Abstract
The majority of infection by the human immunodeficiency virus (HIV-1) across the world occurs by heterosexual transmission and is likely mediated by virus present in genital secretions. In spite of this, infection is followed by clinical markers of the virus present in blood, which may not be representative of the virus involved in transmission. In fact, several studies have demonstrated that the genital tract represents a unique compartment for the virus. We assessed the relationship between immune system integrity, represented by CD4+ T cell counts, and the maintenance of viral compartmentalization between plasma and vaginal fluid virus in treatment naïve women from the Dominican Republic infected by the heterosexual transmission route. We cloned and sequenced cell free virus from plasma and genital fluid samples from six women to assess viral evolution, phylogenetic relatedness, and calculated co-receptor use for the C2V3 region of the envelope. Our analyses demonstrated plasma and vaginal fluid virus compartments remained intact only in samples from women with CD4+ T cell counts over 350 cells/μ1 majority of viral forms were predicted to use the CCR5 co-receptor, although several dual tropic forms were also identified. None of the clones were found to use the CXCR4 co-receptor even though many of the patients showed severe disease. Our findings lend further support to the role of an intact immune system in maintaining compartmentalization across blood and genital quasispecies and provide a compelling rationale to specifically consider genital tract viral forms in therapeutic and vaccine research.
Keywords: HIV-1, env
INTRODUCTION
A salient feature of human immunodeficiency virus type 1 (HIV-1) infection is rapid and continuous accumulation of mutations in different viral genes leading into generation of diverse viral variants within and between HIV-infected individuals. Numerous reports have confirmed the presence of distinct HIV-1 variants, based on the envelop analysis, in different anatomical compartments like brain, lung, lymph nodes, spleen, gastrointestinal and genital tracts, kidney and peripheral blood mononuclear cells (PBMC)(Barnett et al., 1991; Delassus et al., 1992; Dittmar et al., 1997; Donaldson et al., 1994; Epstein et al., 1991; Gratton et al., 2000; Gupta et al., 2000; Haggerty and Stevenson, 1991; Hughes et al., 1997; Itescu et al., 1994; Kemal et al., 2003; Keys et al., 1993; Korber et al., 1994; Marras et al., 2002; Ohagen et al., 2003; Overbaugh et al., 1996; Poss et al., 1995; Poss et al., 1998; Reddy et al., 1996). Likewise pol has been used to define differential evolution of the virus in diverse tissue compartments(Haddad et al., 2000; Potter et al., 2003; Tirado et al., 2004a, b; Wang et al., 2000; Wong et al., 1997).
The exact reasons for virus compartmentalization have not been fully defined. Clearly several factors are involved including immune selective pressures, differential penetration of antiretroviral medications, and distinct target cell populations present in different niche environments throughout the host. These variants in different anatomical compartments pose new challenges for the management of viral infection and vaccine development strategies.
Heterosexual transmission is a major component of the spread of HIV-1 infection with genital mucosa serving as site of the initial contact. Therefore study of the virus genotype in male and female genital compartments is critical to understand the viral pathogenesis, the development of vaccines, treatment strategies and blockage of heterosexual transmission. Furthermore study of the virus genotype in the female genital compartment may offer strategies to block not only the heterosexual transmission but also mother-to-child transmission. The relatively few existing studies, including some by our group, that have attempted to identify and characterize the virus in systemic and genital compartments of HIV-infected female (Andreoletti et al., 2007; Kemal et al., 2003; Overbaugh et al., 1996; Philpott et al., 2005; Poss et al., 1995; Poss et al., 1998; Sullivan et al., 2005; Tirado et al., 2004a, b) have shown distinct variants between vaginal and blood compartments. However these studies did not focus on the effect of CD4+ T cell counts on virus evolution in systemic and genital compartments. In view of our recent findings that higher CD4+ T cell counts causes monophylectic clustering of envelop sequences in patients undergoing highly active antiretroviral therapy (HAART)(Lorenzo et al., 2004), we decided to determine the effect of CD4+ T cell counts on virus evolution in systemic and genital compartments. In this study we examined 6 HIV-infected females who acquired the virus by heterosexual transmission and examined the virus collected from plasma and vaginal fluid for sequence variation (silent as well as non-silent mutation), co-receptor usage, and changes in glycosylation sites.
RESULTS
Six females enrolled for the study comprised two separate groups summarized in Table 1. Two individuals with relatively higher CD4+ counts (001M - 450 and 007M - 368 CD4+ T cells/μl blood) and 4 individuals with very low CD4+ counts (027M - 16; 006M - 38; 003M - 50; and 005M - 178 CD4+ T cells/μl blood). The viral load in the plasma ranged between 3,536 and 750,000 RNA copies/ml whereas in the vaginal fluid it ranged from below the level of detection to a maximum of 10,745 RNA copies/ml. Heterosexual exposure was reported to be only risk factor responsible for HIV transmission in all 6 females. These females were classified for disease status using the CDC 1993 classification(1992).
