevated Ezh2 accumulation and subsequent unchecked cell proliferation and invasion.
Introduction
MicroRNAs (miRNAs) are 20–23 nucleotides-long non-coding RNAs expressed in a cell- and tissue-specific manner, which act by negatively regulating the stability and translational efficiency of their target messenger RNA (mRNAs) (1,2). MiRNAs have been implicated in the initiation and progression of cancer and miRNA loci are frequently located at fragile sites and genomic regions involved in cancer where they exhibit copy number alterations (3,4). Expression of the majority of miRNAs is reduced in human cancers (5–7). However, in some instances, overexpression of specific miRNAs promotes tumorigenesis (8). These observations suggest that miRNAs can function either as tumor suppressors or oncogenes, conferring a predictive diagnostic value to miRNA expression (9). In solid tumors, such as stomach, pancreatic and prostate cancer, alteration of the levels of a small number of miRNAs, including miR-214, has been identified as a signature for these tumors (10,11). In breast cancer, miR-214 expression is reduced, however, the functional relevance of this finding remains unaddressed (10,11).
The Polycomb group (PcG) proteins contribute to the maintenance of cell identity by regulating chromatin function and transcriptional repression (12). Ezh2 is the catalytic component of the Polycomb repressive complex 2 (PRC2) that mediates epigenetic silencing by trimethylating histone H3 lysine 27 (H3K27me3) (13). Ezh2 is preferentially expressed in embryonic tissues and present at low levels in terminally differentiated adult tissues (14), where it acts as a negative regulator of cell differentiation (15–17). In addition, Ezh2 promotes cell proliferation (18,19). Upregulation of Ezh2 mRNA and protein levels were first reported in metastatic prostate cancers (20). Subsequently, Ezh2 overexpression was noted in several neoplasias, including gastric tumors (21), melanoma (22), bladder cancers (23), lymphomas (19) and breast cancers (24). Increased levels of Ezh2 are also observed in non-invasive forms of cancer such as ductal in situ carcinoma (DISC) and atypical hyperplasia. Moreover, Ezh2 was shown to promote neoplastic transformation of breast epithelial cells suggesting that Ezh2 may contribute to the initiation and progression of breast cancer (25). Overexpression of Ezh2 promotes invasion of normal breast cell lines and increased Ezh2 protein levels predict breast cancer aggressiveness and poor clinical outcome (24,26). However, the mechanisms leading to increased expression of Ezh2 in breast tumors are poorly understood.
It has been recently reported that reduced miR-101 levels are associated with increased Ezh2 accumulation in bladder, prostate, gastric and breast cancer (27,28). MiR-101 targets the Ezh2 3′ UTR (untranslated region), promoting its translational inhibition (24). Ezh2 mRNA and protein are upregulated in breast cancer and correlate with tumor aggressiveness. However, deletion of miR-101 was detected in only 55% of the analyzed breast tumors (28). In addition to miR-101, several other miRNAs are predicted to target Ezh2 3′ UTR, including miR-214 (29). MiR-214 expression is reduced in breast cancer and human breast cancer cell lines (10,11). However, the role of miR-214 in this neoplasia is unknown.
Here, we report that miR-214 expression is inversely correlated with Ezh2 mRNA and protein levels in MCF-7 and in the invasive MDA-MB-231 breast cancer cell line and that deletion of at least one copy of the miR-214 genomic alleles occurs in 24% (6/25) cases of breast cancer examined. Overexpression of miR-214, but not that of a mutant miR-214 version, results in inhibition of breast cancer cell proliferation and invasion. Moreover, miR-214 causes inhibition of a reporter construct containing the Ezh2 3′ UTR and reduces endogenous Ezh2 protein levels in both MCF-7 and MDA-MB-231 cells. These results offer a further insight into Ezh2 misregulation and establish a link between miR-214 and Ezh2 in breast cancer cells.
Materials and methods
Plasmids and oligomers
To generate the miR-214 plasmid construct, a genomic fragment of primary miR-214 was amplified by polymerase chain reaction (PCR) and inserted into the BamHI and EcoRI sites of the plasmid vector pcDNA3 (Invitrogen, Carlsbad, CA). The mutated pcDNA-miR-214 construct was generated by site-directed mutagenesis using the QuickChange kit (Stratagene, La Jolla, CA). Human Ezh2 silencing RNA (siRNA) and double-stranded oligomer corresponding to the mature miR-101, miR-214 or miR-214 mutant as well as the anti-miR-214 were purchased (Invitrogen).
Tissues and genomic PCR analyses
Human breast cancer tissues were obtained with Institutional Review Board approval at the University of Singapore/National University Hospital, Singapore (NUS/NUH). MiR-214 and miR-101 gene loci were examined using comparative genomic hybridization and the Agilent oligoarray 244K for copy number analysis, as previously described previously (28).
