Abstract
Aberrant activation of the Wnt/β-catenin signaling axis is a prominent oncogenic mechanism in numerous cancers including cervical cancer. Wnt inhibitory factor-1 (WIF1) is a secreted protein that binds Wnt and antagonizes Wnt activity. While the WIF1 gene is characterized as a target for epigenetic silencing in some tumor types, WIF1 expression has not been examined in human cervical tissue and cervical cancer. Here, we show that WIF1 is unmethylated and its gene product is expressed in normal cervical epithelium and some cultured cervical tumor lines. In contrast, several cervical cancer lines contained dense CpG methylation within the WIF1 gene, and expression of both WIF1 transcript and protein was restored by culturing cells in the presence of the global DNA demethylating agent 5-aza-2′-deoxycytidine. Using single-molecule MAPit methylation footprinting, we observed differences in chromatin structure within the WIF1 promoter region between cell lines that express and those that do not express WIF1, consistent with transcriptional activity and repression, respectively. The WIF1 promoter was aberrantly methylated in ∼60% (10 of 17) high-grade highly undifferentiated squamous cell cervical tumors examined, whereas paired normal tissue showed significantly lower levels of CpG methylation. WIF1 protein was not detectable by immunohistochemistry in tumors with quantitatively high levels of WIF1 methylation. Of note, WIF1 protein was not detectable in two of the seven unmethylated cervical tumors examined, suggesting other mechanisms may contribute WIF1 repression. Our findings establish the WIF1 gene as a frequent target for epigenetic silencing in squamous cell carcinoma of the cervix.
Background
Cervical cancer continues to remain a major public health problem, as it remains a major cause of cancer-related deaths among women worldwide. Infection with oncogenic human papillomavirus (HPV) is the primary etiological factor for cervical cancer and its precursor lesions (1). HPV infections are often transient; however, persistent infections, especially with oncogenic or high-risk HPV types (e.g. HPV-16 and -18), further increase the likelihood of developing cervical cancer (2). HPV-induced oncogenesis in cervical carcinoma is largely attributable to expression of the viral oncoproteins E6 and E7, but HPV infection alone is insufficient to induce malignant transformation of cervical epithelium. Other significant cofactors, such as individual genetic variation and environmental exposures, contribute to the multistep process of tumor formation (3).
Both genetic and epigenetic changes within the genome fuel the process of the tumorigenesis. In the context of normal cellular function, epigenetic DNA modification is a frequent way to regulate chromatin structure and control gene expression. The interplay of DNA methylation, histone modifications and the enzymes that regulate these modifications exerts control over patterns of gene expression in various cell types and developmental stages. Dysregulation of one or more of these mechanisms is commonly found in diverse disease states. In cancer, chromatin reorganization, acting in concert with DNA methylation and transcriptionally repressive histone modifications, has been shown to drive transcriptional silencing of tumor suppressor genes (4).
Wnt was originally identified as a proto-oncogene activated by viral insertion in mouse mammary tumors (5). It is now clear that Wnt proteins comprise a family of secreted signaling molecules that regulate cell–cell interactions during embryogenesis (6). Binding of Wnt to its receptor, termed Frizzled, while complexed with either low-density lipoprotein receptor-related protein-5 (LRP5) or LRP6 co-receptors, activates an intracellular signaling cascade that promotes cytoplasmic accumulation of β-catenin. Consequently, β-catenin translocates to the nucleus where it engages transcription factors such as T-cell factors and lymphoid-enhancing factors (7). Several growth-promoting genes are transcriptionally activated, either directly or indirectly, through this mechanism, including the oncogenic transcription factor c-Myc and the catalytic subunit of telomerase hTERT (8,9).
One mechanism of cellular control over Wnt/β-catenin signaling is the secretion of one or more of a diverse group of secreted inhibitors. Work from several groups has identified numerous extracellular antagonists, such as the soluble Frizzled-related protein (SFRP) family, Cerberus, Wnt inhibitory factor-1 (WIF1), the Dickkopf (DKK) family, Wise and sclerostin (SOST) (10–12). SFRPs and WIF1 function by binding directly to Wnt proteins and inhibiting their ability to interact with Frizzled. Alternatively, DKKs inhibit Wnt signaling by binding to the LRP5/6 co-receptor.
Several human genes that encode functional inhibitors of the Wnt pathway are frequently epigenetically silenced in cervical tumors including CDH1 (E-cadherin), FHIT, APC and the SRFP family (for review, see ref. 13). Silencing of any of these genes either singly or in combination would predictably alter Wnt/β-catenin signaling. In addition to these epigenetic events that promote dysregulation of Wnt/β-catenin signaling, current evidence supports the notion that the HPV E6 oncoprotein indirectly activates Wnt/β-catenin signaling. Specifically, via their PDZ domain, human orthologs of Drosophila Discs large homolog (hDLG) and Scribble (hSCRIB) bind to the E6 oncoprotein (14,15). Through consequential interaction with the human ubiquitin ligase E6-associated protein (E6 AP), E6 promotes proteolysis of hDLG and hSCRIB (16–20). hDLG binds to APC (21) and the APC–hDLG complex negatively regulates cell cycle progression (22). Similarly, hSCRIB binds to APC (23) and has also been characterized as a tumor suppressor gene (24). Taken together, it is clear that dysregulation of the oncogenic Wnt/β-catenin signaling axis can arise due to HPV infection/gene expression and/or aberrant epigenetic/genetic events within the host cell genome.
In this report, we focused our study on expression of the WIF1 protein, an antagonist of Wnt/β-catenin signaling (25). We, and others, have documented that the WIF1 gene is targeted for epigenetic silencing in breast cancer (26), bladder cancer (27), acute lymphoblastic leukemia (28), nasopharyngeal and esophageal carcinoma (29) and non-small-cell lung cancer (30,31). Given the prominent role that dysregulation of Wnt/β-catenin signaling plays in cervical cancer, we sought to determine if WIF1 is epigenetically silenced in this tumor type. Our results demonstrate that WIF1 hypermethylation is a frequent event in cervical cancer and provide another aberrant, cancer-associated event that can dysregulate Wnt/β-catenin signaling during cervical tumorigenesis.
Materials and methods
Cervical specimens
All human tissue samples were obtained from patients under protocols approved by Institutional Review Boards at the Moffitt Cancer Center. Tumor tissue (n = 22) and adjacent matched normal tissue (n = 22) were obtained from archived formalin-fixed paraffin-embedded (FFPE) blocks. The archived tissue came from women diagnosed with squamous cell cervical cancer and having a surgical procedure between 1993 and 1999 at the Moffitt Cancer Center. Cases were restricted to those that did not receive radiation treatment before surgery. Histological diagnosis, tumor stage and grade were conducted at Moffitt Cancer Center using the World Health Organization (WHO) and the Union for International Cancer Control (UICC) classification schemes.
