Abstract
Breast cancer is an estrogen-driven disease. Consequently, hormone replacement therapy correlates with disease incidence. However, increasing male breast cancer rates over the past three decades implicate additional sources of estrogenic exposure including wide spread estrogen-mimicking chemicals or xenoestrogens (XEs), such as bisphenol-A (BPA). By exposing renewable, human, high-risk donor breast epithelial cells (HRBECs) to BPA at concentrations that are detectable in human blood, placenta and milk, we previously identified gene expression profile changes associated with activation of mammalian target of rapamycin (mTOR) pathway genesets likely to trigger prosurvival changes in human breast cells. We now provide functional validation of mTOR activation using pairwise comparisons of 16 independent HRBEC samples with and without BPA exposure. We demonstrate induction of key genes and proteins in the PI3K-mTOR pathway—AKT1, RPS6 and 4EBP1 and a concurrent reduction in the tumor suppressor, phosphatase and tensin homolog gene protein. Altered regulation of mTOR pathway proteins in BPA-treated HRBECs led to marked resistance to rapamycin, the defining mTOR inhibitor. Moreover, HRBECs pretreated with BPA, or the XE, methylparaben (MP), surmounted antiestrogenic effects of tamoxifen showing dose-dependent apoptosis evasion and induction of cell cycling. Overall, XEs, when tested in benign breast cells from multiple human subjects, consistently initiated specific functional changes of the kind that are attributed to malignant onset in breast tissue. Our observations demonstrate the feasibility of studying renewable human samples as surrogates and reinforce the concern that BPA and MP, at low concentrations detected in humans, can have adverse health consequences.
Introduction
Bisphenol-A (BPA) and methylparaben (MP) are xenoestrogens (XEs), i.e. non-steroidal chemicals that act like estrogens (1,2). BPA, a commonly used plasticizer, is so widely dispersed in the environment that 9 of 10 North Americans test positive in random urine samples (3,4). BPA has a short physiologic half-life, but due to continuous environmental exposure, BPA is routinely detected in human blood (5), placenta, cord (fetal) blood (6), fetal liver (7) and breast milk (8). BPA binds to estrogen receptors (ER) α and β (9,10) and reverses antiestrogen- (11) and chemotherapy-induced cytotoxicity in cancer cell lines (12). BPA induces upregulation of AKT (v-Akt murine thymoma viral oncogene homolog 1) in association with increased proliferation and decreased apoptosis of epithelial cells in breast tissue of lactationally exposed rats (13), as well as histological changes associated with mouse mammary carcinogenesis after in utero exposure (14). These effects are specific to breast tissues since BPA treatment of adipocytes (15) and leukemia cells (16) reduces phosphorylation of the serine/threonine protein kinase AKT and promotes terminal differentiation and cell death. MP, a common preservative in medicines, toiletries and skin care products (2), is detected in human breast tumors (2,17) and induces estrogenic signaling in the MCF7 breast cancer cell line (18,19). Because breast cancer incidence is proportional to estrogen exposure (20,21), there is concern that such estrogen mimics have contributed to increased breast cancer in both women and men over the last three decades (22,23).
To test the validity of insights acquired from animal and cancer cell line models, we developed assays based on renewable, early passage, non-malignant, high-risk donor breast epithelial cell (HRBEC) cultures derived from fresh human samples. In global gene expression analysis, HRBECs exposed to a low concentration of BPA exhibited geneset alterations that predicted activation of the mammalian target of rapamycin (mTOR) pathway (24) thereby implicating XE-induced effects in destabilizing a central function in normal cells. For example, downregulation of the mTOR pathway often occurs in a nutrient-poor microenvironment, thereby, limiting cell proliferation and allowing cell death through apoptosis and autophagy (25). However, when activated by hormones and/or abundant nutrition, or when co-opted in cancer development, mTOR signaling initiates protein synthesis, cell proliferation and evasion of apoptosis (25,26). In an independent set of HRBEC samples from similar high-risk individuals, we now demonstrate activation of key mTOR pathway proteins induced by XE exposure and downstream functional consequences. The use of HRBECs sidesteps issues of interspecies variation by testing cells from at-risk humans and bypasses issues of dose, route of delivery and metabolism by examining the effects of XE concentrations found in human tissues and body fluids (27,28). Because live HRBECs are drawn directly from the population of interest, i.e. the heterogeneous population of women at high risk of breast cancer occurrence, they serve well as surrogates for the effects of XEs on this population. The functional changes induced by BPA and MP closely parallel known outcomes of mTOR pathway activation (26) and tumor behavior (29) and reveal an underlying mechanistic basis for restricted effectiveness of breast cancer treatment and prevention strategies.
Materials and methods
Random periareolar fine needle aspirate collection and cytopathology
With Institutional Review Board-approved written informed consent, non-malignant cells were obtained by random periareolar fine needle aspiration from the unaffected contralateral breast of high-risk women undergoing breast surgery. We use the acronym HRBEC to represent ‘high-risk donor breast epithelial cells’. HRBEC donors were recruited based on personal or family history of breast cancer, atypical or neoplastic histopathology on biopsy and/or high mammographic density (30). Specimen size was ∼0.2 ml per volunteer including extraneous fat, blood and debris. Random periareolar fine needle aspirate (RPFNA) cell suspension was divided into aliquots for cytopathology and cell culture.
The cytology aliquot was collected in Cytolyt™, centrifuged at 600 r.p.m. for 10 min, transferred under vacuum to 20 mm circles on ThinPrep™ microscope slides using the ThinPrep™ 2000 processor (Cytec Corp., Boxborough, MA), fixed in 95% alcohol for 10 min and processed through an automated staining and mounting set up (Sakura Finetek USA, Torrance, CA). The entire 20 mm circle was evaluated by a board-certified cytopathologist (I.J.) for cytological atypia, and the number of epithelial and stromal cell clusters was counted.
Aliquots for cell culture were transferred into DME-F12 serum-free growth medium and transported on ice to the laboratory for delivery within 2 h.
Propagation and chemical exposure of HRBECs
Epithelial cells within RPFNA samples were plated and propagated in MCDB 170 medium supplemented with 2% fetal bovine serum as described previously (24). Twenty-three independent samples were expanded in vitro and used in the assays described below. The breast cancer cell lines—T47D, MCF7 and SKBR3—were used as controls and adapted to the same growth medium as HRBEC cultures prior to each assay. After 2–3 weeks of expansion, HRBECs were harvested for a variety of functional response tests.
To determine three-dimensional growth patterns, HRBECs were plated as single cell suspensions in a semisolid growth substrate comprised 3% Matrigel (BD Biosciences, San Jose, CA) and propagated for 10 days. Colonies fixed with 1:1 methanol:acetone were incubated with anti-alpha-6 integrin (BD Pharmingen, San Diego, CA) followed by fluorescence-tagged secondary antibody and propidium iodide (PI) nuclear counterstain. Immunostained colonies were visualized by confocal microscopy.
