SUMMARY
Glycerol-3-phosphate (sn-glycerol-3-P, G3P) acyltransferase catalyzes the first committed step in the biosynthesis of membrane phospholipids, the acylation of G3P to form 1-acyl G3P (lysophosphatidic acid). The paradigm G3P acyltransferase is the Escherichia coli plsB gene product which acylates position-1 of G3P using fatty acids in thioester linkage to either acyl carrier protein (ACP) or CoA as acyl-donors. Although the Escherichia coli plsB gene was discovered about 30 years ago, no evidence for transcriptional control of its expression has been reported. However Kazakov and coworkers (Kazakov, A. E. et al. (2009) J Bacteriol, 191, 52–64) reported the presence of a putative FadR-binding site upstream of the candidate plsB genes of V. cholerae and three other Vibrio species suggesting that plsB might be regulated by FadR, a GntR-family transcription factor thus far known only to regulate fatty acid synthesis and degradation. We report that the V. cholerae plsB homologue restored growth of E. coli strain BB26-36 which is a G3P auxotroph due to an altered G3P acyltransferase activity. The plsB promoter was also mapped and the predicted FadR-binding palindrome was found to span positions -19 to -35, upstream of the transcription start site. Gel shift assays confirmed that both V. cholerae FadR and E. coli FadR bound the V. cholerae plsB promoter region and binding was reversed upon addition of long chain fatty acyl-CoA thioesters. The expression level of the V. cholerae plsB gene was elevated 2–3 fold in an E. coli fadR null mutant strain indicating that FadR acts as a repressor of V. cholerae plsB expression. In both E. coli and V. cholerae the β-galactosidase activity of transcriptional fusions of the V. cholerae plsB promoter to lacZ increased 2–3 fold upon supplementation of growth media with oleic acid. Therefore, V. cholerae coordinates fatty acid metabolism with 1-acyl G3P synthesis.
INTRODUCTION
Our current knowledge of bacterial phospholipid biosynthesis is largely derived from studies of model organisms including E. coli (Cronan & Bell, 1974, Larson et al., 1980, Lightner et al., 1983, Lightner et al., 1980), Streptococcus pneumoniae (Lu et al., 2006) and Bacillus subtilis (Paoletti et al., 2007). The universal precursor of membrane phospholipid formation (Lu et al., 2006, Zhang & Rock, 2008), phosphatidic acid (PtdOH), is synthesized by acylation of sn-glycerol-3-phosphate (G3P) to 1-acyl-G3P followed by a second acylation to give phosphatidic acid (Fig. 1A) (Paoletti et al., 2007). Two different enzyme systems are known to catalyze the first G3P acylation reaction. PlsB (G3P acyltransferase) which acylates the 1-position of G3P using acyl thioesters of either CoA or acyl carrier protein (ACP) as acyl donors, was first identified in E. coli (Fig. 1 A&B) (Cronan & Bell, 1974, Larson et al., 1980, Lightner et al., 1980, Lightner et al., 1983, Zhang & Rock, 2008). The second system is the PlsX/Y two-enzyme system first found in S. pneumonia and B. subtilis (Lu et al., 2006, Paoletti et al., 2007). PlsX activates fatty acids by catalyzing production of fatty acyl-phosphates from fatty acyl-ACP thioesters whereas PlsY transfers the acyl chains of the acyl-phosphates to the 1-position of G3P (Lu et al., 2006, Paoletti et al., 2007). The PlsX-PlsY system is also found in E. coli, although PlsB plays the only essential role in 1-acyl-G3P synthesis (Yoshimura et al., 2007) (Baba et al., 2006). Acylation of position-2 of 1-acyl-G3P is catalyzed by PlsC, an acyltransferase found in most bacteria of known genome sequence (Lu et al., 2006). Like PlsB, E. coli PlsC utilizes either acyl-ACP or acyl-CoA as acyl donor to form phosphatidic acid although the acyl-donor preference of this enzyme varies among different bacteria (Lu et al., 2006, Paoletti et al., 2007, Zhang & Rock, 2008).