Cell culture and 5-aza-2′-deoxycytidine treatment
Cervical cancer cell lines (C33A, CaSki, HeLa 229, SiHa) were purchased from the American Type Culture Collection and cultured according to their specifications at 37°C in a humidified 5% CO2 incubator. Where indicated, cells were seeded, allowed to recover overnight and then 5-aza-2′-deoxycytidine (5-azadC; Sigma–Aldrich, St Louis, MO) was added to complete growth media at a final concentration of 5 μM as previously indicated (32). Fresh drug was subsequently added every 24 h for 5 days. After treatment, cells were washed with phosphate-buffered saline and allowed to recover in their respective medium for 2 days prior to collection and indicated analyses.
RNA and DNA isolation
Cells cultured with and without 5-azadC were harvested for RNA and DNA extraction using 1 ml of TRizol (Invitrogen, Carlsbad, CA) reagent per 10 cm dish as indicated by the manufacturer. RNA was subsequently isolated using the RNAeasy kit (Qiagen, Valencia, CA). DNA was directly isolated from Trizol fractionation according to manufacturer’s instructions. DNA isolated from FFPE samples was extracted using the QIAmp mini DNA kit (Qiagen) according to the manufacturer's protocol. Concentrations of RNA and DNA were determined using a NanoDrop ND-8000 spectrophotometer (Thermo Scientific, Wilmington, DE).
Pyrosequencing analysis
Sodium bisulfite conversion of 1 μg of genomic DNA harvested from FFPE specimens was carried out with the EZ DNA Methylation-Direct kit (Zymo Research Corporation, Orange, CA) according to the manufacturer's protocol. Genomic DNA harvested from cultured cell lines was bisulfite converted using an in-house protocol as described previously (33). Polymerase chain reaction (PCR) amplification of the lower DNA strand was conducted in a 20 μl reaction volume containing 1× Coral buffer (Qiagen), 250 nM deoxynucleotides, 250 nM each of forward (5′-GtAGGtTttTTGGtAtttAGGt-3′; lower case indicating C to T transitions) and reverse (5′-CATaCTaCTCAaaACCTCCT-3′; lower case indicating G to A transitions) primer, 1 U of HotStarTaq Plus DNA polymerase (Qiagen) and 2 μl (50–100 ng) of bisulfite-treated DNA. Amplification conditions were as follows: one cycle at 95°C for 5 min followed by 50 cycles at 95°C for 30 s; 56°C for 30 s; 72°C for 30 s and a final extension step at 72°C for 10 min. Prior to pyrosequencing, PCR products were analyzed by agarose gel electrophoresis/ethidium bromide staining. Pyrosequencing was subsequently conducted as previously outlined (34) using the forward PCR primer as the sequencing primer. Samples and controls were subsequently analyzed using a PyroMark MD system (Biotage, Uppsala, Sweden) and methylation density quantified using PyroMark Pyro Q-CpG (ver 1.0.9) software.
Reverse transcriptase–PCR
Total RNA was used in first-strand complementary DNA synthesis reactions using the GoScript reverse transcriptase system and random hexamer primers (Promega, Madison, WI). WIF1 transcript level was subsequently analyzed using WIF1-specific primers (forward, 5′-CCGAAATGGAGGCTTTTGTA-3′ and reverse, 5′-TGGTTGAGCAGTTTGCTTTG-3′). Amplification conditions were as follows: one cycle at 95°C for 5 min followed by 45 cycles at 95°C for 30 s, 59°C for 30 s, 72°C for 30 s and a final extension step at 72°C for 10 min. PCR amplification of β-actin transcript was amplified using forward (5′-CCCTGGCACCCAGCAC-3′) and reverse (5′-GCCGATCCACACGGAGTAC-3′) primers served as a control for RNA integrity and normalization. Amplification conditions for β-actin were the same as used for WIF1 transcript, except that an annealing temperature of 60°C was used and PCR was conducted for 20 cycles. Following amplification, reactions were analyzed by 1% agarose gel electrophoresis and products visualized by staining with ethidium bromide.
Immunoblotting
Sodium dodecyl sulfate (SDS)–polyacrylamide electrophoresis and immunoblotting procedures were conducted as previously outlined (35). Briefly, cells treated or untreated with 5-azadC were harvested by scraping in ice-cold phosphate-buffered saline and extracts formed by the addition of lysis buffer (125 mM Tris–HCl, pH 7.5; 5 mM ethylenediaminetetraacetic acid; 5 mM ethyleneglycol-bis(aminoethylether)-tetraacetic acid; 10 mM β-glycerolphosphate; 10 mM NaF; 10 mM Na-pyrophosphate; 1.0% SDS wt/vol) for 5 min on ice and then placed in a 95°C hot block for 5 min. Lysates were then sonicated and centrifuged at 3000×g for 3 min. Protein concentrations were determined using the BCA protein assay (Pierce, Rockford, IL) and lysates were stored at −80°C prior to use.
Prior to electrophoresis, an appropriate volume of cell lysate was diluted in 3× SDS sample buffer (150 mM Tris–HCl, pH 6.8; 10% vol/vol β-mercaptoethanol; 20% vol/vol glycerol; 3% wt/vol SDS; 0.01% wt/vol bromophenol blue; 0.01% wt/vol pyronin-Y) and boiled for 5 min. Proteins were resolved on 10% SDS–polyacrylamide gels and electrotransferred to nitrocellulose membranes overnight at 12 V. Membranes were sequentially probed with polyclonal anti-WIF1 antisera (ab71205; Abcam, Cambridge, MA) and horseradish peroxidase-labeled anti-rabbit secondary antibody (Kirkegaard and Perry Labs, Gaithersburg, MD). As a loading control, blots were probed with anti-β-actin (Abcam). Immunoreactivity was visualized using chemiluminescence and recorded on x-ray film.
Methyltransferase accessibility protocol for individual templates (MAPit)
Isolation of nuclei from C33A, CaSki and SiHa cells and probing of chromatin structure with the GC methyltransferase M.CviPI or methyltransferase buffer only (i.e. no enzyme control) were done as described previously (36). Purified genomic DNA was subjected to bisulfite genomic sequencing (37) as described in (33). A 712 bp region of the WIF1 promoter was PCR amplified by forward (5′-TATATACTCGAGATtAttATtAttATtATtAGYAtTtAGTt-3′; XhoI site underlined; where Y is a pYrimidine and lower case indicates C to T transitions) and reverse (5′-ATATATAAGCTTCAaRCACAaaAaaATRCTCCAaA-3′; HindIII site underlined; where R is a puRine and lower case indicates G to A transitions) primers. Three 20 μl reactions with a final concentration of 1× Coral buffer, 250 nM deoxynucleotides, 250 nM each forward and reverse primers, 275 nM MgCl2, 1 U HotStarTaq Plus DNA polymerase (Qiagen) and 2 μl bisulfite-treated DNA were incubated in the thermocycler as follows: one cycle at 95°C for 5 min followed by 40 cycles of 30 s at 95°C, 30 s at 53°C, 2 min at 72°C and a final extension step at 72°C for 10 min. Following PCR, products were pooled, resolved on an agarose gel and bands corresponding to the correct-size product were excised and gel purified using the QiaexII gel extraction kit (Qiagen). Purified PCR products were then cloned by TA cloning (restriction sites were included in the primers to provide a backup directional cloning strategy) using the pGEM-T Vector System (Promega) and TOP10 Escherichia coli cells (Invitrogen). Recombinant plasmids positive for inserts of correct size were identified by colony PCR with Apex Taq (Genesee Scientific, San Diego, CA). Cloned inserts were sequenced using SP6 primer at the University of Florida Interdisciplinary Center for Biotechnology Research DNA sequencing core laboratory. MethylViewer was used to analyze ABI sequencing data files and plot the methylation status of each CpG and GC site (36).