For XE exposure, cells were plated at a density of 100 000 cells per well in six-well plates and exposed to continuous 7 day treatments with 17β-estradiol (5 nM), 100 pM to 100 nM BPA or 10 nM to 1 μM MP in phenol red-free medium supplemented with 0.2% charcoal-stripped fetal bovine serum. Where indicated, cells were exposed to 10 μM 4-hydroxy tamoxifen (OHT) or to 1, 10 or 100 nM rapamycin for 24 h prior to functional analysis. All chemicals were purchased from Sigma–Aldrich (St. Louis, MO).
Protein detection by western blot analysis
Cells were lysed in denaturing lysis buffer in the presence of protease and phosphatase inhibitors and sonicated to disrupt cellular and nuclear membranes. Equal amounts of cell lysates were loaded onto 4–15% gradient gels for sodium dodecyl sulfate–polyacrylamide gel electrophoresis analysis, transferred to PVDF membranes and used for immunodetection with primary antibodies to ERα (Santa Cruz Biotechnology, Santa Cruz, CA), ERβ (Genetex, San Antonio, TX), phosphatase and tensin homolog gene (PTEN) (Cell Signaling Technology, Beverly, MA), PTEN phosphorylated at S380/T382/T383 (Santa Cruz Biotechnology), AKT1 (Santa Cruz Biotechnology), AKT1 phosphorylated at S473 (Santa Cruz Biotechnology), RPS6 (Genetex), RPS6 phosphorylated at S235/S236 (Genetex) and 4EBP1 (Santa Cruz Biotechnology). Actin, served as a loading control.
Real time quantitative polymerase chain reaction analysis
Total RNA was isolated using the RNeasy Kit (Qiagen, Valencia, CA) from untreated control and XE-treated cells. RNA concentration was determined by NanoDrop 2000 (Thermo Scientific, Lafayette, CO). Complementary DNA was synthesized and analyzed as before (31) by an Applied Biosystems 5700 Sequence Detection System (Applied Biosystems, Foster City, CA). The Ct values of test genes were normalized to the expression of the housekeeping genes, ACTB and GAPDH, within each HRBEC sample to represent log base 2-fold increase or decrease in test gene expression over no XE controls. Primer sequences for test genes are listed in supplementary Table S1 (available at Carcinogenesis Online).
Analysis of apoptotic and S-phase cell fractions
For quantitation of apoptotic cells, cultures were harvested and stained with Annexin V-FITC and PI (BD Biosciences) according to the manufacturer’s protocol. Cells diluted in binding buffer were analyzed by FACScan (BD Biosciences) and quantified by CellQuest software (BD Biosciences). Annexin-V-positive cells were measured as ‘early’ (PI-negative) and ‘late’ (PI-positive) apoptotic cell fractions. Each measurement was performed in multiple replicates.
For S-phase quantitation, cells were pulse labeled with 10 μM bromodeoxyuridine for 1 h, stained with anti-bromodeoxyuridine (Santa Cruz Biotechnology), FITC-conjugated secondary antibody (Invitrogen, Carlsbad, CA), counterstained with PI and analyzed by FACScan.
Quantitation of endogenous reactive oxygen species
Endogenous cellular reactive oxygen species (ROS) accrual was determined using live cell cultures loaded with the fluorogenic dye, carboxy-H2DCFDA (5-(and-6)-carboxy-2′7′-dichlorodihydrofluorescein diacetate), also known as C-400 (Invitrogen) in phenol red-free medium for 1 h followed by 30 min incubation in regular growth medium. After dye removal, cells were counterstained with PI, and intracellular oxidation of C-400 was measured by FACScan with the FL1 filter. Experiments were done in triplicate and 10 000 cells were acquired for each sample. ROS activity was expressed as mean fluorescence intensity (MFI) of the C400 dye. MFI of PI-negative (non-necrotic) cells was corrected for autofluorescence of unlabeled cells.
Results
Live HRBEC cultures display non-malignant breast epithelial attributes
In this study, HRBECs were derived from the unafflicted contralateral breast tissue of 23 individual volunteers (mean age—58, range 40–81 years). Sixteen women had a personal history of breast cancer, 4 displayed high-risk histopathology, 20 had mammographically dense breasts and 19 had family history of breast cancer (supplementary Table S2 is available at Carcinogenesis Online). Routine cytology of the samples prior to cell culture showed multiple epithelial clusters (range 4 to >200). No atypia was seen (Figure 1A). RPFNA samples yielded short-term epithelial cultures with variable degrees of growth potential. Generally, the initial cell seeding produced robust growth within 2–3 weeks, enabling culture expansion for up to three passages and providing morphologically homogeneous populations of 106–107 epithelial cells. Due to the low serum concentration of the growth medium, stromal fibroblast expansion did not occur (Figure 1B). There were no selection criteria for RPFNA sampling except that the volunteer present a clinically defined increased risk of breast cancer. HRBECs were used as they grew out in sufficient numbers for one or more assays. Identification of the samples employed is included in the relevant data figures. Our overall cell culture experience with >130 RPFNA samples thus far yielded three spontaneously immortalized (IMM) HRBEC cell lines, designated as IMM-PA024, IMM-PA025 and IMM-PA115, currently at passages 24–26.
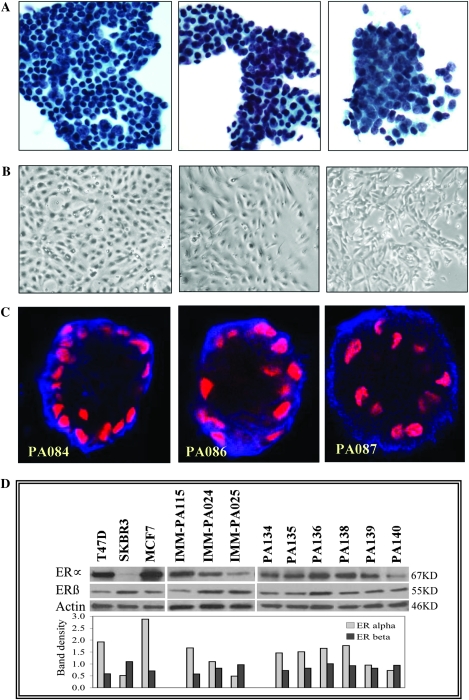
Fig. 1.
RPFNA-derived HRBEC cultures display characteristic non-malignant epithelial phenotypes. (A) Micrographs of cell clusters present in independent representative RPFNA samples. (B) Early passage pure epithelial cultures derived from clusters similar to those shown in A (Brightfield, × 4 objective). (C) Three-dimensional growth pattern of colonies derived from early passage HRBEC cultures in Matrigel. Basal immunolocalization of alpha-6 integrin (blue) and acinar orientation of nuclei (red) in 10 day-old colonies. (D) Relative ERα and ERβ protein levels in breast cancer cell lines (T47D, SKBR3 and MCF7), spontaneously immortalized HRBEC lines (IMM-PA024, IMM-PA025 and IMM-PA115) and early passage HRBECs (PA134, PA135, PA136, PA138, PA139 and PA140) quantitated by western blotting. Numerical values of each protein band representing ERα or ERβ obtained by densitometric scanning were normalized to actin levels in the lysate and plotted. Note low to moderate ERα levels in HRBECs relative to T47D and MCF7, but higher than the SKBR3 cell line, HRBECs display a distinctive ER profile unlike conventional ER-positive or ER-negative breast cancer cell lines.