Table 1.
Virological, immunological and clinical characteristics of therapy naïve women from Domincan Republic
| ID | Clinical1 | Age | Viral Load2 RNA Copy Eq/mL | CD 4+ Cel ls/u 1 | Pattern of HIV-1 | Drug use (6 months) | Reported lifetime partners | HIV Risk Factor | |
|---|---|---|---|---|---|---|---|---|---|
| Plasma | VF/Ultra | ||||||||
|
|
|||||||||
| 001M | D | 29 | 120,148 | <400/555 | 45 | compartmental | none | 2 | Het |
| 0 | |||||||||
|
| |||||||||
| 007M | 30 | 3,536 | <400 | 36 | compartmental | none | Het | ||
| 8 | |||||||||
|
| |||||||||
| 005M | P | 25 | 31,541 | <400/<50 | 17 | intermingled | none | Het | |
| 8 | |||||||||
|
| |||||||||
| 003M | 23 | 750,000 | 842 | 50 | intermingled | none | sex worker | Het/IDU | |
|
| |||||||||
| 006M | P | 23 | 611,092 | <400/10,745 | 38 | intermingled | none | 2 | Het |
|
| |||||||||
| 027M | 30 | 128,037 | <400/292 | 16 | intermingled | none | Het | ||
Het-Known hetrosexual partner with HIV-1 risk, IDU-injection drug use
Clinical symptoms at the time of first visit to clinic, D-diabetic, P-pneumonia.
The viral loads were determined by HIV-1 monitor system for plasma and Ultra sensitive for vaginal fluid.
The CD4 cell count was determined by staining PBMC monoclonal antibodies against CD3 and CD4. The absolute CD4 count was calculated by multiplying lymphocyte numbers (obtained from complete blood count) and percentage of CD4-positive cells in flow cytometry.
Intracompartmental diversity is independent of viral load and CD4+ T cell level
Phylogenetic analyses of C2V3 envelope variants within the plasma compartment and the vaginal fluid compartment were performed by calculation of average sequence divergence within compartment using a maximum composite likelihood model (Tamura et al., 2004) in samples from six heterosexually infected women from the Dominican Republic (Table 2). Examination of both plasma and vaginal fluid compartmental diversity showed variation ranging from less than 1% to greater than 7% for the C2V3 region, which encompasses the co-receptor binding determinants. No correlation was found by comparing intracompartmental diversity in cell free viral sequences against viral loads in either plasma or in vaginal fluid samples or CD4+ T cell counts (Table 1). Participant 001M had the highest T cell count (450cells/μl) and the second highest total plasma diversity of 2.2%, but with a low vaginal fluid diversity of 0.4%. These values are similar to four (003M, 005M, 007M and 027M) of the remaining five participants in plasma and vaginal fluid sample diversity (ranging from 0.2% to 1.7% across all plasma and vaginal fluid compartmental diversity values, except 027M where only a single viral form was identified in vaginal fluid), even while the CD4+ T-cell counts among those four samples ranged as low as 16 cells/μl. In contrast, a single participant exhibited high intracompartmental diversity in both plasma and vaginal fluid (06M at 5.3% and 7.3%, respectively) while having the second lowest CD4+ counts of the group (Table 2). Thus no direct trends between intracompartmental divergence (compartmental diversity) and either viral load or CD4+ counts were observed. Table 1 indicates that viral loads in plasma and vaginal fluid likewise did not correlate with maintenance of CD4+ T cell integrity.
Table 2.
Divergence across compartments and diversity within compartments for untreated female patients
| 001M | 007M | 005M | 003M | 006M | 027M | |
|---|---|---|---|---|---|---|
| Net P-VF divergence1 | 12.5% ±3.1 | 0.9 % ±0.8 | 0.01% ±0.04 | 0.02% ±0.2 | 0.5% ±0.5 | NC2 |
| Plasma Diversity3 | 2.2% ±0.7 | 0.2% ±0.3 | 0.5%±0.4 | 1.5% ±0.7 | 5.3% ±1.2 | 0.8%±0.3 |
| Vaginal Fluid Diversity3 | 0.4% ±0.2 | 1.4% ±0.9 | 1.2%±0.7 | 1.7% ±0.7 | 7.3% ±1.8 | 0%±0 |
Divergence calculated from base substitutions per site of net average between groups.
NC – not calculated
Diversity indicates base substitution per site of average over all sequence pairs for each sample.