Cell culture and transfections
MCF-10A cells were purchased from Lonza (Allendale, NJ) and were grown in mammary epithelial cell growth medium (MEGM) supplemented with hydrocortisone, insulin and human epidermal growth factor (Lonza, Annendale, NJ). MCF-7 and MDA-MB-231 were purchased from American Type Culture Collection (Rockville, MD). Cells were grown in monolayer culture in high glucose Dulbecco's modified Eagle's medium (Invitrogen), containing l-glutamine, 1.5 g/l sodium carbonate and supplemented with 10% fetal bovine serum (Invitrogen) and antibiotic solution (10 000 U Penicillin, 10 000 μg Streptomycin; Invitrogen). For stable cell lines, pcDNA, miR-214 or miR-214 mutant plasmids were transfected in MCF-7 or MDA-MB-231 cells using Lipofectamine 2000 according to the manufacturer’s procedure (Invitrogen). Transfectant cells were selected with 0.6 or 1 mg/ml of geneticin (G-418) for 2 weeks. Pooled clones (n > 50) were employed for subsequent experiments. For transient transfections, MCF-7 and MDA-MB-231 cells were transfected for 48–72 h with either Ezh2 siRNA, green fluorescent protein-siRNA or 1, 10 or 100 nmol of wild-type or mutant miR-214 oligomers. Cells generated from transient or stable expression were processed for RNA extraction and immunoblot performed according to standard procedures.
Real-time PCR and miRNA TaqMan real-time PCR analyses
Total RNA (0.5–1 μg) was reverse transcribed using cDNA synthesis kit (Applied Biosystems, Foster City, CA). About 2 μl of a 1:10 dilution of the synthesized cDNA were used for quantitative real-time PCR, using SyberGreen PCR MasterMix (Applied Biosystems) on the Mx3000P Real-Time PCR thermocycler (Stratagene). Relative mRNA levels were determined by comparing threshold cycles of amplified genes with GAPDH or beta-actin using the ΔCT method. For miRNA quantitative real-time PCR, single-stranded cDNA from RNA samples was prepared using the TaqMan® MicroRNA Reverse Transcription Kit (Applied Biosystems). Real-time PCR was performed using Taqman miRNA probes and TaqMan universal PCR master mix from Applied Biosystems. Data were normalized to the signal obtained from the RNU6B control probe. The oligonucleotide sequences used as primers or probes for reverse transcription (RT)–PCR or real-time PCR are listed in supplementary Table 1, available at Carcinogenesis Online.
Luciferase reporter analysis
The Ezh2-3′ UTR reporter construct (pEzh2-UTR-Luc) consists of the mouse Ezh2-3′ UTR gene (+2577/+3428 from the start of transcription) inserted downstream of the luciferase reporter gene in the pGL3 control vector (Promega, Madison, WI). A mutant form was also generated (mut-Ezh2-UTR) using direct site mutagenesis (Stratagene). Cells were transfected with 200 ng of the pEzh2-3′ UTR reporter construct using Lipofectamine 2000 (Invitrogen). Following 48 h, cells were processed for luciferase activity. In designated experiments, 100 nmol of miR-214 or miR-101 or the combination of both miRNAs were transfected in MCF-7 and MDA-MB-231 in the presence of either pGL3 or pGL3-Ezh2-3′ UTR reporter construct. In all these experiments, 10 ng of Renilla reporter construct (Promega) was cotransfected to normalize for differences in transfection efficiency.
Immunoblots
Cells were lysed, their extracts resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto nitrocellulose filters. Immunoblots were performed with the following antibodies: anti-Ezh2, Bmi1, phosphatase and tensin homolog, E-cadherin (Cell Signaling, Danvers, MA), E7 tubulin antibody (Developmental Studies Hybridoma Bank, Iowa City, IA) and anti-H3K27me3 (Upstate Biotechnology, Temecula, CA), anti-histone H3 (Abcam, Cambridge, MA), anti-cyclin D and E polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Densitometry analyses were performed using NIH image software. Bands from blots were scanned, and signal intensities were calculated by subtracting local background from total intensities and relative intensities were graphed after normalization to the corresponding corrected control intensities for actin.
Cell proliferation analysis
For cell growth experiments, MCF-7 and MDA-MB-231 cells stably expressing the miR-214, the mutant miR-214 form or the empty vector (pcDNA) were plated at 12.5 × 103 cells/cm2 in a 12-well plate in duplicates and cell numbers were determined at 24, 48 and 72 h. For Ezh2 siRNA experiments, cells were counted 24, 48 and 72 h posttransfection. For 5′-bromo-2′-deoxyuridine (BrdU) staining, cells were pulsed with BrdU for 4 h, then washed with phosphate-buffered saline and fixed in 4% paraformaldehyde, permeabilized for 15 min in 0.2% Triton-1× phosphate-buffered saline. Cellular DNA was denatured in 1.5N HCl for 25 min at 37°C and cells were neutralized in 0.1 M borate buffer, pH 8.5. Cells were incubated overnight with the anti-BrdU mouse monoclonal antibody (GE Healthcare, Piscataway, NJ) followed by incubation with anti-mouse IgG-Alexa 488 (Molecular Probes). Cells were counted in 10 random fields and the ratio of BrdU/DAPI stained cells was calculated.