Generation and hierarchical clustering of three-color MAPit representation
Tab-delimited text files summarizing the sequence of each M.CviPI target site in each sequenced molecule were exported from MethylViewer. For this application, these sites can be GCH (only M.CviPI target sites, where H is A, C or T), HCG (only endogenous sites) or GCG (ambiguous). GCG information was discarded and HCG information was discarded temporarily. Using a series of calculations performed in Microsoft Excel, we expanded the MethylViewer text file to ‘color’ all bases in the sequence. All methylated sites were assigned a value of 1, as were all bases between two successive methylated sites. Unmethylated sites were assigned a value of –1, as were all bases between two successive unmethylated sites. All other bases present in the original sequence were assigned a value of 0 and exported as a text file. The resultant text file was analyzed by Cluster (http://rana.lbl.gov/EisenSoftware.htm) to generate hierarchical clusters and a map of patches of G-m5C (accessible sites with value of 1, shaded yellow), GC (protected sites with value of –1, shaded blue) and the borders between sites (shaded gray). Clustered data was graphed with the program MapleTree (http://mapletree.sourceforge.net).
Immunohistochemistry
A tissue microarray was assembled from archival FFPE blocks of 22 unique cervical squamous cell carcinoma (SCC) and patient-matched adjacent normal cervical tissues. Prior to staining, sections were dewaxed in xylene, rehydrated in graded alcohol and rinsed in deionized H2O. Following this, samples were blocked using the avidin/biotin blocking kit (Vector Laboratories, Burlingame, CA) following the manufacturer’s recommended protocol. Antigen retrieval was then conducted by steam heat for 20 min with 10 mM sodium citrate, pH 6.0. Subsequently, an overnight incubation at 4°C with rabbit anti-WIF1 antibody (Abcam) diluted 1:50 was conducted. Samples were then washed and incubated with biotinylated-goat anti-rabbit antibody (Vector Laboratories). Antigen detection was done using avidin-conjugated horseradish peroxidase followed by colorimetric development using 3,3′-diaminobenzidine tetrahydrochloride. Sections were then counterstained with hematoxylin and mounted using aqueous mounting medium. As a control, parallel sections were incubated with purified non-immune rabbit IgG. Sections were photographed using a Leica DM6000B microscope (Leica Microsystems, Bannockburn, IL) and microphotograpy conducted with a Retiga SRV Fast 1394 digital camera (QImaging, Surrey, British Columbia, Canada).
Results
WIF1 is epigenetically silenced in cervical cancer cell lines
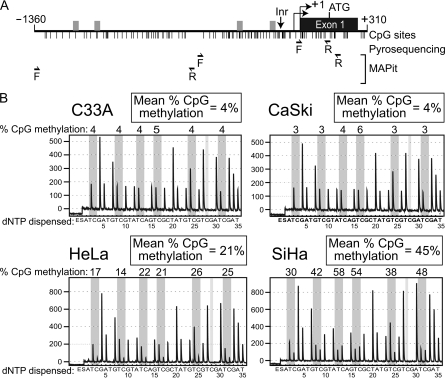
To determine if the WIF1 gene is targeted for epigenetic silencing in cervical cancer, we first analyzed the methylation status of WIF1 by pyrosequencing genomic DNA harvested from four cervical cancer lines (C33A, CaSki, HeLa and SiHa). Pyrosequencing allows for quantitative analysis of methylation at individual CpG dinucleotides after PCR amplification of bisulfite-converted DNA (38). We designed primers to amplify a 164 bp region of the WIF1 promoter that encompasses the first seven CpGs downstream of the transcription start site (TSS; Figure 1A). The resulting pyrograms showed that C33A and CaSki cells exhibited very low levels (4%) of CpG methylation (m5CpG) in this region (Figure 1B). Conversely, HeLa and SiHa cells exhibited overall methylation levels of 21 and 45%, respectively, of the same seven CpG sites.
Fig. 1.
The WIF1 promoter is hypermethylated in cultured cervical carcinoma cells. (A) Diagram of the WIF1 promoter. Vertical lines represent the location of CpG sites and grey boxes represent possible SP1 binding sites. Shown below are the positions of the forward (F) and reverse (R) primers for pyrosequencing and MAPit. (B) Pyrograms from the four cell lines before 5-azadC treatment analyzing seven CpGs in the CpG island spanning the transcription start site of WIF1. The y-axis represents the signal intensity in arbitrary units, whereas the x-axis shows the dispensation order (E, enzyme; S, substrate). Dispensations corresponding to potentially methylated cytosines (C or T after bisulfite treatment) are highlighted in gray. The percentage of methylation at individual CpG positions is shown as the percentage of methylation above the respective positions. The average methylation for the area analyzed is shown in the box on top of the pyrogram.
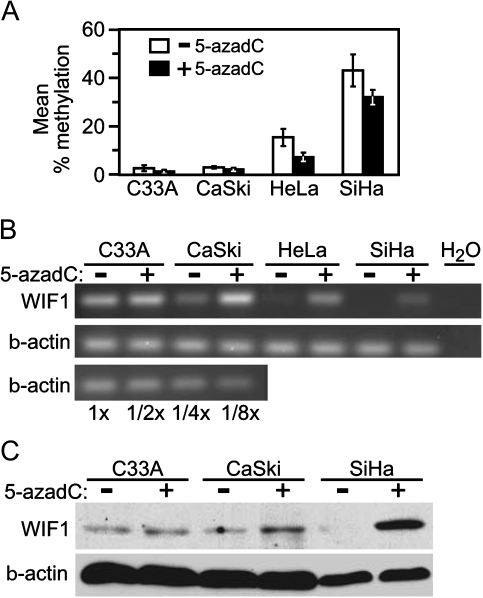
To examine the effects of global genome demethylation on WIF1 expression, we cultured each cell line in the absence or presence of 5 μM of the DNA demethylating drug 5-azadC for 5 days. To confirm demethylation by 5-azadC, we used pyrosequencing to measure methylation of seven CpG sites near the WIF1 TSS. By this assay, HeLa and SiHa cells displayed a 10–15% decrease in average methylation within the portion of the WIF1 promoter assayed, whereas 5-azadC did not significantly affect WIF1 methylation levels in either C33A or CaSki cells (Figure 2A).