Integrity of compartmentalization correlates directly with CD4+ T cell counts
In contrast to our analysis of intracompartmental diversity vs. CD4+ T cells, the between compartment divergence showed a direct relationship to CD4+ levels. Table 2 shows that the net evolutionary divergence between compartments, calculated using the maximum composite likelihood model with 500 bootstrap replicates to determine standard error, trends toward higher values with higher CD4+ T cell counts. Samples from participant 001M, with the highest CD4+ T cell counts of 450 cells/μl also had the largest net divergence between plasma and vaginal fluid virus of 12.5% +/- 3.1%. Participant 007M had the second highest net divergence (0.9% +/- 0.8%) and the second highest CD4+ T cell count (368 cells/μl). All other participants had net divergence between compartment values of well under 1%. Due to presence of a single clone identified in vaginal fluid, net divergence was not calculated for participant 027M.
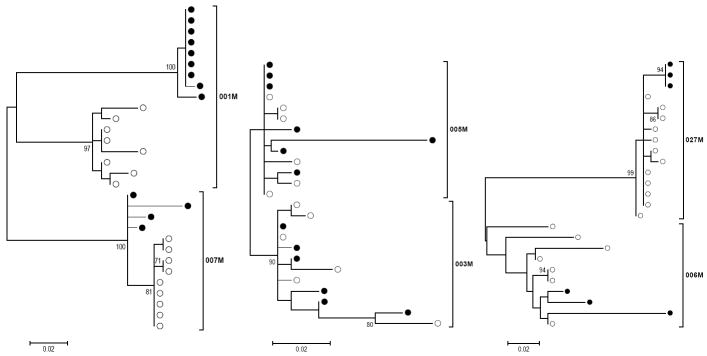
In addition to Table 2, phylogenetic trees were constructed to illustrate the extent of compartmental separation between virus derived from plasma and vaginal fluid. Maximum likelihood evolutionary history for sequences within each patient were calculated and trees were generated (Figure 1) and statistically assessed by bootstrapping (500 replicates). Trees for participant 001M and 007M, 450 cells/μl and 368 cells/μl, clearly show separation of plasma (closed circles) and vaginal fluid (open circles) clones. The other four participants, with CD4+ T cell counts under 200 fail to demonstrate compartmentalization, but rather show mixing of plasma and vaginal fluid forms within the same branches. These data more clearly indicate the integrity of compartmentalization of viral forms for CD4+ counts over 350 cells/μl and the loss at lower levels of CD4+ T cells. Further examination of synonymous and non-synonymous substitution rates (Table 4) across virus in plasma and vaginal fluid using the SNAP program provide an indication of statistically significant selective forces only in the participant with the highest CD4+ counts (001M), in agreement with the net-divergence (Table 2) and phylogenetic predictions in Figure 1. Each of the other samples for which dS/dN ratios could be calculated, failed to show significant mean ratios (Table 4), and notably had very low CD4+ counts. Sample 007M, with the second highest CD4+ counts failed to demonstrate sufficient levels of synonymous changes in plasma to perform this calculation.
Figure 1. Maximum Likelihood Model Phylogenetic Analysis shows compartmentalization in patients with higher CD4+ T cell counts.
Maximum likelihood model was employed to infer evolutionary history. Trees indicate phylogenetic relationship of clones isolated from vaginal fluid (closed circles) or plasma (open circles) from simultaneous samples. Patients 001M and 007M trees indicate separation of clones from plasma and vaginal fluid (leftmost tree). The phylogenetic arrangements of clones from 005M, 003M (middle tree) and 027M, 006M (rightmost tree) fail to show separate clusters of plasma and vaginal fluid clones. Scale bars indicate 2 nucleotide changes per 1000 positions. Bootstrap values greater than 70 are shown to indicate strength of clustering patterns.
Table 4.
Mean Synonymous and non-synonymous nucleotide substitution changes in different cell-free compartments of therapy naïve females.
| Sample | Mean ds ± SD | P value* | Mean dn ± SD | P value* | Mean ds/dn ± SD | P value* | |
|---|---|---|---|---|---|---|---|
| 001M | Plasma | 0.0292±0.0104 | 0.08 | 0.0238±0.0120 | 0.02 | 1.3369±0.4575 | 0.01 |
| Vaginal | 0.0425±0.0071 | 0.0100± 0.0000 | 7.3713±1.2763 | ||||
|
| |||||||
| 007M | Plasma | 0.0000±0.0000 | 0.83 | 0.0067±0.0048 | 0.11 | 0.0000±0.0000 | ϕ |
| Vaginal | 0.0400±.0.0000 | 0.0300±0.0100 | 1.4444±0.5092 | ||||
|
| |||||||
| 005M | Plasma | 0.0257±0.0098 | 0.16 | 0.0114±0.0038 | 1.00 | 3.6014±2.4277 | 0.50 |
| Vaginal | 0.0200±0.0000 | 0.0100±0.0000 | 2.9800±0.8600 | ||||
|
| |||||||
| 003M | Plasma | 0.0660±0.0260 | 0.63 | 0.0210±0.0129 | 0.89 | 5.3200±4.7624 | 0.65 |
| Vaginal | 0.0700±0.0263 | 0.0183±0.0112 | 6.2383±3.6943 | ||||
|
| |||||||
| 006M | Plasma | 0.0935±0.0448 | 0.11 | 0.0460±0.0274 | 0.18 | 3.5145±3.2352 | 0.59 |
| Vaginal | 0.1267±0.0513 | 0.0567±0.0322 | 2.3100±0.5200 | ||||
|
| |||||||
| 027M | Plasma | 0.0221±0.0063 | 0.10 | 0.0111±0.0032 | 0.08 | 2.1053±0.7375 | ϕ |
| Vaginal | 0.0000±0.0000 | 0.0000±0.0000 | undefined | ||||
P values calculated by Wilcoxon’s signed rank test
There are not enough valid cases to perform the Wilcoxon Signed Rank Test for dsdnplasma vs dsdnvaginal.