Cell invasion analysis
MDA-MB-231 cells stably expressing the miR-214, miR-214 mutant or the empty vector pcDNA were plated at 5 × 103 cells/cm2 in a 10 cm plate and cultured overnight in Dulbecco's modified Eagle's medium medium containing 10% fetal bovine serum then cells were serum starved. MCF-7 cells were transfected with 100 nM of anti-miR-214 oligomers using Lipofectamine (Invitrogen). Following 24 h of transfection, cells were serum starved for another 24 h. MCF-7 and MDA-MB-231 cells were collected following trypsin treatment. Cells were then seeded on a laminin-coated plate (Calbiochem-EMD Chemicals, Gibbstown, NJ). Fetal bovine serum containing medium was added to the lower chamber as a chemoattractant. After 24 h, the invading cells were dissociated from the lower side of the insert and stained in a cell detachment solution containing calcein. Fluorescence was measured as recommended (Calbiochem, La Jolla, CA). Alternatively, cells were seeded on a laminin-coated 24-well insert (Calbiochem-EMD Chemicals) and incubated for 48–72 h. Non-invading cells were gently removed from the top of the insert with a cotton swab. Invasive cells on the lower side of the inserts and in the 24-well plate were stained with crystal violet, air-dried and photographed.
Statistical analysis
For all experiments, statistical analyses were performed using three independent experiments conducted in triplicates and the mean ± SD (P ≤ 0.05) was determined by analysis of variance.
Results
miR-214 levels are reduced in breast cancer cells and miR-214 locus is deleted a subset of breast cancer
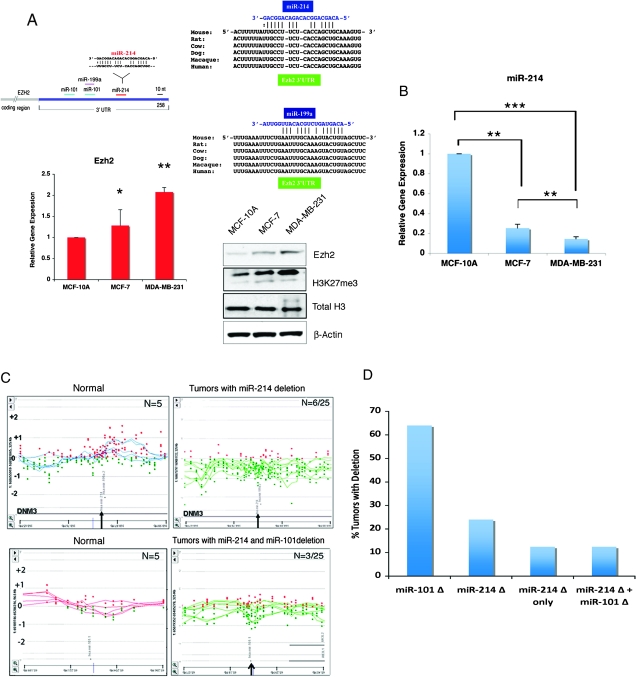
The Ezh2 3′ UTR contains an atypical miR-214 site which is highly conserved in mammals (Figure 1A). To determine whether miR-214 has a role in breast tumors, we first correlated miR-214 expression with Ezh2 mRNA and protein levels in the benign breast epithelial cells MCF-10A and in MCF-7 and MBA-MB-231 cancer cell lines. These cell lines are of luminal (MCF-10A and MCF-7) and basal origin (MDA-MB-231) and are associated with very low (MCF-7) and high (MDA-MB-231) invasiveness potential, respectively. Quantitative real-time RT–PCR (qPCR) and immunoblot of Ezh2 showed increased Ezh2 mRNA and protein levels in the MCF-7 and MBA-MB-231 cancer cells, when compared with the benign breast epithelial cells MCF-10A (Figure 1A). Consistent with Ezh2 upregulation, its enzymatic activity was also increased, as revealed by increased H3K27me3 levels (Figure 1A). The Ezh2 upregulation was inversely correlated with ∼4- and 7-fold reduction of mature miR-214 levels in the human MCF-7 and MDA-MB-231 breast cancer cell lines, respectively, when compared with the benign immortalized breast epithelial cells MCF-10A (Figure 1B). These differences were maintained when MCF-10A cells were grown in identical culture conditions as those of MCF-7 and MDA-MB-231 (data not shown).
Fig. 1.