Non-malignant epithelial characteristics of HRBEC cultures included:
a. Polarized growth in Matrigel—unlike breast cancer cell lines propagated in a three-dimensional matrix (which develop apolar colonies with random orientation of nuclei and the basement membrane protein, alpha-6 integrin), HRBEC colonies were distinctive. Polarized colonies, which displayed an acinar pattern of nuclear orientation and characteristic basal immunolocalization of alpha-6 integrin, were observed in all HRBEC cases tested (Figure 1C).
b. ER expression—distinct from ER-positive breast cancer cell lines, represented by T47D and MCF7, which express abundant ERα, and the ER-negative cancer cell line, SKBR3 in which ERα is undetectable, HRBEC lines (IMM-PA024, IMM-PA025 and IMM-PA115) displayed a low to moderate range of receptor protein (Figure 1D). On the other hand, levels of ERβ were relatively similar between HRBEC and cancer cell lines. Protein levels of both ER isoforms in six independent early passage HRBEC cultures (PA134, PA135, PA136, PA138, PA139 and PA140) were closely similar to those of spontaneously immortalized counterparts. We conclude that unlike many cancer cell lines, HRBECs represent an ERα-low/moderate status. Levels of both ERα and ERβ proteins are consistent with the characteristics of non-malignant human breast cells (32,33).
Early passage HRBEC cultures (passage 2 or 3) of 23 independent cases were used to generate subsets of data described below. HRBECs from 16 cases were used for pairwise analysis in functional assays (supplementary Table S2 is available at Carcinogenesis Online). Like normal human epithelial cells, HRBECs display a finite life and senesce after three passages, providing a limited amount of starting experimental material. For this reason, it is usually possible to pursue only one or two whole cell-based assays from a single sample, such as determination of apoptotic fraction, ROS estimation or assays based on RNA amplification, for example by quantitative polymerase chain reaction (QPCR) or by microarrays (reported by us previously). Similarly, early passage HRBEC-derived protein lysates are often insufficient for quantitative sodium dodecyl sulfate–polyacrylamide gel electrophoresis and western blot analysis. Therefore, we have included immortalized-HRBEC lines in protein quantitation studies.
BPA exposure modulates the signal transduction cascade of the PI3K–mTOR pathway within non-malignant breast epithelial cells
By QPCR analysis of the subset of critical mTOR pathway genes, early passage HRBECs derived from six individuals demonstrated identifiable shifts in expression associated with BPA exposure (Figure 2A). Considerable inter-subject variability in relative transcript levels was associated with the cellular BPA response, due to which dose–response relationships were not detectable in this small sample size. However, normalized to housekeeping genes, a consistent decline was noted in transcript levels of the suppressor genes, PTEN (P = 0.02, two-tailed t-test against theoretical ratio = 1.0), TSC1 and TSC2 (P = 0.03). Conversely, downstream activators of the mTOR pathway, e1F4B and e1F4E, were upregulated in the presence of BPA (P = 0.003). Expression of PIK3R1, another mTOR activator, was elevated (P = 0.02), transcript levels of RPS6, a downstream effector of mTOR that is activated by phosphorylation, were unexpectedly lower (P = 0.03) and there was a trend (P = 0.07) toward increased mTOR transcripts.
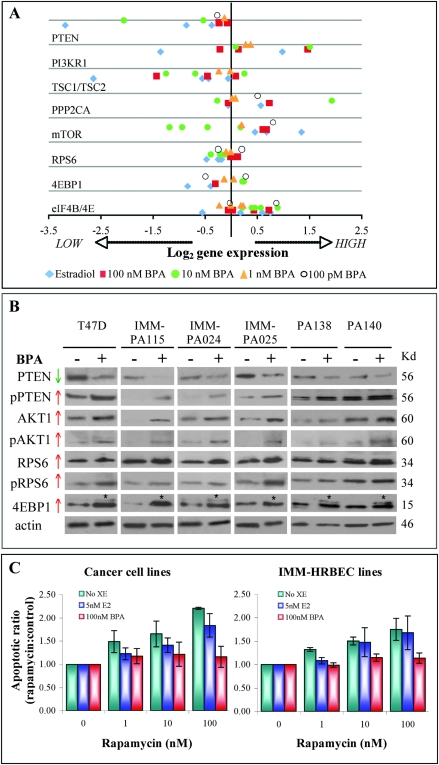
Fig. 2.
BPA exposure modulates expression of mTOR pathway components and induces functional changes in breast epithelial cells. (A) QPCR measurements of relative transcript levels of mTOR pathway genes in early passage HRBEC cultures (PA024, PA025, PA072, PA075, PA081 and PA112) normalized to housekeeping genes. Data represent exposure to a low dose range of BPA or to a luteal serum estradiol (E2) level. Plotted values represent fold change for each gene in treated samples, relative to the corresponding untreated control sample. Each data point is an average of triplicate QPCRs. (B) BPA-induced alterations in mTOR pathway proteins in early passage (PA138 and PA140), immortalized HRBECs (IMM-PA024, IMM-PA025 and IMM-115) and breast cancer cells (T47D). Pretreatment with BPA reduces steady-state PTEN protein levels and promotes functional inactivation by increased phosphorylation (indicated by ‘P’) at sites S380/T382/T383 compared with untreated controls. Increased expression of total and phosphorylated AKT1 (at S473), RPS6 (at S235/236) and 4EBP1 (at multiple sites) is detectable in both non-malignant and malignant breast cells. Asterisks indicate shift in the molecular weight of phosphorylated 4EBP1. (C) Effects of BPA pretreatment in the induction of resistance to the mTOR inhibitor, rapamycin. Data plotted to represent Annexin V-positive apoptotic populations within breast cancer cell lines (T47D, SKBR3—left panel) and HRBEC lines (IMM-PA024, IMM-PA025, IMM-PA115—right panel). Each bar represents the mean and standard deviation of triplicate values. Increased apoptotic ratios were observed with increasing doses of rapamycin in all cultures without BPA pretreatment. Values demonstrating the effect of BPA in reducing rapamycin-induced apoptosis were statistically significant in all cell lines (P > 0.001).
Confirmation of the observed transcriptional alterations was subsequently pursued at the protein level comparing two early passage HRBEC cultures, three immortalized HRBEC lines and an ERα-positive breast cancer cell line, T47D (Figure 2B). As predicted by gene transcript quantitation, BPA exposure led to the following: a significant reduction in total PTEN protein levels; an increase in the inactive phosphorylated form of PTEN (PTENSer380/2/3) and increased total and phosphorylated (activated) forms of the mTOR activating kinase, AKT1 and two major downstream targets of mTOR: RPS6 and of 4EBP1 (at multiple sites, as shown by the increased molecular weight of the phosphorylated protein). Unlike RPS6 transcript data, pRSP6 levels were representative of mTOR pathway activation. Increased phosphorylation observed for upregulated proteins is strongly indicative of increased mTOR activity and consistent with the finding of higher mTOR transcript levels.