Patient 1 showed a difference in the non-synonymous nucleotide substitutions (p= 0.02) and ds/dn (P = 0.01) between plasma and vaginal samples.
No statistical significance differences were found in the patients 3, 5, 6, 7 and 27.
Co-receptor switch with progression is more prevalent in the vaginal fluid viral compartment
We did not obtain information related to the length of time since initial infection for any of the women in this study, although four of six have CD4+ counts below 200cells/μl, an immunological criterion for AIDS diagnosis. Furthermore, two of the participants presented pneumonia at time of first clinic visit, again supporting the advanced stage of HIV infection (Table 1). Interestingly, analysis of the co-receptor binding motif in the C2V3 amino acid sequence provided almost no evidence of an R5 to X4 co-receptor switch that is frequently associated with progression to AIDS Table 3 and Figure 2. Co-receptor prediction was nearly uniform in defining sequences with the R5 genotype among the entire cohort. Several clones with potentially dual tropic viral forms were defined. Though relatively few in plasma, 66% of the 03M plasma clones, several samples indicated the presence of dual tropic viral envelopes in vaginal fluid, ~20% of 003M, 66% of 006M, and all 027M vaginal fluid clones were predicted as dual tropic.
Table 3.
Predicted co-receptor use of clones isolated from plasma and vaginal fluid of treatment naïve females
| Patient | R5 | Dual | X4 | |
|---|---|---|---|---|
| 001M | Plasma | 8 | - | - |
| VF | 9 | - | - | |
| 007M | Plasma | 9 | - | - |
| VF | 4 | - | - | |
| 005M | Plasma | 6 | - | - |
| VF | 6 | - | - | |
| 003M | Plasma | 2 | 4 | - |
| VF | 5 | 1 | - | |
| 006M | Plasma | 7 | - | - |
| VF | 1 | 2 | - | |
| 027M | Plasma | 12 | - | - |
| VF | - | 3 | - |
Co-receptor use predicted using Web PSSM
Figure 2. Inferred amino acid alignment of C2V3 HIV-1 env sequences from plasma and vaginal fluid derived clones in therapy naïve females.
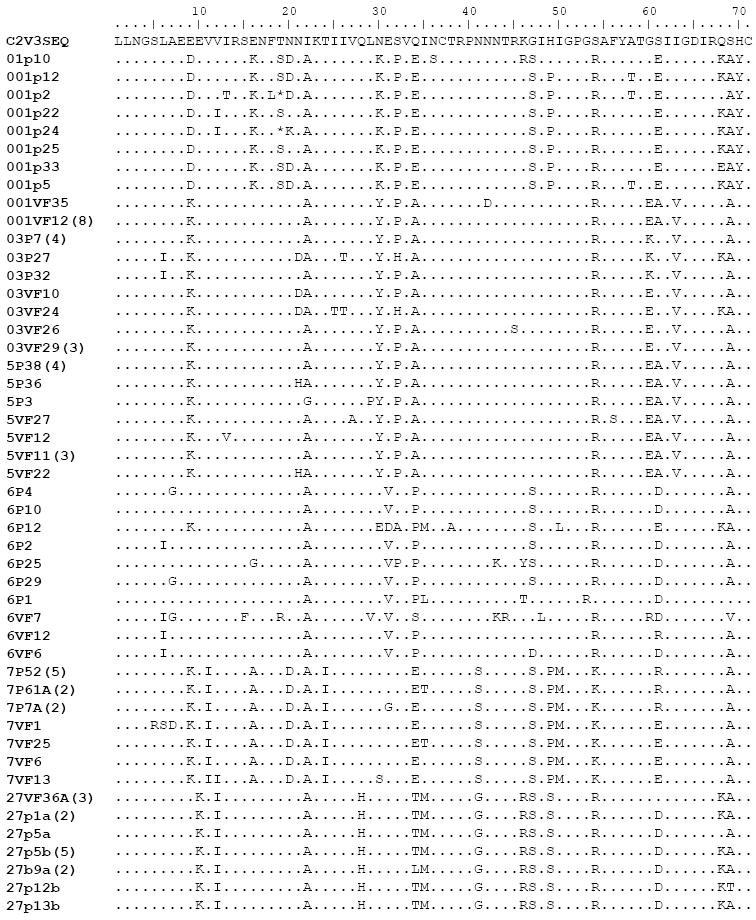
Amino acid changes are indicated relative to the C2V3 sequence as a reference sequence generated by the Bio Edit. Identical clones within each patient are not shown, but clones appearing multiple times are indicated in parentheses after clone names. Dots indicate identical amino acid sequences, dashes indicate deletions.