Ezh2 mRNA and protein levels in breast cancer cell lines and miR-214 expression and genomic locus deletion in breast tumors. (A) Schematic of miR-214 site on the Ezh2 3′UTR (top left panel) and its conservation across mammalian species (top right panel), Ezh2 RT–qPCR (lower left panel) and Ezh2, H3K27me3 and total histone H3 immunoblot (lower right panel) of the benign immortalized breast epithelial cell line MCF-10A and the MCF-7 and MDA-MB-231 breast cancer cell lines. (B) miRNA TaqMan RT–PCR of miR-214 in MCF-10A, MCF-7 and MDA-MB-231 breast cell lines. The three cell lines were cultured in the same growth medium. Shown are levels of miR-214 corrected to RNU6B used as an internal control (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0002). (C) High resolution comparative genomic hybridization analysis of breast cancers show a non-focal deletion in a region containing miR-214 in a subset of tumors (right panel) but not in the benign samples (left panel). (D) Tumor samples from (C) harboring a deletion in miR-214 show focal deletion for miR-101 in 3/6 tumor samples (left panel) but not in the benign samples (right panel). Horizontal green and blue lines correspond to the ratio profile for copy number changes in each sample and the red and green dots indicate the probes represented on the array platform (Agilent 244K array CGH). (D) Graph represents miR-214 deletion frequency in primary breast tumors and their distribution as a function of the presence or absence of a concomitant miR-101 deletion.
Since miR-101 is known to target Ezh2 and was also found to be deleted in a large number of breast cancers (28), we investigated the genomic organization of the region encompassing the miR-214 locus. DNA obtained from 25 primary breast tumors and 5 matched normal samples was examined by comparative genomic hybridization using the Agilent oligoarray 244K for copy number determination. Six of the 25 tumor samples (24%) revealed loss of at least one copy of the miR-214 locus, as compared with matched normal breast tissue (Figure 1C). Since deletion of miR-101 was found in 55% of the tumors (28), we evaluated the status of miR-101 gene in the cancers with a miR-214 deletion. Of the six tumors harboring deletion of miR-214 locus, only three (50%) revealed deletion of the miR-101 locus as well (Figure 1D), whereas three of the six (50%) positive tumors displayed a selective miR-214 deletion (Figure 1C). These data indicate that, in some breast cancers, loss of miR-214 is not accompanied by concomitant deletion of miR-101 and suggest that increased Ezh2 levels in breast cancer and cell lines may result from reduction of miR-214 in a miR-101-independent manner.
miR-214 targets the Polycomb Ezh2 methyltransferase
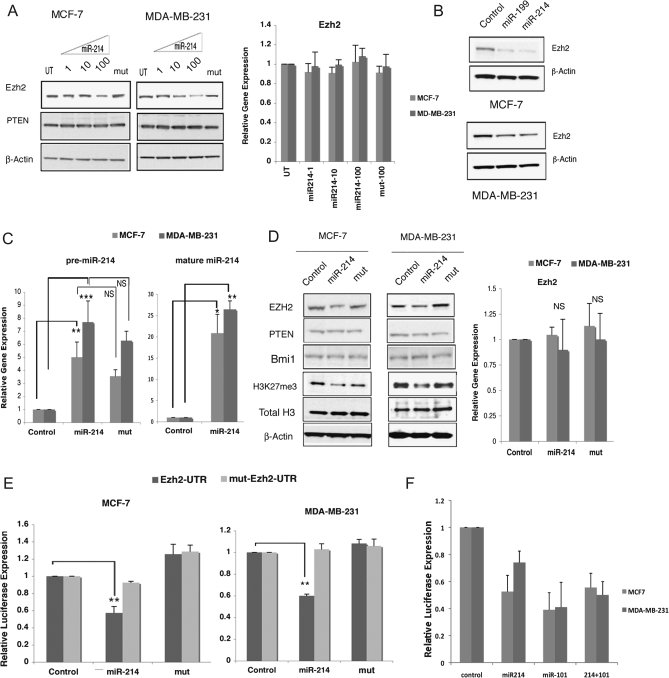
To examine the effect of miR-214 on Ezh2, MCF-7 and MDA-MB-231 cells were transiently transfected with oligonucleotides corresponding to mature miR-214 or its mutant counterpart. The results of these experiments showed that miR-214, but not its mutant form, specifically reduced the Ezh2 protein levels in a dose-dependent manner (Figure 2A, left panel). However, no change in Ezh2 mRNA was detected, as measured by RT–qPCR (Figure 2A, right panel). In addition to miR-214, a highly conserved miR-199 putative site is located within the Ezh2 3′ UTR (Figure 1A). Since miR-199a/a* is clustered with miR-214 and the two miRNAs are cotranscribed in several tissues and cell lines (30,31), we reasoned that miR-199 may also regulate Ezh2. Indeed, stable expression of a precursor miR-199 in MDA-MB-231 and MCF-7 cells resulted in a reduction of Ezh2 protein levels (Figure 2B). Since we have shown previously that miR-214 targets Ezh2 in skeletal muscle and embryonic stem cells, we decided to focus on the miR-214 role on breast cancer cells. MCF-7 and MDA-MB-231 cells were stably transfected with either a construct expressing precursor miR-214, a mutant form of miR-214 (mut) or control plasmid. RT–qPCR showed that in addition to the premature wild-type miR-214 and mut forms (Figure 2C, right panel), Dicer-processed mature miR-214 was also efficiently overexpressed in MCF-7 and MDA-MB-231 