Fig. 2.
WIF1 expression in cultured cervical carcinoma cells is upregulated in response to DNA demethylation. (A) Graph of average methylation of WIF1 before and after 5-azadC treatment. The y-axis represents the average methylation of the seven CpG sites analyzed by pyrosequencing while the x-axis represents the four cell lines analyzed before (white) and after 5-azadC treatment (black). (B) Ethidium bromide-stained acrylamide gels of RT-PCR analysis of WIF1 expression (top) and β-actin (bottom). A dilution series of input cDNA demonstrates linearity of the reactions. (C) Immunoblot analysis of cervical cell lines before and after 5-aza treatment were visualized with anti-WIF1 (top) and β-actin (bottom).
Reverse transcriptase–PCR analysis of total RNA isolated from these cultures indicated that untreated C33A, CaSki and HeLa cells expressed detectable levels of WIF1 transcript, which was undetectable in SiHa cells (Figure 2B). However, when cultured in the presence of 5 μM 5-azadC for 5 days, WIF1 transcript levels increased in HeLa and SiHa cells were slightly upregulated in CaSki cells and were not detectably changed in C33A cells.
We next sought to test if 5-azadC treatment also resulted in increased abundance of WIF1 protein. Thus, in a parallel experiment, whole cell extracts were prepared from 5-azadC-treated and -untreated cells. Protein extracts from C33A cells displayed no significant increase in WIF1 protein level in response to 5-azadC, and CaSki cells showed a slight increase in WIF1 abundance following drug exposure (Figure 2C). In contrast, SiHa cells that exhibited high endogenous DNA methylation downstream of the TSS displayed a pronounced increase in WIF1 protein following 5-azadC treatment.
These data indicate that both SiHa and HeLa cells display CpG hypermethylation within the WIF1 promoter and exhibit heightened expression of WIF1 following 5-azadC-induced genome demethylation. Although this drug treatment did result in measurable restoration of WIF1 expression, we observed that 5-azadC modestly reduced WIF1 promoter methylation in both cell lines. While both lines were relatively resistant to the 5-azadC regimen used, they showed a modest reduction in proliferation rates that may have contributed to the observed partial demethylation. The partial nature of demethylation induced by 5-azadC could also be attributable to a variety of factors such as relative DNMT1 expression levels (39,40). Nevertheless, we conclude that the WIF1 gene is epigenetically silenced through DNA methylation in HeLa and SiHa cervical carcinoma cells.
Distinct chromatin architectures in different cervical cancer cell lines
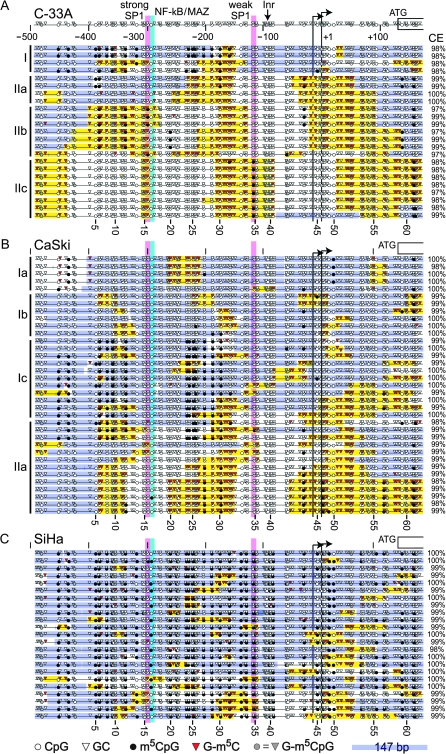
Epigenetic silencing by DNA methylation is often linked to changes in chromatin structure. To simultaneously map chromatin structure and m5CpG of a larger region of the WIF1 promoter (see Figure 1A), we performed single-molecule MAPit methylation footprinting (41,42) in the C33A, CaSki and SiHa cell lines (Figure 3). In this experiment, nuclei were probed with and without M.CviPI, a DNA methyltransferase that methylates only GC dinucleotides (43). Accessibility in chromatin to DNA methyltransferase is influenced by the affinity and span of protein–DNA interactions (44). The methylation status of every C along single strands of DNA was determined by bisulfite genomic sequencing (37) in conjunction with MethylViewer (36). Because M.CviPI methylates GC, a non-averaged single-molecule view of both chromatin accessibility at GC sites and endogenous methylation at CpG sites is obtained (45).
Fig. 3.
Diverse chromatin structures of the WIF1 gene promoter in cervical cancer cell lines. Nuclei from (A) C33A, (B) CaSki and (C) SiHa cells were subjected to single-molecule MAPit methylation footprinting with the GC methyltransferase M.CviPI, as described in the Materials and Methods. WIF1 promoter coordinates (to scale) relative to the downstream TSS are shown over the top dataset. Each individually cloned and sequenced molecule is indicated by a horizontal line, with the bisulfite conversion efficiency (CE) for cytosines not in CpG or GC sites indicated at the right. Symbols indicating the status of methylation at all CpG and GC sites (numbered at the bottom of each dataset) are defined in the legend at the bottom of C. Putative binding sites for SP1 (pink) and both nuclear factor-kappaB (NF-κB) and MYC-associated zinc finger (MAZ) (cyan) are indicated as well as two annotated TSSs (bent arrows), putative upstream Inr and first coding exon (open-ended rectangle) are indicated at the top of A. Hierarchical clusters of similar molecules in A and B are labeled at the left. Consecutive G-m5C (more than or equal to two sites) flanked by more than or equal to two unmethylated GC sites indicate patches of chromatin accessibility (yellow). Similarly, protected patches of 147 bp (or less at either end of each molecule) are shaded blue and inferred to correspond to nucleosomes. Sub-nucleosomal protections of <147 bp (unshaded) are likely to correspond to sites of protein–DNA interaction. Methylated GCpG sites (G-m5CpG; gray triangles) were excluded from patch designations as methylation by endogenous enzymes versus M.CviPI cannot be discerned.
Two main patterns of CpG methylation were observed in both C33A and CaSki cells harboring transcribed WIF1 (in the absence of 5-azadC). Endogenous m5CpG was nearly absent from many of the promoter molecules, whereas many others were methylated at CpGs from −453 to −319 and −234 to −160 (Figure 3; CpG sites 2–13 and 23–30). In addition, m5CpG was observed in CaSki cells in a few molecules near the TSS and within the 5′ untranslated region (Figure 3B, bottom). Furthermore, about one-third (8/23) of WIF1 promoter copies in C33A cells were methylated at C −119 (site 34) located within a weak consensus Sp1 site (Figure 3A, top). In contrast, SiHa cells were densely methylated throughout the body of the promoter in all 23 sequenced clones (Figure 3C), consistent with the epigenetic silencing of WIF1 in this cell line (Figure 2). In addition, dense CpG methylation was observed from −1356 to −572 in all three cell lines (supplementaryFigure 1 is available at Carcinogenesis Online).