DISCUSSION
Our results suggest a positive relationship between maintenance of viral compartments at the C2V3 region of the envelope gene within the host and the integrity of the CD4+ T cell population. Our group previously found a loss of viral compartmentalization between vaginal fluid and plasma for the pol gene in a report of an individual with a CD4+ T cell count under 50 cells/μl that showed drug resistance mutations in the absence of ART(Tirado et al., 2004a). Several other studies have identified interesting patterns of viral load concordance between vaginal fluid and plasma. For example, analysis of over seventy women found detectable virus in 85% of plasma samples, while virus was detected in only 39% of the same individuals at coincident vaginal fluid sampling(Uvin and Caliendo, 1997). However, when the data were stratified by CD4+ counts, 67% of patients exhibited concordant detection of virus in plasma and vaginal fluid from a single time point. This study was unable to differentiate the role of loss of immune control of viral replication within compartments versus mixing of virus across compartments in driving the concordant detection of virus in plasma and vaginal fluid with lower CD4+ counts. However the findings are consistent with our data showing loss of compartmentalization with lower CD4+ T cells. In addition, only 50% of compartmentalized patients versus 75% of intermingled participants, showed detectable viral loads across plasma and vaginal fluid samples (Table 1).
Several other groups have reported a direct relationship between higher CD4+ counts and viral compartmentalization in the female genital tract compared with plasma. Kemal, et al. found correlation between higher CD4+ levels and distinct viral populations in plasma and vaginal tract cell-free virus(Kemal et al., 2003). Their study used cervico-vaginal lavage, which obtains virus from endocervix, vagina and fornix and represents a larger portion of the lower genital tract than our vaginal swab sampling assessed, indicating a greater extent of tissue is compartmentalized. A later study by the same group analyzed full length RNA genomes from plasma and female genital tract and found strong evidence for compartmentalization(Philpott et al., 2005). Interestingly, the association between CD4+ T cell levels and compartmentalization was less clear, as only a single study subject showed intermingled viral forms in the two compartments and this study subject had counts of 306 cells/μl. In contrast, three participants had lower counts and showed compartmentalization. There was evidence for recombination between plasma and vaginal tract virus in the patient with the lowest CD4 counts (6 cells/μl). This suggested mixing of virus between plasma and the female genital tract had occurred and perhaps the viral forms represented influence of recent replication as this individual had a genital tract viral load of 2.5million copies/mL(Philpott et al., 2005). A different study of clade A infection demonstrated clear blood and genital fluid viral compartmentalization in four women with clinically asymptomatic HIV-1 infection, again arguing for a strong role of the intact immune response in maintaining divergence of virus across compartments(Andreoletti et al., 2007). Sullivan et al., have also reported an association between CD4+ levels and viral divergence using heteroduplex mobility shift analysis of env C2V4, which they suggested was evidence for immune status in maintenance of compartmentalization(Sullivan et al., 2005). Thus our data further support the suggestion that the immune response impacts the compartmentalization of viral forms in genital and plasma fluids.
The important role of an intact immune system in maintaining selective pressure on HIV is a clear function of the CD4+ T cell sub-set; however, the role of the CD4+ T cells in this phenomenon in our dataset is more complex. Only a single participant, 001M, had evidence for immune selection according to calculation of the synonymous:non-synonymous change ratio. Yet the viral load for this patient demonstrated loss of control of the virus at time of sampling, suggesting the selection either occurred prior to sampling or was driven by other factors like target cell repertoire within the compartment. Alternatively, this may represent emergence of an archival form.
Initial comparison of plasma-vaginal fluid compartment divergence showed a rough correlation with CD4+ T cell level; however, patient 006M clearly showed high intracompartmental diversity even with a very low cell count, again indicating a complex role. Upon closer examination, this patient was remarkable in having high diversity in both plasma and vaginal fluid, particularly in comparison with the overall plasma-vaginal fluid divergence, which was quite low at 0.5%. In Figure 1 and Table 2, we clearly observe a direct relationship between compartmental separation and CD4+ T cells over 350 cells/μl. Thus divergence between compartments is only maintained at higher CD4+ levels. This may suggest that T cells are important for establishing or at least maintaining distinctions in the niches of viral replication in different anatomical regions of the host, whether or not the diversity within those niches is controlled. Thus the breakdown in the CD4+ T cell compartment by loss below 200 may result in loss of host compartmental selective forces thus allowing the systemic viral forms to become more uniform. In the case of 006M, since the intracompartment diversity is already high, the intercompartmental divergence represents greater diversity of forms within the entire host, rather than maintenance of unique viral niches, thus the net divergence is quite low. In contrast, no other patient has high diversity in both plasma and vaginal fluid. Among these five study participants, the intercompartmental divergence more closely follows a direct comparison with CD4+ levels.