XE exposure induces evasion of apoptotic cell death in HRBEC cultures
In order to establish additional functional association with mTOR pathway regulation, we asked whether BPA pretreatment altered the sensitivity of IMM-HRBEC lines to the antitumor drug—rapamycin, a potent mTOR inhibitor. A significant reduction in rapamycin-induced apoptotic cell death was observed after BPA exposure of immortalized HRBECs and in ERα-positive as well as ERα-negative breast cancer cells (Figure 2C). This effect of BPA in circumventing cell death was most prominent at 100 nM rapamycin, a concentration at which the apoptotic ratio of drug-treated versus untreated control was >2-fold in the absence of prior BPA exposure.
To further assess a role for XEs in the evasion of apoptotic cell death, the antiestrogenic OHT was used to initiate cell death. Early passage HRBECs from eight subjects were compared with IMM-HRBEC lines and with breast cancer cell lines. Percent apoptotic cells were estimated in untreated control cells, those pretreated with 100 nM BPA or 1 μM MP prior to a 24 h OHT exposure or those exposed to OHT alone. Although the percentage of apoptotic cells was doubled in the presence of OHT alone in all cell cultures tested, the effect of OHT was almost undetectable in XE-treated cultures (Figure 3A). Representative FACS data are illustrated in Figure 3B.
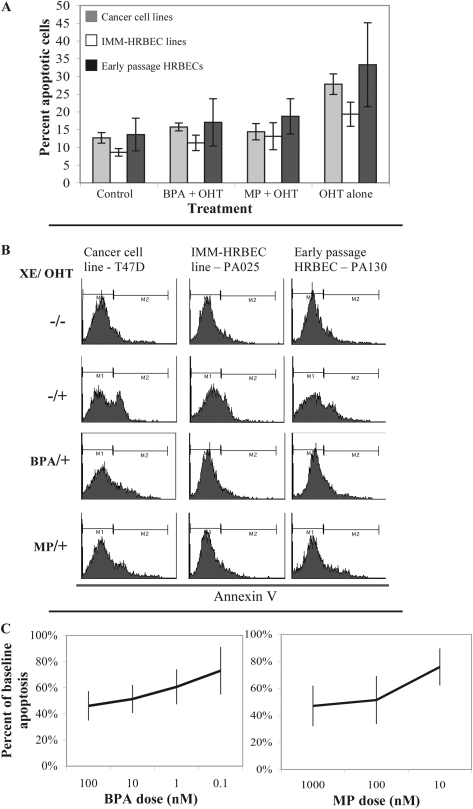
Fig. 3.
XE exposure promotes apoptosis evasion in HRBEC cultures. (A) Potential for XE-induced apoptosis evasion measured as percent reduction in Annexin V-positive cells by FACS analysis. Breast cancer cell lines (T47D and SKBR3), HRBEC cell lines (IMM-PA024, IMM-PA025 and IMM-PA115) and early passage HRBEC (PA094, PA099, PA103, PA106, PA107 and PA130) exposed to BPA or MP were treated with OHT for 24 h prior to Annexin V staining and compared with untreated controls. All experiments were performed in triplicate. Plots illustrate average values and the standard deviation for each culture group under conditions of no treatment, XE exposure followed by OHT or OHT treatment alone. Values demonstrating the effect of XEs in reducing OHT-induced apoptosis are statistically significant in all cases (P < 0.002). (B) FACS profiles of representative samples. M1 fraction—autofluorescence; M2—Annexin V-positive cells (which increase with OHT treatment). (C) Dose–response measurements (shown as decreasing XE concentrations from left to right) in early passage HRBECs. In all cases, the protection from OHT-induced apoptosis (apoptotic evasion) was calculated as a fraction of the apoptotic response in the absence of XEs (set to 1). Each data point represents an average of eight independent HRBEC samples (shown individually in supplementary Figure S1 is available at Carcinogenesis Online). Error bars display the variation between cases (triplicate values for each case). Note a striking dose–response effect for both XEs, despite variability between samples in the protection from OHT-induced apoptosis.
In separate experiments, early passage HRBECs were evaluated for the potential to evade apoptosis after exposure to a wide range of BPA and MP concentrations (Figure 3C as averaged data and supplementary Figure S1 is available at Carcinogenesis Online for each sample). Maximum protection from apoptotic death was conferred by exposure to 100 nM BPA (58.39 ± 6.8%). An appreciable degree of apoptosis reduction was also observed at 10- to 100-fold lower BPA doses: 52.84 ± 6.8% at 10 nM, 41.24 ± 10.4% at 1 nM and 19.93 ± 7.9% at 100 pM (Figure 3C, left panel). A significant dose–response was observed when log BPA concentration was regressed on log percent reduction in apoptosis (P = 0.002, two-sided test for logistic regression). MP exposure of HRBECs also dramatically reduced the fraction of OHT-induced apoptotic cells at all three doses tested: 57.82 ± 6.77% at 1 μM (Figure 4A), 55.93 ± 10.54% at 100 nM and 28.14 ± 11.3% at 10 nM (Figure 3C, right panel). Similar to the BPA dose response, MP-induced changes in HRBECs were also concentration dependent (P = 0.001).
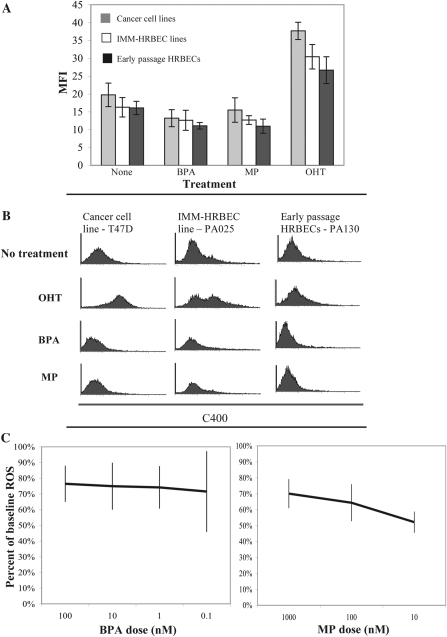
Fig. 4.