Next, we asked if single WIF1 promoters populated distinct classes of accessibility to M.CviPI chromatin probe and the extent to which accessibility to M.CviPI correlated with endogenous CpG methylation. For this analysis, sequence information of all methylated and unmethylated GC sites as well as all other residues in each molecule (to maintain spacing between GC sites) was numerically converted (see Materials and Methods). These maps were then subjected to hierarchical clustering to identify coexisting molecules with distinct patterns of chromatin accessibility (supplementaryFigure 2 is available at Carcinogenesis Online).
Distinct subclasses of chromatin architecture were found within and between each cell line. In the C33A and CaSki lines supporting WIF1 transcription (Figure 3A and B), promoter molecules populated two major clusters (supplementaryFigure 2 is available at Carcinogenesis Online). On the whole, these clusters differed in nucleosome occupancy, as shown by blue bars representing 147 bp, and the amount of sequence that appeared to be depleted or free of nucleosomes. In C33A cells, individual WIF1 promoters also contained a variable number of protected regions that were too short to accommodate a nucleosome as well as hypersensitivity to M.CviPI at the TSS. Methylation by M.CviPI is indicative of nucleosome depletion at TSSs of actively transcribed genes (36). In CaSki cells (Figure 3B), similar, but not identical (see Discussion), patterns of protection at the SP1 sites and accessibility at the TSS were observed. In contrast, SiHa cells contained considerably less hypersensitivity at the analyzed WIF1 promoters (Figure 3C), again consistent with the dense m5CpG and silencing in this cultured line. We conclude that markedly diverse states of chromatin co-exist among the population of cells in each cultured cervical cancer line. Moreover, chromatin states specific to C33A and CaSki lines that express the WIF1 gene were also observed, suggesting that transcription itself is not solely responsible for generating chromatin structural diversity.
WIF1 promoter is aberrantly methylated in cervical squamous cell carcinoma
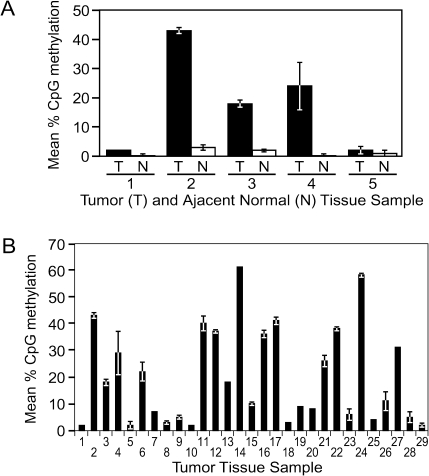
To determine if CpG methylation within the WIF1 gene promoter occurs in primary cervical tumors, we analyzed WIF1 methylation in a panel of primary SCC tumors. Matched adjacent normal cervical epithelial tissue was available for analysis for five of these patient tumor samples. Pyrosequencing analysis of the five normal cervical tissue samples each showed very low levels of WIF1 methylation (mean m5CpG < 5%) (Figure 4A). Analysis of additional normal cervical epithelium samples further supported the conclusion that the WIF1 gene is relatively unmethylated in normal cervical tissue (data not shown).
Fig. 4.
The WIF1 promoter is aberrantly hypermethylated in primary cervical squamous cell carcinoma tumors. (A) WIF1 promoter methylation was measured in five primary cervical SCC tumors (T) and adjacent normal tissue (N) tissue samples. (B) Graph of WIF1 promoter methylation of all tumor samples analyzed.
Analysis of the five SCC tumors indicated that three of the tumors displayed increased levels of WIF1 methylation compared with matched normal tissue (Figure 4A). Specifically, the mean percent CpG methylation in tumors 2, 3 and 4 was measured as 43, 18 and 24%, respectively. In tumors 1 and 5, 2% mean CpG methylation was measured, similar to the matched normal tissue. These results suggest that the WIF1 promoter was targeted for hypermethylation in cervical SCC; however, to obtain an accurate view of occurrence rate, we analyzed a panel of 29 cervical SCC samples by pyrosequencing (Figure 4B). We arbitrarily categorized these specimens based on their average methylation of six CpG sites within the amplicon, three categories were established: low methylation (0–9%), moderate methylation (10–19%) and heavy methylation (>20%). Of the 29 tumors, 12 (41.4%) displayed low methylation, five (17.2%) displayed moderate methylation, and 12 (41.4%) were heavily methylated.
From these findings, we conclude that hypermethylation of the WIF1 promoter is not present in any of the normal cervical epithelium samples analyzed and thus is likely to be a molecular event specific to cancer cells. Furthermore, analysis of a sizable panel of cervical SCC tumors indicates that dense m5CpG at the WIF1 promoter is a common occurrence in this tumor type.
Cervical SCC displays dysregulated WIF1 expression
To examine expression and localization of the WIF1 protein in normal cervical tissue and cervical SCC, we assembled a tissue microarray consisting of 22 archived (FFPE) SCC tumors and patient-matched adjacent normal tissue. This tissue microarray was subsequently used in immunohistochemical (IHC) analyses using WIF1 antibody and hematoxylin counterstaining.
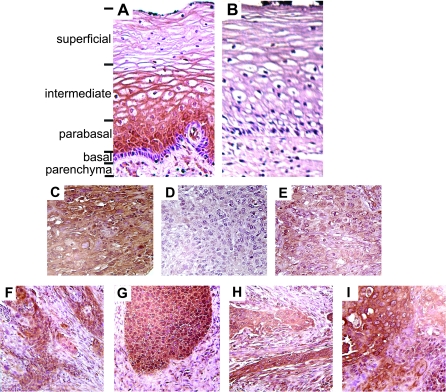
Immunostaining of normal tissue with anti-WIF1 antibody indicated expression in the stratified squamous epithelium of the ectocervix (Figure 5A). No staining of the underlying cervical parenchyma was observed, except within the endothelium of blood vessels, which immunostained intensely. Close examination of WIF1 staining within the stratified epithelium indicated a gradient of WIF1 expression within this tissue. Specifically, no detectable staining was observed in the basal epithelium or underlying basement membrane. However, strong stain was evident within the parabasal epithelial layer. We observed notable extracellular staining in the differentiated non-keratinized epithelium within the intermediate layer and diminished staining in cells at the apical surface of the ectocervix. A parallel tissue section stained with non-immune rabbit IgG clearly demonstrated the specificity of WIF1 staining within the cervical epithelium (Figure 5B).
Fig. 5.
Immunohistochemical analysis of WIF1 expression in normal and cancerous stratified cervical epithelium. A sample of normal ectocervix tissue was sectioned and stained with (A) anti-WIF1 or (B) non-specific rabbit IgG. Various tissue layers within the ectocervix are indicated. (C) Primary cervical tumor with intense WIF1 staining and low (4%) promoter methylation. (D) Primary cervical tumor with weak WIF1 staining and high (30%) promoter methylation. (E) Primary cervical tumor with moderate WIF1 staining and low (4%) promoter methylation. (F–I) Cervical tumors displaying heterogenous WIF1 staining.