cells (Figure 2C, left panel). Immunoblot analysis of MCF-7 and MDA-MB-231 cell lines stably expressing precursor miR-214 showed reduced Ezh2 protein when compared with control plasmid, but not in cells expressing the miR-214 mutant form (Figure 2D). Ezh2 reduction well correlated with reduced H3K27 me3 levels (Figure 2D, left panel). However, Ezh2 RNA levels were not significantly changed in both MCF-7 and MDA-MB-231 cell lines stably expressing precursor miR-214 (Figure 2D, right panel), consistent with a role of miR-214 in posttranscriptional regulation of Ezh2 (29). We examined phosphatase and tensin homolog, a reported miR-214 target in ovarian cancer (6). In both MCF-7 and MDA-MB-231, miR-214 overexpression had no effect on phosphatase and tensin homolog, indicating that this protein is not regulated by miR-214 in breast cancer cell lines (Figure 2A and D). In addition, miR-214 expression did not affect the levels of Bmi1, a component of the PRC1 complex, indicating targeting specificity of miR-214 for Ezh2 (Figure 2D).
Fig. 2.
miR-214 overexpression reduces Ezh2 levels by targeting the 3′ UTR of Ezh2 in breast cancer cell lines. (A) Immunoblot of Ezh2 and phosphatase and tensin homolog (left panel) and Ezh2 mRNA by RT–qPCR (right panel in MCF-7 and MDA-MB-231 cells transiently transfected with increasing amounts (1,10 or 100 pmol, respectively) of wild-type or mutant (mut) miR-214 oligomers. (B) Immunoblot of Ezh2 in MCF-7 and MDA-MB-231 cells stably transfected with either a construct encoding the pre-miR-199 or the pre-miR-214. (C) miRNA TaqMan RT–PCR of mature miR-214 (right panel), and RT–qPCR of primary miR-214 and mutant miR-214 (mut) relative to control (left panel), in MCF-7 and MDA-MB-231 cells stably expressing either miR-214, mut or the control plasmid. (*P ≤ 0.05; **P ≤ 0.005; ***P ≤ 0.0002; NS, not significant. (D) Immunoblot (left panel) of Ezh2, phosphatase and tensin homolog, Bmi1, H3K27me3 and total histone H3 and Ezh2 mRNA RT–PCR (right panel) of MCF-7 and MDA-MB-231 cells stably transfected with either a construct encoding the pre-miR-214, the mutant form or an empty control vector. Shown is a representative experiment of three sets of stable transfectants, with β-actin as a control for equal loading of protein cell extract. (E) Luciferase activity of wild-type Ezh2-3′ UTR or the mutant, mut-Ezh2-3′ UTR reporter constructs in MCF-7 or MDA-MB-231 cells stably expressing either the wild-type pre-miR-214, the mutant form, mut or the control vector. The data represent the mean ± SD (**P ≤ 0.005) as determined from three independent experiments performed in triplicate. (F) Luciferase activity of wild-type Ezh2-3′ UTR or the pGl3 control vector in the presence of miR-214 or miR-101 or both oligomers in MCF-7 and MDA-MB-231. The data represent the mean ± SD as determined from two independent experiments done in triplicate.
To examine whether miR-214 directly targets the 3′ UTR of Ezh2, luciferase reporter constructs containing the wild-type or mutated form of the mouse Ezh2 3′ UTR were transfected in cells stably overexpressing either wild-type or mutant miR-214. In cells expressing miR-214, luciferase activity from the construct containing the Ezh2 3′ UTR was decreased by >40% in both MCF-7 and MDA-MB-231 cells (Figure 2E). Expression of the mutated miR-214 form did not induce any significant change in luciferase activity when compared with control cells (Figure 2E). Luciferase activity from a construct harboring a mutation in the miR-214 site of Ezh2 3′UTR was unaffected in MCF-7 and MDA-MB-231 cells expressing either miR-214 or its mutant form (Figure 2E). Collectively, these results indicate that miR-214 regulates Ezh2 protein accumulation in breast cancer cells by directly targeting Ezh2 3′ UTR. To test whether miR-101 and miR-214 can co-operate in reducing Ezh2 levels, miR-214, miR-101 or the combination of both miRNAs were transfected in MCF-7 and MDA-MB-231 cells in the presence of either pGL3 (control) or pGL3-Ezh2-3′ UTR reporter construct. Figure 2F shows that while either miR-214 or miR-101 reduced luciferase activity, their coexpression did not further downregulate luciferase activity.