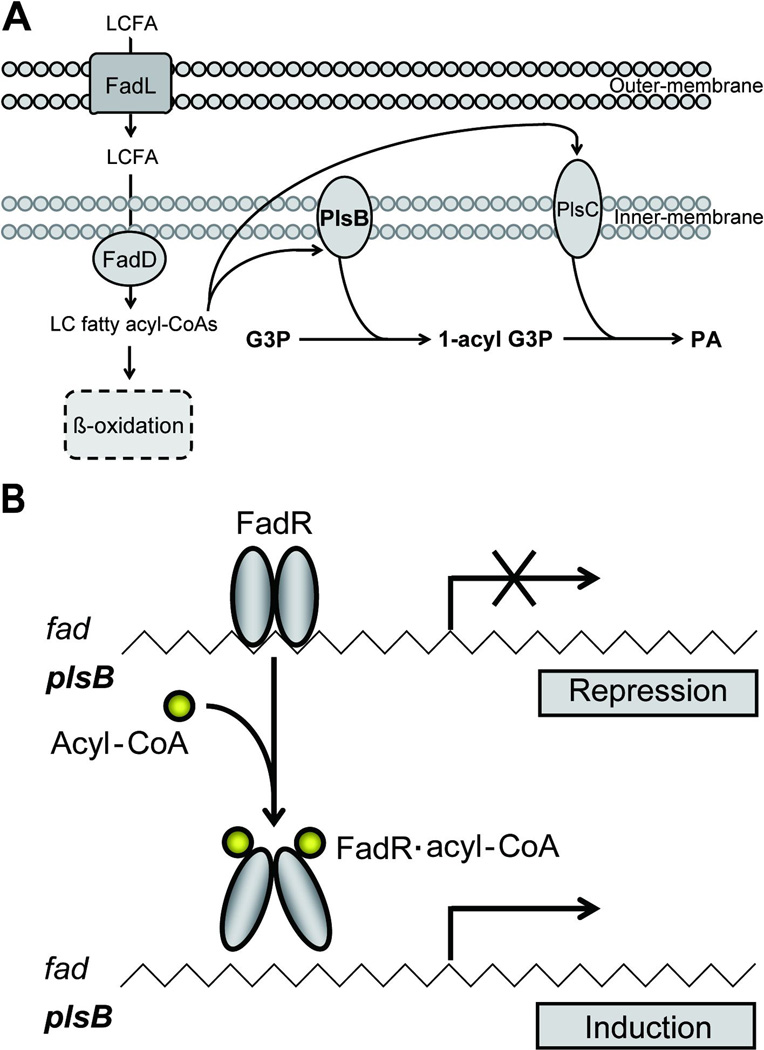
Fig. 1. Working model for the regulation of membrane phospholipid biosynthesis in V. cholerae.
A. Phosphatidic acid biosynthesis from exogenous acyl chains in V. cholerae. Phosphatidic acid synthesis is initiated by the PlsB-catalyzed transfer of fatty acyl chains to G3P from acyl-CoA, and subsequent PlsC-catalyzed acylation of acyl-G3P. LCFA: Long chain fatty acids; G3P: sn-glycerol-3-phosphate; LPA: 1-acylG3P; PA: phosphatidic acid.
B. Cartoon of the model for regulation of plsB expression in V. cholerae (the fad regulon would be similarly regulated). Binding of acyl-CoA results in dramatic rearrangement of the FadR C-terminal domain which drives the N-terminal DNA binding domains into a conformation which precludes cooperative DNA binding (van Aalten et al., 2001).
PlsB is a well-characterized membrane-bound enzyme (Fig. 1) (Cronan & Bell, 1974, Bell, 1975, Larson et al., 1980, Lightner et al., 1980, Lightner et al., 1983, Zhang & Rock, 2008). PlsB was discovered by isolation of E. coli strain BB26, a G3P auxotroph having a G3P acyltransferase with an elevated Km for G3P (Cronan & Bell, 1974, Bell, 1975). Further studies described the biochemical and enzymatic properties of PlsB (Bell, 1975, Larson et al., 1980, Lightner et al., 1980) and the plsB gene (Lightner et al., 1980, Lightner et al., 1983). However, comparative genomics indicates that plsB is confined to a subset of gram-negative bacteria, the Enterobacteriales, Vibrionales, and Burkholderiales (Lu et al., 2006). To date there have been no data demonstrating transcriptional regulation of expression of any bacterial G3P or 1-acyl-G3P acyltransferase, although due to the essential nature of phospholipid synthesis, expression was assumed to be constitutive. However, Kazakov et al. (Kazakov et al., 2009) recently conducted an extensive bioinformatics study of γ-protebacteria and reported a putative FadR biding site upstream of plsB homologues of four Vibrio species, V. cholerae, V. parahaemolyticus, V. vulnificus, and V. fischeri. FadR is a transcription factor of the GntR family having an N-terminal helix-turn-helix DNA-binding domain and a C-terminal ligand-binding domain (van Aalten et al., 2000, van Aalten et al., 2001, Xu et al., 2001, Iram & Cronan, 2005). Long chain fatty acyl-CoA species have been demonstrated to be regulatory ligands for FadR DNA-binding activity. Upon binding of long chain fatty acyl-CoA species the dimeric FadR undergoes dramatic conformational changes that results in rearrangement of the DNA recognition helices and dissociation of FadR protein from its operator sites (van Aalten et al., 2001, Henry & Cronan, 1992, Cronan, 1997). FadR, is known to play plays central roles in modulating lipid metabolism in E. coli (Henry & Cronan, 1991, Cronan & Subrahmanyam, 1998). Not only does it serve as a repressor for fatty acid degradation (fad) regulon (Iram & Cronan, 2005, Feng & Cronan, 2009b, Cronan & Subrahmanyam, 1998, Campbell & Cronan, 2002), but also functions as an activator of fabA and fabB, two key genes required for unsaturated fatty acids biosynthesis (Henry & Cronan, 1992, Campbell & Cronan, 2001). In addition, FadR also positively regulates the iclR gene which encodes a repressor for glyoxylate bypass operon (Gui et al., 1996). We report that in V. cholerae FadR acts as a repressor of plsB expression (Fig. 1C). To our knowledge this is the first example of transcriptional control of a membrane phospholipid acyltransferase.