When IHC was conducted on SCC samples with this WIF1 antibody, we observed intense staining in tumor samples in which pyrosequencing measured low levels of CpG methylation. For example, Tumor #10, in which pyrosequencing measured mean WIF1 m5CpG of 2% (see Figure 4B), exhibited high levels of immunoreactivity throughout the tumor (Figure 5C). Coordinately, tumors with high methylation values showed limited anti-WIF1 staining. As an example, Tumor #17, with a measured mean WIF1 methylation of 41%, displayed no detectable immunostaining (Figure 5D). These results support the association between aberrant hypermethylation of the WIF1 promoter and silencing of WIF1 expression in cervical tumors.
Of note, two of the tumor specimens in which pyrosequencing indicated low levels of WIF1 methylation showed a general lack of staining with WIF1 antibody. For example, pyrosequencing scored a mean m5CpG of 2% in DNA harvested from Tumor #1, but sections from this tumor showed a clear reduction in immunostaining (Figure 5E) when compared with other unmethylated tumors present (and simultaneously stained) on the microarray. Such findings seemingly indicate that promoter hypermethylation may not be the only mechanism linked to reduced WIF1 expression in cervical SCC.
Finally, while conducting IHC analyses on stained SCC specimens, we observed several tumors with heterogenous staining; specifically, we noted areas within the same tumor that stained intensely directly adjacent to regions which showed reduced or absent staining (Figure 5F–I). Upon close examination, we noted that the intense WIF1 staining was in regions of the tumor composed of well-differentiated lower-grade invasive SCC tumor cells. Conversely, regions of reduced staining principally comprised tumor cells displaying poorly differentiated, high-grade tumor cells suggesting that loss of WIF1 expression is associated with a more aggressive cervical SCC phenotype. A tumor recurrence rate of 25% was found among patients with high levels of WIF1 methylation and 12% in patients with low to moderate WIF1 methylation (P = 0.62); however, this study lacks sufficient sample size to examine WIF1 methylation as an independent prognostic factor.
Discussion
We were compelled to examine the WIF1 gene as a potential target for epigenetic silencing in cervical cancer based on three principal criteria: Firstly, dysregulation of the Wnt/β-catenin signaling axis is a common event in numerous tumor types, cervical cancer being among these. Uncontrolled β-catenin signaling, either from disruption of its regulatory proteins (i.e. Axin, APC, WIF1) or genetic mutations that prevent β-catenin degradation lead to excessive proliferation that predisposes cells to tumorigenesis. Secondly, while WIF1 is a component of a broad group of extracellular Wnt antagonists, expression of this molecule in normal cervical tissue and cervical cancer remained uninvestigated. Thirdly, WIF1 has been shown to be downregulated in a variety of cancer types, and this aberrant event has been linked to promoter hypermethylation.
Using both cervical cancer cell lines and primary cervical SCC tissues, we established that the WIF1 gene is commonly subjected to DNA hypermethylation in cervical cancer. Studies in cervical cell lines indicated that WIF1 expression correlated with promoter methylation. This conclusion is supported, in part, by our documentation that the DNA-demethylating agent 5-azadC increased WIF1 expression at both the messenger RNA and protein level. Furthermore, we observed that hypermethylation of the WIF1 promoter was associated with low or undetectable levels of WIF1 staining. Taken together, these findings support our conclusion that WIF1 is a target for epigenetic silencing in SCC of the cervix. Moreover, since analysis of a panel of cervical SCC samples indicates that 41% of these tumors show high levels of WIF1 promoter methylation, epigenetic silencing of WIF1 is a frequent molecular event in cervical SCC.
CpG methylation as determined by MAPit in cervical SCC lines is strikingly reminiscent of the mosaic patterns observed previously by us and others at the WIF1 promoter in breast, nasopharyngeal, esophageal and non-small-cell lung cancers (26–31). In particular, cell lines that transcribe WIF1 either confine endogenous methylation to sequences 5′ of the strong SP1 site (−916 to −319; cf. A549 and U1752 in (31)) and/or contain methylation between the two putative SP1 binding sites (Figure 3A and B; C33A and CaSki). Moreover, the strong consensus SP1 site was protected from CpG methylation to varying extents even in cell lines and patient tumor samples in which all examined copies were otherwise densely methylated over the full length of the WIF1 promoter (Figure 3C) (26,29,31,46). CpG methylation was also reversed at the strong SP1 site in MDA-MB-231 breast carcinoma cells upon WIF1 promoter activation by the adipokine adiponectin (46). Therefore, in cervical SCC cell lines, the defect in WIF1 expression is correlated with dense m5CpG 3′ of −160 relative to the TSS (CpG site 31).
By taking advantage of the additional capability of MAPit to footprint protein–DNA interactions (42), we show protection for the first time of the upstream SP1 and surrounding sequences against exogenously added M.CviPI in cervical SCC cells expressing the WIF1 gene (Figure 3A and B, cluster II). While this protection is presumably due to SP1 binding, it is possible that other DNA-bound proteins may contribute. For example, we observed additional footprints coinciding with a putative weak SP1 site (lacking flanking G residues), a potential initiator region (Inr) and bona fide transcription initiation regions (i.e. TSSs). The Myc-interacting protein-1 (Miz-1) is a candidate for binding to WIF1 TSSs and putative upstream Inr element (47). Miz-1 counteracts c-Myc transcriptional repressive activity by stimulating transcription through minimal promoter elements (48) and was recently show to associate directly with the WIF1 promoter in cultured non-small-cell lung cancer cells (31). Initiating or paused RNA polymerase II and/or other chromatin-associated proteins may also contribute to the observed footprints. Identification of the protein(s) responsible for specific footprints will require further study.
Nonetheless, our findings are consistent with previous reports that SP1 binding can block CpG island methylation (49,50). Moreover, several features underscore a remarkable epigenetic heterogeneity at the WIF1 promoter. Firstly, as MAPit can detect multiple footprints along the length of at single promoters (45), footprints increased in number while progressing from cluster IIa to IIc in C33A cells showing the highest abundance of WIF1 messenger RNA. This strongly suggests a model whereby factors are sequentially loaded onto the WIF1 promoter CpG island during activation, as previously reported for the GRP78 promoter (51). Secondly, sequential loading of factors at the WIF1 promoter was accompanied by three main regions of hypersensitivity to M.CviPI that increasingly extended in only the 3′ (CaSki) or both 5′ and 3′ directions (C33A) at the expense of protection by nucleosomes. Thirdly, there was a trend among molecules from C33A and CaSki cells toward excluding or changing the distribution of m5CpG. Fourthly, there were substantial qualitative and quantitative differences in chromatin accessibility and endogenous methylation suggesting association of a different constellation of factors and/or level of promoter activation in each cell line. A plausible interpretation of these results is that DNA-bound factor(s) corresponding to each footprint recruit chromatin remodelers and lead to varying extents of nucleosome sliding in the 5′ and/or 3′ direction, as we previously observed in simpler yeast and biochemical systems (41,52). Positive correlation of an increased fraction of nucleosome-depleted WIF1 promoters and transcription level from C33A to CaSki to SiHa cells lends strong support to this model. It is tempting to speculate that the partition between open and closed promoters in the C33A to CaSki lines explains the heterogenous WIF1 staining in tissue samples (Figure 5F–I).