miR-214 overexpression inhibits breast cancer cell proliferation
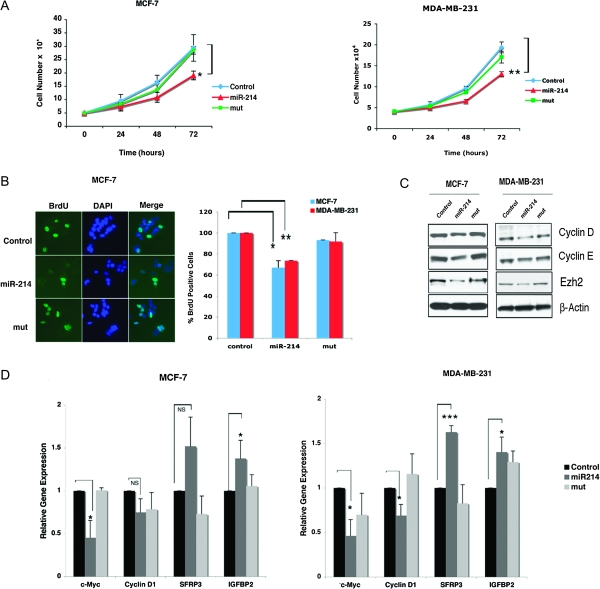
MiR-214 stable expression reduced cell growth of MCF-7 and MDA-MB-231 cells by >20 and 30% (P = 0.03 and P = 0.004), respectively, as measured by cell counts taken over a 72 h time course. However, no significant growth reduction was observed in mut-miR-214-expressing cells relative to control (Figure 3A). If the effects of miR-214 are mediated by Ezh2, reducing Ezh2 is expected to recapitulate the effects of miR-214 overexpression. To directly test this hypothesis, we transfected Ezh2 siRNA in MCF-7 and MDA-MB-231 cells. The results of these experiments are reported in supplementary Figure 1A, available at Carcinogenesis Online and indicate that, similarly to miR-214 overexpression (Figure 3A), Ezh2 reduction diminished cell proliferation, thus suggesting specificity for miR-214 effects on cell growth via Ezh2 down-modulation. To determine whether miR-214-mediated growth reduction was due to inhibition of cell proliferation, MCF-7 and MDA-MB-231 cells were subjected to BrdU incorporation analysis and examined by immunofluorescence (Figure 3B, left panel). MiR-214 expression reduced the proportion of BrdU-positive cells by 33% in MCF-7 (P = 0.019) and by 26% in MDA-MB-231 cells (P = 0.006), when compared with control cells In contrast, expression of miR-214 mut had no significant effect on the proliferation rate of MCF-7 and MDA-MB-231 (Figure 3B, right panel). MiR-214-mediated cell growth inhibition was further increased to 40% in both MCF-7 and MDA-MB-231 cells-expressing miR-214, when cells were synchronized by serum starvation prior to BrdU treatment (data not shown). To evaluate whether miR-214 inhibition of cell proliferation was associated with blockade of cell cycle mediators, we examined expression of cyclins D1 and E as well as Ezh2. Immunoblot showed that overexpression of miR-214 resulted in a reduction of cyclin D1, cyclin E and Ezh2 protein levels in both cell lines (Figure 3C). To further explore the regulation of Ezh2 by miR-214, we examined the expression of the Ezh2 downstream target genes cyclin D1, IGFBP2, SFRP3 and c-Myc (20). In MDA-MB-231 cells, overexpression of miR-214 resulted in a reduction of c-Myc and cyclin D1 transcripts as measured by RT–qPCR (Figure 3D). Inversely, expression of Ezh2 target genes IGFBP2 and SFRP3 (20,32), that are associated with inhibition of cell proliferation, was upregulated in miR-214-expressing cells relative to control, but was unaffected by miR-214 mutant (Figure 3D). Transfection of Ezh2 siRNA in either MCF-7 or MDA-MB-231 cells resulted in a similar profile of regulation of Ezh2 downstream target genes (supplementary Figure 1B is available at Carcinogenesis Online), suggesting specificity for miR-214 effects on mediators of cell proliferation via Ezh2 down-modulation. In MCF-7 cells, miR-214 overexpression resulted in decreased c-Myc and upregulation of IGFBP2, but no significant change in the levels of cyclin D1 transcripts was observed. The latter result points at a potential posttranscriptional regulation of cyclin D1 in miR-214 transfected MCF-7 cells (Figure 3D).