MATERIALS AND METHODS
Study cohort and sample preparation
Six 23-30 year-old females from Dominican Republic were enrolled in the study. An informed consent was obtained from these individuals. The clinical profile of these individuals is shown in Table-1. Paired blood and vaginal swab samples were collected. Twenty ml blood was collected from each female of which 2 aliquots of 200μl were used for complete blood count (CBC) and CD4+ T-cell count, respectively. The remaining blood was centrifuged at 3,000 rpm for 10 min to separate the plasma. Plasma was frozen at -85° and an aliquot of that was used for viral RNA extraction. Vaginal secretion was collected by using 2 Dacron-tipped applicators and taking swabs from the cervico-vaginal area. The swabs were vigorously washed in 2 ml of R-10 (RPMI-1640, HEPES and 10% human AB plasma) to release the virus and loosen cells before being removed from tubes. The amount of vaginal secretions retained by each tip was calculated to be 133 μl by a dye dilution method. The vaginal cells were separated by centrifugation at 1,000 rpm for 10 minutes and supernatants were filtered through a 0.2 μm pore membrane filter. All vaginal fluid aliquots were frozen at -85°C until used for virus load determination and sequencing.
Viral load in plasma and vaginal fluid and CD4+ counts in the blood
Plasma and vaginal HIV-1 RNA viral loads were assessed using the Amplicor HIV-1 Monitor test standard specimen processing (Roche Diagnostics Corp., NJ). In case the viral load in a particular sample was found below the level of detection (<400 copies/ml), an ultra-sensitive specimen processing procedure was used to determine the viral loads. During sample preparation, since the vaginal fluids were diluted 8.5-fold (i.e., 2 × 133μl vaginal fluid diluted in the final volume of 2,266μl medium), the viral load in vaginal fluid was, therefore, expressed as (Assay Value) × 8.5. The CD4+ T-cell percentage was determined by flow cytometry and the absolute CD4+ T cell number was calculated using percentage CD4+ T cell count and lymphocyte numbers per μl blood, obtained from CBC.
RNA Extraction and RT-PCR
HIV-1 RNA was extracted from plasma and vaginal fluid using the QIAmp Viral RNA Mini Kit (QIAGEN, Valencia, CA) as described previously(Tirado et al., 2004b). In patient 005M with undetectable viral load (<50 copies/ml), approximately 2 ml of vaginal fluid sample was applied to the same column followed by other steps as recommended by manufacturer for extraction of very low viral load samples. A 541 base pair fragment (nucleotides 7001-7541, HXB2) was amplified using a nested PCR protocol. The first amplification utilized a RT-PCR reaction with 100 ng RNA, ready-to-go RT-PCR beads (Amersham Bioscience, Piscataway, NJ), forward (F 5’-CCTCAGTCATTACACAGGCCTGTCCAAAG-3’) and reverse primers (5’-AGTGCTTCCTGCTGCTCCCAAGAACCCAA-3’) in a 50 μl reaction mixture. The PCR conditions included 1 cycle of reverse transcription (42°C for 30 min), followed by denaturation at 95° for 5 min, 35 cycles of amplification (95°-30 sec, 60°-30 sec and 72°-1 min) and final elongation at 72° for 5 minute. One μl of 1st round PCR product was amplified in a nested PCR reaction using a forward (5’-CTGTTAAATGGCAGTCTAGC-3’) and reverse primer (5’-TGATGGGAGGGGCATACATT-3’) following the PCR conditions above (except reverse transcription step). The PCR product was resolved on 1.5% agarose, stained with ethidium bromide and visualized under ultraviolet light for size confirmation.
Cloning and sequencing of the PCR product
The PCR products were cloned into a TA vector according to TOPO TA Cloning Kit protocol (Invitrogen, Carlsbad, CA). The plasmid DNA was purified (QIAprep Spin column, QIAGEN) and sequenced using a 7-Deaza-dGTP Cy5/CY5.5 DYE primer kit and automated DNA sequencer (Visible Genetics, Toronto, ON, Canada).
C2-V3 sequence analysis
A 213 base pair sequence covering the C2-V3 segment was amplified from plasma and vaginal fluid (VF) viruses for analysis. The nucleotide sequences were aligned with BioEdit version 5.0.9 (Hall, 1999) followed by manual editing. The evolutionary analyses were made by maximum composite likelihood model (Tamura et al., 2004) and trees were inferred using 500 bootstrap replicates for statistical analysis. The trees were visualized using MEGA 5(Tamura et al., 2011). Potential N-linked glycosylation sites were determined by using the N-glycosite tool from Los Alamos sequence database.