A significant fraction of cluster Ia-b molecules in CaSki cells appeared to lack upstream m5CpG and contained randomly positioned nucleosomes punctuated by short accessible linker regions. Interestingly, hypermethylated copies of the silent WIF1 promoter from SiHa cells contained a similar random nucleosomal organization but contained significantly less and shorter regions of hypersensitivity adjacent to footprinted factors (Figure 3C). Taking this into account, we hypothesize that occasional promoter occupancy and chromatin remodeling by SP1 or other factors is sufficient to insulate against DNA methylation and/or drive local demethylation.
The role of the Wnt pathway in the development of cervical cancer is not well understood and mutations in β-catenin are rare in this type of cancer (53). Nevertheless, Uren et al. (54) observed that Wnt was required for transformation of cultured keratinocytes expressing HPV. Analysis of cervical tumors commonly showed heightened levels of β-catenin, clearly suggestive of dysregulation of Wnt/β-catenin signaling (55,56). High β-catenin immunoreactivity in cervical adenocarcinoma is associated with reduced disease-free survival (57), and altered β-catenin expression has been documented in cervical SCC as well (55). Considering the role of WIF1 as an antagonist of Wnt/β-catenin signaling, epigenetic silencing of its gene would result, at least in part, in dysregulation of this oncogenic mechanism. To address this possibility, we immunostained our cervical SCC tissue microarray with anti-β-catenin. Increased β-catenin staining was commonly observed in these cervical tumors, however, we observed no correlation between β-catenin staining and WIF1 methylation (supplementaryFigure 3 is available at Carcinogenesis Online). We conclude from these studies that WIF1 methylation is not a necessary molecular event to drive β-catenin dysregulation in SCC of the cervix.
While WIF1 is a well-established antagonist of oncogenic Wnt signaling, other molecular components of this signaling axis are also targets for epigenetic dysregulation in tumor cells. We have conducted pyrosequencing analysis on three of these genes, CDH1, FHIT and APC and unlike WIF1 promoter methylation, aberrant methylation of the promoter region of these genes was infrequently observed in our panel of cervical tumor samples. However, we have not analyzed the SRFP gene family, members of which are often epigenetically silenced in cervical cancer (13), for hypermethylation in our tumor panel. Thus, it remains undetermined if methylation of one or more of these genes coincides with WIF1 silencing. Nevertheless, given the prominent role that Wnt signaling plays in cervical carcinogenesis, it is probably that multiple distinct genetic and/or epigenetic events serve to dysregulate this signaling axis in cervical cancer.
Finally, in normal cervical tissue, we established that the WIF1 promoter is unmethylated and WIF1 protein is expressed in the parabasal layer of the squamous epithelium of the ectocervix. This IHC also suggests that as the squamous epithelium differentiates and migrates toward the apical surface of the ectocervix that WIF1 expression diminishes. Various Wnt family members have been implicated in the development of the female reproductive tract (58). Specifically, Wnt9b and Wnt4 are essential in the early development of the Müllerian duct, the embryonic structure that later develops into the Fallopian tubes, uterus, cervix and andvagina (59,60). Another family member, Wnt7a, has been implicated in controlling developmental patterning in the developing female reproductive tract (61) and has been implicated as a suppressor of cell death during uterine development (62). Thus, given the prominent roles that Wnt signaling plays in FRT development, expression of the WIF1 antagonist may be important in maintenance of differentiated cervical squamous epithelium.
Supplementary material
Supplementary Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (R03 CA143980); University of Florida start-up funds to M.P.K.
Supplementary Material
Acknowledgments
We thank Dr Nancy Nabilsi for critical reading of the manuscript and are grateful for support from the National Institutes of Health (R03 CA143980) and a University of Florida-Moffitt Collaborative Research Award to K.D.B. and E.M.S. as well as University of Florida start-up funds to M.P.K.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- 5-azadC
5-aza-2′-deoxycytidine
- FFPE
formalin-fixed paraffin-embedded
- HPV
human papillomavirus
- IHC
immunohistochemical
- PCR
polymerase chain reaction
- SCC
squamous cell carcinoma
- SDS
sodium dodecyl sulfate
- TSS
transcription start site
- WIF1
Wnt inhibitory factor-1
References
- 1.Walboomers JM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Morris SM, et al. p53, mutations, and apoptosis in genistein-exposed human lymphoblastoid cells. Mutat. Res. 1998;405:41–56. doi: 10.1016/s0027-5107(98)00126-2. [DOI] [PubMed] [Google Scholar]
- 3.Subramanya D, et al. HPV and cervical cancer: updates on an established relationship. Postgrad. Med. 2008;120:7–13. doi: 10.3810/pgm.2008.11.1928. [DOI] [PubMed] [Google Scholar]
- 4.Jones PA, et al. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nusse R, et al. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 6.Peifer M, et al. Wnt signaling in oncogenesis and embryogenesis—a look outside the nucleus. Science. 2000;287:1606–1609. doi: 10.1126/science.287.5458.1606. [DOI] [PubMed] [Google Scholar]
- 7.Polakis P. Wnt signaling and cancer. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 8.He TC, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 9.Wu KJ, et al. Direct activation of TERT transcription by c-MYC. Nat. Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 10.Kawano Y, et al. Secreted antagonists of the Wnt signalling pathway. J. Cell. Sci. 2003;116:2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. Sclerostin binds to LRP5/6 and antagonizes canonical Wnt signaling. J. Biol. Chem. 2005;280:19883–19887. doi: 10.1074/jbc.M413274200. [DOI] [PubMed] [Google Scholar]
- 12.Itasaki N, et al. Wise, a context-dependent activator and inhibitor of Wnt signalling. Development. 2003;130:4295–4305. doi: 10.1242/dev.00674. [DOI] [PubMed] [Google Scholar]
- 13.Szalmas A, et al. Epigenetic alterations in cervical carcinogenesis. Semin. Cancer Biol. 2009;19:144–152. doi: 10.1016/j.semcancer.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Kiyono T, et al. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the Drosophila discs large tumor suppressor protein. Proc. Natl Acad. Sci. USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SS, et al. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc. Natl Acad. Sci. USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa S, et al. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantovani F, et al. Proteasome-mediated regulation of the hDlg tumour suppressor protein. J. Cell. Sci. 2001;114:4285–4292. doi: 10.1242/jcs.114.23.4285. [DOI] [PubMed] [Google Scholar]
- 18.Handa K, et al. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J. Virol. 2007;81:1379–1389. doi: 10.1128/JVI.01712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuballa P, et al. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J. Biol. Chem. 2007;282:65–71. doi: 10.1074/jbc.M605117200. [DOI] [PubMed] [Google Scholar]