Fig. 3.
miR-214 overexpression reduces proliferation of breast cancer cell lines. (A) Cell growth curves of MCF-7 MDA-MB-231 cells stably expressing miR-214, mut-miR-214 (mut) or control vector. The data represent average cell numbers ± SD determined at 24, 48 and 72 h from five experiments done in duplicates (*P ≤ 0.05, **P ≤ 0.005). (B) BrdU staining of MCF-7 (left panel) and MDA-MB-231 (data not shown). The ratio of BrdU/DAPI-stained cells calculated using counts from 10 random fields is presented as an average of three independent experiments ± SD (*P ≤ 0.05, **P ≤ 0.005) expressed as a percentage of the control vector cells (right panel). (C) Immunoblot of cyclin D,E, Ezh2 and β-actin in MCF-7 and MDA-MB-321 breast cancer cells stably expressing miR-214, mut-miR-214 (mut) or control vector. (D) RT–qPCR of Ezh2 downstream targets was performed in MCF-7 and MDA-MB-231 cells stably expressing miR-214, mut-miR-214 (mut) or control vector (*P ≤ 0.05, ***P ≤ 0.0002).
miR-214 expression blocks in vitro cell invasion
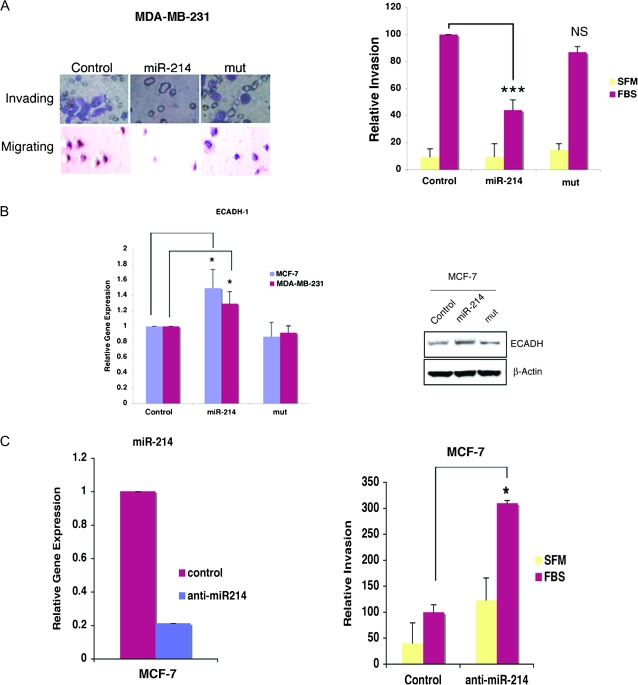
To determine the effect of miR-214 on the invasive behavior of MDA-MB-231 cells, we used a laminin basement membrane invasion chamber assay. Expression of miR-214 markedly decreased the invasion potential of MDA-MB-231 cells when compared with cells expressing the miR-214 mutant or control cells (Figure 4A, left panel). Quantitative analysis indicated that miR-214 expression resulted in a 56% reduction of cell invasion (Figure 4A, right panel). However, this reduction was not achieved with cells expressing the mutant miR-214 or transfected with control plasmid (Figure 4A, right panel). Importantly, in both MCF-7 and MDA-MB-231 cells, miR-214 expression correlated with increased expression of E-cadherin (ECADH-1) (Figure 4B), a transcript whose expression is inversely correlated with cell invasion (24,26). Upregulation of ECADH-1 by miR-214 overexpression was confirmed by immunoblot in MCF-7 (Figure 4B). ECADH-1 protein was below detection levels in MDA-MB-231 (data not shown). We next evaluated whether reducing the miR-214 levels in MCF-7 cells could alter their weakly invasive behavior. MCF-7 cells transfected with anti-miR-214 had reduced miR-214 levels (Figure 4C, left panel) and displayed increased cell invasion, when compared with control-transfected MCF-7 cells (Figure 4C, right panel).
Fig. 4.
miR-214 expression blocks cell invasion. (A) Photographs of crystal violet-stained MDA-MB-231cells invading a laminin-coated membrane and migrating into the lower chamber containing fetal bovine serum (FBS). Shown is the reduced number of invading cells in MDA-MB-231 cells stably expressing miR-214 relative to mut or the control vector expressing-cells (left panel). Quantitative analysis of MDA-MB-231 invasion activity measured as fluorescence units in response to FBS (right panel). ***P ≤ 0.0002; NS, not significant. (B) RT–qPCR in MCF-7 and MDA-MB-231 cells for ECADH-1 transcript (left panel). Right panel shows ECAD immunoblot of MCF-7 cells-expressing miR-214 or mut. (C) RT–qPCR analyses of endogenous mature miR-214 in MCF-7 cells transfected with anti-miR-214 (left panel right panel) and increased invasion of MCF-7 overexpressing anti-miR-214 oligomer (right panel).