The V3 sequences were also used for the prediction of coreceptor usage as described previously (Jensen et al., 2003). This program reliably predicts CXCR4 usage (sensitivity, 84% and specificity, 96%) by scoring V3 amino acids. This score is based on the basis of position-specific scoring matrices (PSSM) that predicts the co-receptor type. A low score suggests presence of CCR5 binding motif whereas an intermediate score predicts R5X4 virus and a high score suggests X4 virus.
Nucleotide sequence accession numbers
The sequence discussed in this work can be found in Genbank under accession numbers AY621091 to AY621092, AY624587 to AY624601, AY626150 to AY626155, AY623936 to AY623964, DQ060305 to DQ060317 and DQ205915 to DQ205929.
Acknowledgments
The study was supported by grants from NCRR (RR003050) and National Institute on Drug Abuse (DA015013, DA025011). We also acknowledge the support of the RCMI Molecular Biology Core.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreoletti L, Skrabal K, Perrin V, Chomont N, Saragosti S, Gresenguet G, Moret H, Jacques J, Longo JdD, Matta M, Mammano F, Belec L. Genetic and Phenotypic Features of Blood and Genital Viral Populations of Clinically Asymptomatic and Antiretroviral-Treatment-Naive Clade A Human Immunodeficiency Virus Type 1-Infected Women. J Clin Microbiol. 2007;45:1838–1842. doi: 10.1128/JCM.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett SW, Barboza A, Wilcox CM, Forsmark CE, Levy JA. Characterization of human immunodeficiency virus type 1 strains recovered from the bowel of infected individuals. Virology. 1991;182:802–809. doi: 10.1016/0042-6822(91)90621-h. [DOI] [PubMed] [Google Scholar]
- CDC. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- Delassus S, Cheynier R, Wain-Hobson S. Nonhomogeneous distribution of human immunodeficiency virus type 1 proviruses in the spleen. J Virol. 1992;66:5642–5645. doi: 10.1128/jvi.66.9.5642-5645.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar MT, Simmons G, Donaldson Y, Simmonds P, Clapham PR, Schulz TF, Weiss RA. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson YK, Bell JE, Holmes EC, Hughes ES, Brown HK, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68:5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LG, Kuiken C, Blumberg BM, Hartman S, Sharer LR, Clement M, Goudsmit J. HIV-1 V3 domain variation in brain and spleen of children with AIDS: tissue-specific evolution within host-determined quasispecies. Virology. 1991;180:583–590. doi: 10.1016/0042-6822(91)90072-j. [DOI] [PubMed] [Google Scholar]
- Gratton S, Cheynier R, Dumaurier MJ, Oksenhendler E, Wain-Hobson S. Highly restricted spread of HIV-1 and multiply infected cells within splenic germinal centers. Proc Natl Acad Sci U S A. 2000;97:14566–14571. doi: 10.1073/pnas.97.26.14566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta P, Leroux C, Patterson BK, Kingsley L, Rinaldo C, Ding M, Chen Y, Kulka K, Buchanan W, McKeon B, Montelaro R. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. J Infect Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- Haddad DN, Birch C, Middleton T, Dwyer DE, Cunningham AL, Saksena NK. Evidence for late stage compartmentalization of HIV-1 resistance mutations between lymph node and peripheral blood mononuclear cells. AIDS. 2000;14:2273–2281. doi: 10.1097/00002030-200010200-00008. [DOI] [PubMed] [Google Scholar]
- Haggerty S, Stevenson M. Predominance of distinct viral genotypes in brain and lymph node compartments of HIV-1-infected individuals. Viral Immunol. 1991;4:123–131. doi: 10.1089/vim.1991.4.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- Hughes ES, Bell JE, Simmonds P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J Virol. 1997;71:1272–1280. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itescu S, Simonelli PF, Winchester RJ, Ginsberg HS. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci U S A. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MA, Li FS, t Wout AB, Nickle DC, Shriner D, He HX, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. Improved Coreceptor Usage Prediction and Genotypic Monitoring of R5-to-X4 Transition by Motif Analysis of Human Immunodeficiency Virus Type 1 env V3 Loop Sequences. J Virol. 2003;77:13376–13388. doi: 10.1128/JVI.77.24.13376-13388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemal KS, Foley B, Burger H, Anastos K, Minkoff H, Kitchen C, Philpott SM, Gao W, Robison E, Holman S, Dehner C, Beck S, Meyer WA, Landay A, Kovacs A, Bremer J, Weiser B. HIV-1 in genital tract and plasma of women: Compartmentalization of viral sequences, coreceptor usage, and glycosylation. Proc Natl Acad Sci U S A. 2003;100:12972–12977. doi: 10.1073/pnas.2134064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys B, Karis J, Fadeel B, Valentin A, Norkrans G, Hagberg L, Chiodi F. V3 sequences of paired HIV-1 isolates from blood and cerebrospinal fluid cluster according to host and show variation related to the clinical stage of disease. Virology. 1993;196:475–483. doi: 10.1006/viro.1993.1503. [DOI] [PubMed] [Google Scholar]