XE exposure alters oxidative stress levels in breast epithelial cells. (A) Comparative analysis of intracellular ROS levels measured and quantified by FACS analysis of C400-stained cancer cell lines (T47D and SKBR3), HRBEC lines (IMM-PA024, IMM-PA025 and IMM-PA115) and early passage HRBEC cultures (PA094, PA099, PA103, PA106, PA107 and PA130) exposed to BPA or MP. A post-XE 24 h treatment with tamoxifen (OHT) was used to induce ROS. All experiments were performed in triplicate. Averaged data representing the MFI of C400 are plotted, and standard deviations are shown. Values demonstrating the effect of XEs in reducing OHT-induced ROS are statistically significant in all cases (P < 0.0001). (B) FACS profiles of representative samples. The area under each curve reflects C400-positive cells. Note the right shift of the C400 peak in OHT-treated samples (indicating higher MFI) when compared with untreated control populations (top two panels) and the mild reduction of MFI in cells exposed to XEs (bottom two panels). (C) ROS levels measured as C400 MFI in early passage HRBECs exposed to various concentrations of BPA or MP. Results are expressed as percent reduction from baseline MFI of no XE controls. Each data point represents an average of six independent HRBEC samples (shown individually in supplementary Figure S2, available at Carcinogenesis Online). Error bars display the variation between cases.
XE exposure modulates cellular oxidative stress in HRBEC cultures
To determine whether functional changes in apoptosis evasion and cell survival were with a reflection of oxidative stress reduction, levels of endogenous ROS were quantified in early passage HRBECs, IMM-HRBEC lines and breast cancer cell lines. The MFI of C400-stained cells was used as an indicator of ROS. OHT treatment was used as a positive control for ROS induction. Both BPA and MP induced a detectable decline in endogenously accumulated ROS in all cell cultures during the 7 day exposure period (Figure 4A). Representative FACS data are illustrated in Figure 4B.
In further experiments, neutralization of ROS was measured in response to increasing log concentrations of BPA and MP exclusively in early passage HRBECs (Figure 4C as averaged data and supplementary Figure S2 is available at Carcinogenesis Online for each sample). The average reduction in ROS by BPA treatment was 26% (95% CI 24–28%; P < 0.001, two-sided t-test against theoretical ratio = 1.0, Figure 4C, left panel). Similarly, the average ROS reduction by MP treatment was 38% (95% CI 24–48%; P < 0.02, Figure 4C, right panel). A dose–response relationship was not observed for this end point.
XE-mediated cell cycle changes surmount drug-induced HRBEC growth inhibition
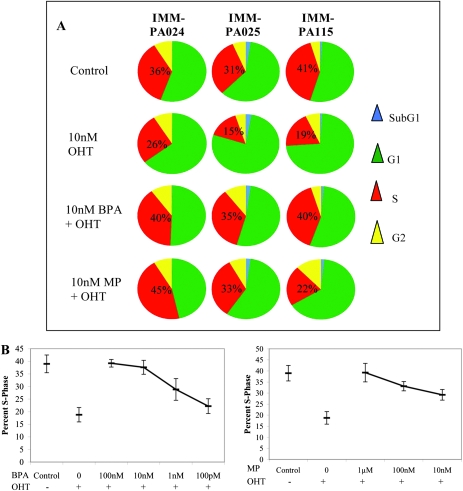
To determine whether resistance to OHT-induced apoptosis in BPA or MP-pretreated cells was accompanied by the maintenance of proliferative potential, we measured the efficiency of bromodeoxyuridine incorporation during the S-phase of the cell cycle in IMM-HRBEC lines. Prior exposure to a dose range of either BPA or MP resulted in a dramatic, concentration-dependent complete to partial evasion from the G1-phase arrest induced by 10 μM OHT and a concurrent increase in the S-phase fraction (Figure 5A and B). In contrast, the growth inhibitory effects of OHT were not reversed by a simulated physiological scenario of a combination of luteal phase serum levels of 17β-estradiol and progesterone in the absence of BPA (data not shown). As with apoptosis evasion, maintenance of S-phase in OHT-treated cells was significantly correlated with increasing concentrations of BPA and MP (P < 0.001 for BPA; P < 0.001 for MP, two-sided test for mixed effects linear regression).
Fig. 5.
XE exposure compromises tamoxifen-mediated cell cycle arrest in breast epithelial cells—(A) Cell cycle data derived from bromodeoxyuridine labeling of HRBEC lines (IMM-PA024, IMM-PA025 and IMM-PA115). Representative pie charts illustrate percent cells in different phases of the cell cycle (red—S-phase, yellow—G2, green—G1 and blue—sub G1). Note reversal of OHT-induced G1 arrest and subsequent S-phase decline in XE-pretreated cells. IMM-PA115 is relatively insensitive to the MP concentration shown compared with the other two HRBEC lines. (B) Graphical summary of S-phase reduction in OHT-treated cells and maintenance of S-phase by BPA (left panel) and MP (right panel) pretreatment in response to decreasing concentrations (from left to right). Plots represent an average of the combined bromodeoxyuridine-positive populations and standard deviations around the mean within independent HRBEC lines exposed to either BPA or MP (IMM-PA024, IMM-PA025 and IMM-PA115), shown individually in supplementary Figure S3 (available at Carcinogenesis Online).
Discussion
We demonstrate that BPA activates the mTOR pathway in non-malignant HRBECs. Both transcript and protein quantitation analyses reflected changes in the most important representative elements of this signaling pathway. In live cells, BPA suppresses apoptosis, enhances S-phase and decreases ROS levels, a well-known prelude to apoptosis evasion. Live cells respond in a similar manner to the XE, MP, suggesting that mTOR activation might be a general effect of XEs. These functional assays serve to test and confirm the consequences of mTOR activation predicted by global expression profiling of a previous independent set of BPA-exposed HRBECs (24).
Chemicals tested here are so common in bodily fluids (3–8) as to make unexposed control subjects functionally, if not literally, unavailable. We provide a pragmatic approach to overcome this problem by using live, renewable breast epithelial cell samples from high-risk donors (HRBECs) propagated in vitro with and without the chemicals of interest for pairwise comparisons of known end points of mTOR activation. HRBECs are samples from women who, based on extensive epidemiology, are identifiable as predisposed to malignant progression, even before overt cytopathological alterations are present. The opportunity to demonstrate a concordant set of multiple end points portraying the status of the mTOR metabolic pathway—despite small sample size—underscores the suitability of this source of target human cells for recapitulating early functional changes induced by carcinogen exposure. There is, and always will be, a gap between human biology in vivo and its representation by surrogate in vitro models, but unlike the limited genotypic variation represented by common immortalized cell lines, HRBECs more closely reflect the diversity of genetics, exogenous hormone use, life events such as pregnancy, etc. of women encountered in clinical practice. Our demonstration that HRBECs from all subjects displayed prosurvival changes after XE exposure does not claim that all exposed persons will develop cancer, only that such exposure can cause changes that may facilitate malignant progression.
Human cell samples that maintain phenotypes relevant to clinical targets of interest are essential for carcinogenicity testing. For example, to evaluate the role of XEs in the initiation of breast cancer, HRBECs are particularly well suited because they consistently express low/moderate levels of both ERα and ERβ. Earlier studies that did not distinguish ER isoforms (as continues in current clinical practice where primarily ERα is measured) typically reported low ER positivity in normal or benign breast tissue (32–34), suggesting that this phenotype is sufficient for agonistic activity of natural and synthetic estrogens, as well as for antagonistic effects of tamoxifen in reducing breast cancer incidence in randomized trials (35,36), even in patients harboring ER-negative atypia (37). Thus, high ERα levels are not a prerequisite for XEs to exert their biological effects either clinically or in vitro. Similarly, reversal of tamoxifen toxicity by exposure to the common XEs, BPA and MP, implies that the ER profile of HRBECs is also sufficient for the induction of cancer-associated phenotypes by estrogenic chemicals.