- 20.Lichtig H, et al. HPV16 E6 augments Wnt signaling in an E6AP-dependent manner. Virology. 2010;396:47–58. doi: 10.1016/j.virol.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Matsumine A, et al. Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 22.Ishidate T, et al. The APC-hDLG complex negatively regulates cell cycle progression from the G0/G1 to S phase. Oncogene. 2000;19:365–372. doi: 10.1038/sj.onc.1203309. [DOI] [PubMed] [Google Scholar]
- 23.Takizawa S, et al. Human scribble, a novel tumor suppressor identified as a target of high-risk HPV E6 for ubiquitin-mediated degradation, interacts with adenomatous polyposis coli. Genes Cells. 2006;11:453–464. doi: 10.1111/j.1365-2443.2006.00954.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhan L, et al. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsieh JC, et al. A new secreted protein that binds to Wnt proteins and inhibits their activities. Nature. 1999;398:431–436. doi: 10.1038/18899. [DOI] [PubMed] [Google Scholar]
- 26.Ai L, et al. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–1348. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 27.Urakami S, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin. Cancer Res. 2006;12:383–391. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 28.Roman-Gomez J, et al. Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood. 2007;109:3462–3469. doi: 10.1182/blood-2006-09-047043. [DOI] [PubMed] [Google Scholar]
- 29.Chan SL, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab. Invest. 2007;87:644–650. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 30.Mazieres J, et al. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–4720. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 31.Licchesi JD, et al. Transcriptional regulation of Wnt inhibitory factor-1 by Miz-1/c-Myc. Oncogene. 2010;29:5923–5934. doi: 10.1038/onc.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ai L, et al. Epigenetic silencing of the tumor suppressor Cystatin M occurs during breast cancer progression. Cancer Res. 2006;66:7899–7909. doi: 10.1158/0008-5472.CAN-06-0576. [DOI] [PubMed] [Google Scholar]
- 33.Darst RP, et al. Bisulfite sequencing of DNA. Curr. Protoc. Mol. Biol., Chapter 7, Unit 7. 2010;9:1–17. doi: 10.1002/0471142727.mb0709s91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demircan B, et al. Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer. 2009;48:83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adamson AW, et al. ATM is activated in response to MNNG-induced DNA alkylation. J. Biol. Chem. 2002;31:31. doi: 10.1074/jbc.M204409200. [DOI] [PubMed] [Google Scholar]
- 36.Pardo CE, et al. MethylViewer: computational analysis and editing for bisulfite sequencing and methyltransferase accessibility protocol for individual templates (MAPit) projects. Nucleic Acids Res. 2010;39:e5. doi: 10.1093/nar/gkq716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colella S, et al. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 39.Datta J, et al. A new class of quinoline-based DNA hypomethylating agents reactivates tumor suppressor genes by blocking DNA methyltransferase 1 activity and inducing its degradation. Cancer Res. 2009;69:4277–4285. doi: 10.1158/0008-5472.CAN-08-3669. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Issa JP, et al. Targeting DNA methylation. Clin. Cancer Res. 2009;15:3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jessen WJ, et al. Active PHO5 chromatin encompasses variable numbers of nucleosomes at individual promoters. Nat. Struct. Mol. Biol. 2006;13:256–263. doi: 10.1038/nsmb1062. [DOI] [PubMed] [Google Scholar]
- 42.Pardo C, et al. DNA methyltransferase probing of chromatin structure within populations and on single molecules. Methods Mol. Biol. 2009;523:41–65. doi: 10.1007/978-1-59745-190-1_4. [DOI] [PubMed] [Google Scholar]
- 43.Xu M, et al. Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 1998;26:3961–3966. doi: 10.1093/nar/26.17.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoose SA, et al. DNA methyltransferase probing of DNA-protein interactions. Methods Mol. Biol. 2006;338:225–244. doi: 10.1385/1-59745-097-9:225. [DOI] [PubMed] [Google Scholar]
- 45.Pondugula S, et al. Single-molecule analysis of chromatin: changing the view of genomes one molecule at a time. J. Cell. Biochem. 2008;105:330–337. doi: 10.1002/jcb.21849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, et al. Adiponectin stimulates Wnt inhibitory factor-1 expression through epigenetic regulations involving the transcription factor specificity protein 1. Carcinogenesis. 2008;29:2195–2202. doi: 10.1093/carcin/bgn194. [DOI] [PubMed] [Google Scholar]
- 47.Smale ST, et al. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 48.Peukert K, et al. An alternative pathway for gene regulation by Myc. EMBO J. 1997;16:5672–5686. doi: 10.1093/emboj/16.18.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macleod D, et al. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 50.Brandeis M, et al. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 51.Gal-Yam EN, et al. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dechassa ML, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ueda M, et al. Mutations of the beta- and gamma-catenin genes are uncommon in human lung, breast, kidney, cervical and ovarian carcinomas. Br. J. Cancer. 2001;85:64–68. doi: 10.1054/bjoc.2001.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uren A, et al. Activation of the canonical Wnt pathway during genital keratinocyte transformation: a model for cervical cancer progression. Cancer Res. 2005;65:6199–6206. doi: 10.1158/0008-5472.CAN-05-0455. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Sastre MA, et al. Abnormal distribution of E-cadherin and beta-catenin in different histologic types of cancer of the uterine cervix. Gynecol. Oncol. 2005;97:330–336. doi: 10.1016/j.ygyno.2004.12.062. [DOI] [PubMed] [Google Scholar]
- 56.Pereira-Suarez AL, et al. Frequent alterations of the beta-catenin protein in cancer of the uterine cervix. Tumour Biol. 2002;23:45–53. doi: 10.1159/000048688. [DOI] [PubMed] [Google Scholar]
- 57.Imura J, et al. Beta-catenin expression as a prognostic indicator in cervical adenocarcinoma. Int. J. Mol. Med. 2001;8:353–358. [PubMed] [Google Scholar]
- 58.Miller C, et al. Differential expression patterns of Wnt genes in the murine female reproductive tract during development and the estrous cycle. Mech. Dev. 1998;76:91–99. doi: 10.1016/s0925-4773(98)00112-9. [DOI] [PubMed] [Google Scholar]
- 59.Vainio S, et al. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- 60.Carroll TJ, et al. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev. Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 61.Parr BA, et al. Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature. 1998;395:707–710. doi: 10.1038/27221. [DOI] [PubMed] [Google Scholar]
- 62.Carta L, et al. Wnt7a is a suppressor of cell death in the female reproductive tract and is required for postnatal and estrogen-mediated growth. Biol. Reprod. 2004;71:444–454. doi: 10.1095/biolreprod.103.026534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.