- Korber BT, Kunstman KJ, Patterson BK, Furtado M, McEvilly MM, Levy R, Wolinsky SM. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo E, Colon MC, Almodovar S, Maldonado IM, Gonzalez S, Costa SE, Hill MD, Mendoza R, Sepulveda G, Yanagihara R, Nerurkar V, Kumar R, Yamamura Y, Scott WA, Kumar A. Influence of CD4+ T cell counts on viral evolution in HIV-infected individuals undergoing suppressive HAART. Virology. 2004;330:116–126. doi: 10.1016/j.virol.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, Ross MD, Gusella GL, Benson G, D’Agati VD, Hahn BH, Klotman ME, Klotman PE. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nature Med. 2002;8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- Ohagen A, Devitt A, Kunstman KJ, Gorry PR, Rose PP, Korber B, Taylor J, Levy R, Murphy RL, Wolinsky SM, Gabuzda D. Genetic and Functional Analysis of Full-Length Human Immunodeficiency Virus Type 1 env Genes Derived from Brain and Blood of Patients with AIDS. J Virol. 2003;77:12336–12345. doi: 10.1128/JVI.77.22.12336-12345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbaugh J, Anderson RJ, Ndinya-Achola JO, Kreiss JK. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–115. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- Philpott S, Burger H, Tsoukas C, Foley B, Anastos K, Kitchen C, Weiser B. Human Immunodeficiency Virus Type 1 Genomic RNA Sequences in the Female Genital Tract and Blood: Compartmentalization and Intrapatient Recombination. J Virol. 2005;79:353–363. doi: 10.1128/JVI.79.1.353-363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Martin HL, Kreiss JK, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–8122. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss M, Rodrigo AG, Gosink JJ, Learn GH, Vange Panteleeff D, Martin HL, Bwayo J, Kreiss JK, Overbaugh J. Evolution of Envelope Sequences from the Genital Tract and Peripheral Blood of Women Infected with Clade A Human Immunodeficiency Virus Type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter SJ, Dwyer DE, Saksena NK. Differential cellular distribution of HIV-1 drug resistance in vivo: evidence for infection of CD8+ T cells during HAART. Virology. 2003;305:339–352. doi: 10.1006/viro.2002.1703. [DOI] [PubMed] [Google Scholar]
- Reddy RT, Achim CL, Sirko DA, Tehranchi S, Kraus FG, Wong-Staal F, Wiley CA. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res Hum Retroviruses. 1996;12:477–482. doi: 10.1089/aid.1996.12.477. [DOI] [PubMed] [Google Scholar]
- Sullivan ST, Mandava U, Evans-Strickfaden T, Lennox JL, Ellerbrock TV, Hart CE. Diversity, Divergence, and Evolution of Cell-Free Human Immunodeficiency Virus Type 1 in Vaginal Secretions and Blood of Chronically Infected Women: Associations with Immune Status. J Virol. 2005;79:9799–9809. doi: 10.1128/JVI.79.15.9799-9809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular biology and evolution. 2011 doi: 10.1093/molbev/msr121. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirado G, Jove G, Kumar R, Noel RJ, Reyes E, Sepulveda G, Yamamura Y, Kumar A. Compartmentalization of drug resistance-associated mutations in a treatment-naive HIV-infected female. AIDS Res Hum Retroviruses. 2004a;20:684–686. doi: 10.1089/0889222041217509. [DOI] [PubMed] [Google Scholar]
- Tirado G, Jove G, Kumar R, Noel RJ, Reyes E, Sepulveda G, Yamamura Y, Kumar A. Differential virus evolution in blood and genital tract of HIV-infected females: evidence for the involvement of drug and non-drug resistance-associated mutations. Virology. 2004b;324:577–586. doi: 10.1016/j.virol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Uvin SC, Caliendo AM. Cervicovaginal human immunodeficiency virus secretion and plasma viral load in human immunodeficiency virus-seropositive women. Obstet Gynecol. 1997;90:739–743. doi: 10.1016/S0029-7844(97)00411-0. [DOI] [PubMed] [Google Scholar]
- Wang YM, Dyer WB, Workman C, Wang B, Sullivan JS, Saksena NK. Molecular evidence for drug-induced compartmentalization of HIV-1 quasispecies in a patient with periodic changes to HAART. AIDS. 2000;14:2265–2272. doi: 10.1097/00002030-200010200-00007. [DOI] [PubMed] [Google Scholar]
- Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]