Against the backdrop of non-malignant ER levels simulated by HRBECs, BPA-induced transcriptional and protein alterations characteristic of breast cancer were discernible. For example, activation by phosphorylation of AKT is a key regulatory step in subsequent mTOR activation and induction of cell growth (25). During in vitro morphogenesis and differentiation of acini from non-malignant cells in Matrigel, pAKT localizes to peripheral cells, away from the central apoptotic region (38). Moreover, constitutive pAKT expression prevents lactogenic differentiation (39). Expectedly therefore, pAKT is increased during RAS-induced carcinogenesis of MCF10 xenografts (40) and pAKT is necessary for carcinogenic progression in this model since blocking pAKT results in apoptotic cell death (40). Clinically, a similar progression is observed whereby pAKT is significantly higher in malignant than benign breast tissue (41) and pAKT levels in cancer correlate directly with poor prognosis (42,43). Conversely, in vitro PTEN inactivation by phosphorylation and reduction in protein levels parallel Cowden’s syndrome where PTEN loss of function is associated with an increase in breast and other cancers (44,45). Downstream, mTOR acts by phosphorylating (activating) 4EBP1 and RPS6 (indirectly through p70S6K activation). Both p4EBP1 and its target eIF4E increase during experimental carcinogenesis (40); p4EBP1 is higher in cancer than in benign biopsies (41) and associated with higher tumor grade (46); and consequently, p4EBP1 correlates directly with tumor recurrence in women (46). In vitro, pRPS6 is downregulated by exposure to rapamycin (a defining mTOR inhibitor) in 12/12 cell lines (46,47). In one series, pRPS6 did not correlate with prognosis, but it was upregulated in the majority (77%) of breast cancers (46). mTOR activation by BPA exposure is further defined by inhibition of rapamycin-induced apoptosis (48), which is the converse of the therapeutic benefit of rapamycin analogs achieved through suppression of the mTOR pathway (49,50). In vitro pretreatment of HRBECs with BPA essentially replicates the continuous environmental exposure underlying positive urine tests in the general population. The failure of rapamycin to induce apoptosis after BPA treatment raises concern that activation of mTOR by continuous XE exposure could limit the effectiveness of this drug and its analogs that are being tested in clinical trials (49,50). Thus, a single estrogen mimic undermines multiple cell control mechanisms, as demonstrated here for tamoxifen and rapamycin and by others for the chemotherapeutic agents, doxorubicin and cisplatin (12).
Evasion of apoptosis and increased S-phase, induced by test XEs in HRBECs, are accepted hallmarks of cancer (51) with wide applicability in clinical cancer management. Apoptosis occurs rapidly after initiation of successful chemotherapy (52,53). Tamoxifen induces apoptosis within the first 24 h of treatment in animal models (54,55). Radiation therapy works by triggering apoptosis (56). High S-phase is an established indicator of poor tumor outcome (57), and maintenance of proliferation in the face of therapy indicates inadequate treatment response (58). Moreover, a drop in the ratio of proliferating to apoptotic cells (Cell Turnover Index) is indicative of a response to hormone-based therapy in benign (59) and malignant breast tissue (60). Thus, the ability of HRBECs to evade apoptosis and continue DNA replication after XE exposure portrays the acquisition of two fundamental phenotypes of cancer. Another fact supporting the causative role of BPA and MP is that both evasion of apoptosis and maintenance of S-phase are induced in a dose-dependent manner in HRBECs.
In this study, BPA and MP exposures also led to reduced ROS in live HRBECs. This is consistent with the finding that inhibition of the AKT/mTOR complex correlates with a marked increase in ROS production (61), and conversely, induction of the S6 kinase promotes resistance to oxygen and glucose deprivation and reduction of ROS levels (62). Being certain of the effects of free radicals is complicated because, depending on the level of ROS, cellular oxidative stress promotes either apoptosis or DNA damage (63,64). Low levels of ROS that accumulate during normal metabolism cause repairable DNA damage, whereas high levels of ROS are necessary for induction of apoptosis by hormone therapy (65), chemotherapy (66) and ionizing radiation (67).
Expansion of clinical samples with limited cellularity undoubtedly requires additional effort compared with the convenience of infinite cell yields from immortalized lines. However, renewable samples, such as HRBECs obtained by RPFNA, are readily available and represent a significantly wider population than rare immortalized human cell lines. Even in just this study, if confined to cell lines alone, the majority of test samples would have been unavailable to reveal the consistent XE effects that occurred despite variable baseline levels of proteins such as PTEN, AKT and 4EBP1 (Figure 2B). HRBECs are uniquely suited to capture and ultimately to investigate the basis of this variability among humans. We conclude: (i) testing ‘renewable’ samples from the at-risk population allows a wider sampling of different genetics, hormonal history, etc. than occur in a limited class of experimental models, (ii) employing HRBECs in experimental studies facilitates hypothesis generation and confirmation in independent sample sets (as we have done) and (iii) the complexity of xenoestrogenic effects requires evaluation of multiple cellular end points at this time for impact assessment instead of relying on a single receptor or functional assay.
Funding
This study was funded by the California Breast Cancer Research Program (Grant 12IB-0115) and by donations to the California Pacific Medical Center Foundation. None of the donations were from a commercial entity.