Discussion
In this study, we show that miR-214 targets Ezh2 in breast cancer cell lines and that one miR-214 allele is deleted in 24% (6/25) of primary breast tumors. Although miR-101 deletion is associated with increased Ezh2 accumulation and consequent disease progression in breast cancers (27,28), our data suggest that Ezh2 accumulation may also result from deletion of miR-214 without concomitant loss of miR-101 in a subset of breast tumors. Our data are consistent with a number of studies reporting that miR-214 levels are reduced in breast cancer (10,11). In one study, reduction of miR-214 levels was associated with a subset of tumors that are basal, estrogen receptor-negative, human epidermal growth factor-2-positive, of histological high grade (10) and aggressive behavior. This observation correlates well with the reported poor prognosis of breast cancers expressing high level of Ezh2 (24,26). In addition, Ezh2 expression is upregulated in breast ductal carcinoma in situ and atypical ductal hyperplasia (20), two conditions that are not characterized by invasiveness, suggesting that Polycomb proteins control several steps of breast tumorigenesis. Ezh2 overexpression in breast cancer has been shown to repress a large number of genes, including specific Polycomb group proteins, transcription factors and cell cycle regulators (20). Inhibition of cell proliferation by miR-214 in MCF-7 and MDA-MB-231 cells in our study is consistent with this hypothesis and is supported by downregulation of cyclin D1 and E in both cell lines and relief of the repression of the Ezh2 downstream target IGFBP2 (20,32). Moreover, downregulation of c-Myc, which has been shown to control Ezh2 gene expression by controlling its negative regulator miR-26a (33), is consistent with this model and indicates that miR-214 specifically interferes with Ezh2-mediated activity on cell proliferation. Importantly, the correlation of blockade of the invasive potential of MDA-MB-231 in vitro by miR-214 expression with increased expression of ECADH-1 further indicates that miR-214 is also able to repress Ezh2-mediated invasion. However, it is clear that breast metastasis is a very complex and regulated phenomenon (34) and other factors may contribute to the different invasive behavior of MCF-7 and MDA-MB-231 cells.
Overall, our data suggest that miR-214 (and possibly co-transcribed miR-199a/a*) may function as a tumor suppressor and its deletion in a subset of breast tumors is an independent event from deletion of miR-101, thus providing an additional mechanism for increased Ezh2 levels. It is formally possible that miR-214 levels may be reduced in tumors that do not harbor deletions of the miR-214 alleles. In these cases, DNA methylation of miR-214 regulatory regions may silence expression, as reported for several tumor suppressor genes (35). Reduction in miRNA levels resulting from genomic loss or other mechanisms coupled with increased Ezh2 levels may provide new prognostic markers for the management of cancer.
Supplementary material
Supplementary Figure 1 and Table 1 can be found at http://carcin.oxfordjournals.org/
Funding
Intramural Research Program of the National Institute of Arthritis, Musculoskeletal and Skin Diseases of the National Institutes of Health.
Supplementary Material
Acknowledgments
We would like to thank Dr A.M.Chinnaiyan for help with the comparative genomic hybridization data of primary breast cancers.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- BrdU
5′-bromo-2′-deoxyuridine
- miRNA
microRNA
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- RT
reverse transcription
- siRNA
silencing RNA
- UTR
untranslated region
References
- 1.Flynt AS, et al. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stefani G, et al. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 3.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl Acad. Sci. USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, et al. microRNAs exhibit high frequency genomic alterations in human cancer. Proc. Natl Acad. Sci. USA. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 6.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 7.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 8.Garzon R, et al. MicroRNAs in normal and malignant hematopoiesis. Curr. Opin. Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 9.Hagan JP, et al. MicroRNAs in carcinogenesis. Cytogenet. Genome Res. 2007;118:252–259. doi: 10.1159/000108308. [DOI] [PubMed] [Google Scholar]
- 10.Blenkiron C, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuettengruber B, et al. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 14.O'Carroll D, et al. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caretti G, et al. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong CF, et al. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 17.Ezhkova E, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller H, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Matsukawa Y, et al. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachmann IM, et al. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006;24:268–273. doi: 10.1200/JCO.2005.01.5180. [DOI] [PubMed] [Google Scholar]
- 23.Weikert S, et al. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int. J. Mol. Med. 2005;16:349–353. [PubMed] [Google Scholar]
- 24.Kleer CG, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl Acad. Sci. USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding L, et al. Enhancer of Zeste 2 as a marker of preneoplastic progression in the breast. Cancer Res. 2006;66:9352–9355. doi: 10.1158/0008-5472.CAN-06-2384. [DOI] [PubMed] [Google Scholar]
- 26.Cao Q, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedman JM, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 28.Varambally S, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juan AH, et al. Mir-214-dependent regulation of the polycomb protein ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shingara J, et al. An optimized isolation and labeling platform for accurate microRNA expression profiling. RNA. 2005;11:1461–1470. doi: 10.1261/rna.2610405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu J, et al. Integrative genomics analysis reveals silencing of beta-adrenergic signaling by polycomb in prostate cancer. Cancer Cell. 2007;12:419–431. doi: 10.1016/j.ccr.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 33.Sander S, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 34.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feinberg AP, et al. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.