Supplementary Material
Acknowledgments
W.H.G. and S.H.D. equally developed resources, directed research plan, analyzed data and prepared manuscript. M.G.L. and S.A.S. performed laboratory experiments and data analysis. D.H.M.—biostatistics. I.M.J.–cytopathology. All authors approved the final manuscript.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- AKT
v-Akt murine thymoma viral oncogene homolog 1
- BPA
bisphenol-A
- ER
estrogen receptor
- HRBEC
high-risk donor breast epithelial cell
- MFI
mean fluorescence intensity
- MP
methylparaben
- mTOR
mammalian target of rapamycin
- OHT
4-hydroxy tamoxifen
- PI
propidium iodide
- PTEN
phosphatase and tensin homolog gene
- QPCR
quantitative polymerase chain reaction
- ROS
reactive oxygen species
- RPFNA
random periareolar fine needle aspirate
- XE
xenoestrogens
References
- 1.Vom Saal FS, et al. An extensive new literature concerning low-does effects of bisphenol-A shows the need for a new risk assessment. Environ. Health Perspec. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darbre PD, et al. Paraben esters: review of recent studies of endocrine toxicity, absorption, esterase and human exposure, and discussion of potential human risks. J. Appl. Toxicol. 2008;28:561–578. doi: 10.1002/jat.1358. [DOI] [PubMed] [Google Scholar]
- 3.Calafat AM, et al. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003-2004. Environ. Health Perspect. 2008;116:39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Statistics Canada. Bisphenol A concentrations in the Canadian population, 2007 to 2009. http://www.statcan.gc.ca/pub/82-003-x/2010003/article/11324/tbl/tbl1-eng.htm (17 April 2011, date last accessed)
- 5.Padmanabhan V, et al. Maternal bisphenol-A levels at delivery: a looming problem? J. Perinatol. 2008;28:258–263. doi: 10.1038/sj.jp.7211913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schonfelder G, et al. Parent bisphenol A accumulation in the maternal-fetal-placental unit. Environ. Health Perspect. 2002;110:A703–A707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, et al. GC-MS analysis of bisphenol A in human placental and fetal liver samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011;879:209–214. doi: 10.1016/j.jchromb.2010.11.031. [DOI] [PubMed] [Google Scholar]
- 8.Sun Y, et al. Determination of bisphenol A in human breast milk by HPLC with column-switching and fluorescence detection. Biomed. Chromotogr. 2004;18:501–507. doi: 10.1002/bmc.345. [DOI] [PubMed] [Google Scholar]
- 9.Hiroi H, et al. Differential interactions of bisphenol A and 17 beta-estradiol with estrogen receptor alpha and ER-beta. Endocr. J. 1999;46:773–778. doi: 10.1507/endocrj.46.773. [DOI] [PubMed] [Google Scholar]
- 10.Matthews JB, et al. In vitro and in vivo interactions of bisphenol A and its metabolite, bisphenol A glucuronide, with estrogen receptors α and ß. Chem. Res. Toxicol. 2011;14:149–157. doi: 10.1021/tx0001833. [DOI] [PubMed] [Google Scholar]
- 11.Lewis JB, et al. 4-Hydroxytamoxifen-induced cytotoxicity and bisphenol A: competition for estrogen receptors in human breast cancer cell lines. In Vitro Cell. Dev. Biol. Anim. 2000;36:320–326. doi: 10.1290/1071-2690(2000)036<0320:HICABA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.LaPensee EW, et al. Bisphenol-A at low nanomolar does confers chemoresistance in estrogen receptor alpha positive and negative breast cancer cells. Environ. Health Perspect. 2009;117:175–180. doi: 10.1289/ehp.11788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins S, et al. Oral exposure to bisphenol A increases dimethylbenzanthracene-induced mammary cancer in rats. Environ. Health Perspect. 2009;117:910–915. doi: 10.1289/ehp.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Markey CM, et al. In utero exposure to bisphenol A alters the development and tissue organization of the mouse mammary gland. Biol. Reprod. 2001;65:1215–1223. doi: 10.1093/biolreprod/65.4.1215. [DOI] [PubMed] [Google Scholar]
- 15.Masuno H, et al. Bisphenol A accelerates terminal differentiation of 3T3-L1 cells into adipocytes through the phosphatidylinositol 3-kinase pathway. Toxicol. Sci. 2005;84:319–327. doi: 10.1093/toxsci/kfi088. [DOI] [PubMed] [Google Scholar]
- 16.Bontempo P, et al. Molecular analysis of the apoptotic effects of BPA in acute myeloid leukemia cells. J. Transl. Med. 2009;18:48. doi: 10.1186/1479-5876-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darbre PD, et al. Concentrations of parabens in human breast tumors. J. Appl. Toxicol. 2004;24:5–13. doi: 10.1002/jat.958. [DOI] [PubMed] [Google Scholar]
- 18.Byford JR, et al. Oestrogenic activity of parabens in MCF7 human breast cancer cells. J. Steroid Biochem. Mol. Biol. 2002;80:49–60. doi: 10.1016/s0960-0760(01)00174-1. [DOI] [PubMed] [Google Scholar]
- 19.Pugazhendhi D, et al. Comparison of the global gene expression profiles produced by methylparaben, n-butylparaben, and 17-beta estradiol in MCF7 human breast cancer cells. J. Appl. Toxicol. 2007;27:67–77. doi: 10.1002/jat.1200. [DOI] [PubMed] [Google Scholar]
- 20.Ravdin PM, et al. The decrease in breast-cancer incidence in 2003 in the United States. N. Engl. J. Med. 2007;356:1670–1673. doi: 10.1056/NEJMsr070105. [DOI] [PubMed] [Google Scholar]
- 21.Jungheim ES, et al. Short-term use of unopposed estrogen. A balance of inferred risks and benefits. JAMA. 2011;305:1354–1355. doi: 10.1001/jama.2011.405. [DOI] [PubMed] [Google Scholar]
- 22.Dey S, et al. Xenestrogens may be the cause of high and increasing rates of hormone receptor positive breast cancer in the world. Med. Hypotheses. 2009;72:652–656. doi: 10.1016/j.mehy.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Anderson WF, et al. Male breast cancer: a population-based comparison with female breast cancer. J. Clin. Oncol. 2010;28:232–239. doi: 10.1200/JCO.2009.23.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dairkee SH, et al. Bisphenol A induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res. 2008;68:2076–2080. doi: 10.1158/0008-5472.CAN-07-6526. [DOI] [PubMed] [Google Scholar]
- 25.Hay N, et al. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 26.Strimpakos AS, et al. The role of mTOR in the management of solid tumors. Cancer Treat. Rev. 2009;35:148–159. doi: 10.1016/j.ctrv.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Ginsberg G, et al. Does rapid metabolism ensure negligible risk from bisphenol A? Environ. Health Perspect. 2009;117:1639–1643. doi: 10.1289/ehp.0901010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengstler JG, et al. Critical evaluation of key evidence on the human health hazards of exposure to bisphenol A. Crit. Rev. Toxicol. 2011;41:263–291. doi: 10.3109/10408444.2011.558487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armengol G, et al. 4E-binding protein 1: a key molecular “Funnel Factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–7555. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 30.Ursin G, et al. Mammographic density and breast cancer in three ethnic groups. Cancer Epidemiol Biomarkers Prev. 2003;12:332–338. [PubMed] [Google Scholar]
- 31.Dairkee SH, et al. Oxidative stress pathways are highlighted in an immortalization signature in breast cancer. Oncogene. 2007;26:6269–6279. doi: 10.1038/sj.onc.1210452. [DOI] [PubMed] [Google Scholar]
- 32.Shoker BS, et al. Oestrogen receptor expression in the normal and pre-cancerous breast. J. Pathol. 1999;188:237–244. doi: 10.1002/(SICI)1096-9896(199907)188:3<237::AID-PATH343>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 33.Warner M, et al. The normal and malignant mammary gland: a fresh look with ERß on board. J. Mammary Gland Biol. Neoplasia. 2000;5:289–294. doi: 10.1023/a:1009598828267. [DOI] [PubMed] [Google Scholar]
- 34.Shaaban AM, et al. Declining estrogen receptor-ß expression defines malignant progression of human breast neoplasia. Am. J. Surg. Pathol. 2003;27:1502–1512. doi: 10.1097/00000478-200312000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Fisher B, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant breast and Bowel Project P-1 study. J. Natl. Cancer Inst. 1998;90:1371–1388. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 36.Poweles TJ, et al. Twenty-year follow-up of the Royal Marsden randomized, double-blinded tamoxifen breast cancer prevention trial. J. Natl. Cancer Inst. 2007;99:283–290. doi: 10.1093/jnci/djk050. [DOI] [PubMed] [Google Scholar]
- 37.Baker JC, et al. ESR1 promoter hypermethylation does not predict atypia in RPFNA nor persistent atypia after 12 months of tamoxifen. Cancer Epidemiol. Biomarkers Prev. 2008;17:1884–1890. doi: 10.1158/1055-9965.EPI-07-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Debnath J, et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 39.Galbaugh T, et al. EGF-induced activation of Akt results in mTOR-dependent p70S6 kinase phosphorylation and inhibition of HC11 cell lactogenic differentiation. BMC Cell Biol. 2006;7:34. doi: 10.1186/1471-2121-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SH, et al. Proteomic and phosphoproteomic alterations in benign, premalignant and tumor human breast epithelial cells and xenograft lesions: biomarkers of progression. Int. J. Cancer. 2009;124:2813–2828. doi: 10.1002/ijc.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou X, et al. Activation of the Akt/mammalian target of rapamycin/4E-BP1 pathway by ErbB2 overexpression predicts tumor progression in breast cancers. Clin. Cancer Res. 2004;10:6779–6788. doi: 10.1158/1078-0432.CCR-04-0112. [DOI] [PubMed] [Google Scholar]
- 42.Perezz-Tenorio G, et al. Activation of SKT/PBK in breast cancer predicts a worse outcome among endocrine treated patients. Br. J. Cancer. 2002;86:s40–S45. doi: 10.1038/sj.bjc.6600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkegaard T, et al. AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J. Pathol. 2005;207:139–146. doi: 10.1002/path.1829. [DOI] [PubMed] [Google Scholar]
- 44.De Vivo I, et al. Novel germline mutations in the PTEN tumour suppressor gene found in women with multiple cancers. J. Med. Genet. 2000;37:336–341. doi: 10.1136/jmg.37.5.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agrawal S, et al. Differential expression of novel naturally occurring splice variants of PTEN and their functional consequences in Cowden syndrome and sporadic breast cancer. Hum. Mol. Genet. 2006;15:777–787. doi: 10.1093/hmg/ddi492. [DOI] [PubMed] [Google Scholar]
- 46.Rojo F, et al. 4E-binding protein 1, a cell signaling hallmark in breast cancer that correlates with pathologic grade and prognosis. Clin. Cancer Res. 2007;13:81–89. doi: 10.1158/1078-0432.CCR-06-1560. [DOI] [PubMed] [Google Scholar]
- 47.Noh WC, et al. Determinants of rapamycin sensitivity in breast cancer cells. Clin. Cancer Res. 2004;10:1013–1023. doi: 10.1158/1078-0432.ccr-03-0043. [DOI] [PubMed] [Google Scholar]
- 48.Price DJ, et al. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 49.Baselga J, et al. Phase II randomized study of neoadjuvant everolimus plus letrozole compared with placebo plus letrozole in patients with estrogen receptor-positive breast cancer. J. Clin. Oncol. 2009;27:2630–2637. doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 50.Macaskil EJ, et al. The mammalian target of rapamycin inhibitor everolimus (RAD001) in early breast cancer: results of a pre-operative study. Breast Cancer Res. Treat. 2011;128:725–734. doi: 10.1007/s10549-010-0967-z. [DOI] [PubMed] [Google Scholar]
- 51.Hanahan D, et al. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 52.Buchholz TA, et al. Chemotherapy-induced apoptosis and Bcl-2 levels correlate with breast cancer response to chemotherapy. Cancer J. 2003;9:33–41. doi: 10.1097/00130404-200301000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Davis DW, et al. Automated quantification of apoptosis after neoadjuvant chemotherapy for breast cancer: early assessment predicts clinical response. Clin. Cancer Res. 2003;9:955–960. [PubMed] [Google Scholar]
- 54.Ellis PA, et al. Induction of apoptosis by tamoxifen and ICI 182780 in primary breast cancer. Int. J. Cancer. 1997;72:608–613. doi: 10.1002/(sici)1097-0215(19970807)72:4<608::aid-ijc10>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 55.Kallio A, et al. Role of mitochondria in tamoxifen-induced rapid cell death of MCF-7 breast cancer cells. Apoptosis. 2005;10:1395–1410. doi: 10.1007/s10495-005-2137-z. [DOI] [PubMed] [Google Scholar]
- 56.Eriksson D, et al. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31:363–372. doi: 10.1007/s13277-010-0042-8. [DOI] [PubMed] [Google Scholar]
- 57.Goodson WH, 3rd, et al. The prognostic value of proliferation indices: a study with in vivo bromodeoxyuridine and Ki-67. Breast Cancer Res. Treat. 1997;59:113–123. doi: 10.1023/a:1006344010050. [DOI] [PubMed] [Google Scholar]
- 58.Ellis MJ, et al. Outcome prediction for estrogen receptor—positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J. Natl. Cancer Inst. 2008;100:1380–1388. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.deLima GR, et al. Effects of low dose tamoxifen on normal breast tissue from premenopausal women. Eur. J. Cancer. 2003;39:891–898. doi: 10.1016/s0959-8049(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 60.Bundred NJ, et al. Fulvestrant, an estrogen receptor downregulator, reduces cell turnover index more effectively than tamoxifen. Anticancer Res. 2002;22:2317–2320. [PubMed] [Google Scholar]
- 61.Sun H, et al. Co-administration of perifosine with paclitaxel synergistically induces apoptosis in ovarian cancer cells: more than just AKT inhibition. Cancer Lett. 2011;310:118–128. doi: 10.1016/j.canlet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 62.Pastor MD, et al. mTOR/S6 kinase pathway contributes to astrocyte survival during ischemia. J. Biol. Chem. 2009;284:22067–22078. doi: 10.1074/jbc.M109.033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storz P. Reactive oxygen species in tumor progression. Front. Biosci. 2005;10:1881–1896. doi: 10.2741/1667. [DOI] [PubMed] [Google Scholar]
- 64.Liou GY, et al. Reactive oxygen species in cancer. Free Radic. Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nazarawicz RR, et al. Tamoxifen induces oxidative stress and mitochondrial apoptosis via stimulating mitochondrial nitric oxide synthesis. Cancer Res. 2007;67:1282–1290. doi: 10.1158/0008-5472.CAN-06-3099. [DOI] [PubMed] [Google Scholar]
- 66.Ramanathan B, et al. Resistance to paclitaxel in proportional to cellular total antioxidant capacity. Cancer Res. 2005;65:8455–8460. doi: 10.1158/0008-5472.CAN-05-1162. [DOI] [PubMed] [Google Scholar]
- 67.Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–785. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.