RESULTS
Characterization of V. cholerae FadR
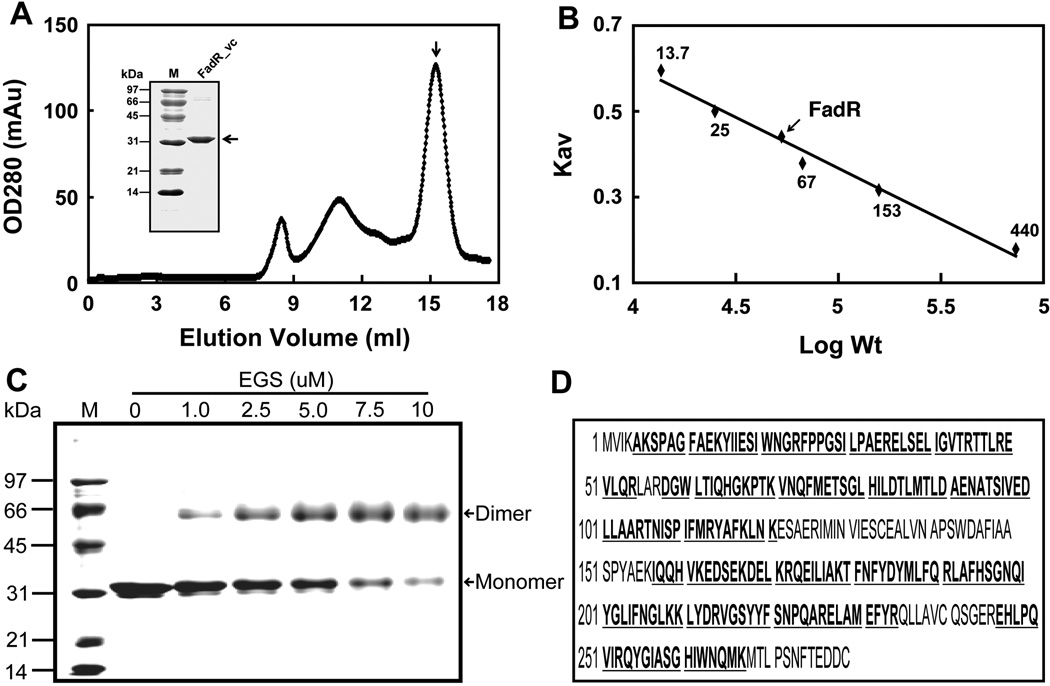
The V. cholerae fadR encodes a 279 residue polypeptide (Heidelberg et al., 2000) 40 residues longer than that of E. coli FadR, the best studied FadR protein (Fig. 2 A&E). Sequence alignment of these two FadR proteins indictaes they share 50.5% sequence identity. As previously noted the V. cholerae FadR (FadR_vc) contains a centrally located 40 residue insert (residues 138–177) relative to E. coli FadR and other FadR proteins (Fig. 2A) (Iram & Cronan, 2005). Recombinant FadR_vc protein was overexpressed, purified to homogeneity (Fig. 2B) essentially as previously described (Iram & Cronan, 2005) and verified by LC mass-spectrometry with a coverage score of 76% (Fig. 2E). Analysis by size exclusion chromatography (Fig. 2B) indicated that the FadR_vc solution structure was predominantly a dimer with an apparent molecular weight of ~62 kDa, although some larger forms were also present (Fig. 2C). Dimerization was also detected by chemical cross-linking assays (Fig. 2D). Modeling based on the E. coli FadR structure indicates that the 40-residue insertion basically extends a loop present in the E. coli protein (data not shown).
Fig. 2. Biochemical and structural characterization of V. cholerae FadR.
A. Gel exclusion chromatographic profile of recombinant V. cholerae FadR run on a Superdex 200HR 10/30 column (GE Healthcare). The expected peak of purified V. cholerae FadR was eluted at the position of 15.2 ml (indicated with an arrow). The inset gel is the SDS-PAGE analysis of the purified V. cholerae FadR. The apparent molecular weight of recombinant V. cholerae FadR is about 31 kDa. OD280, optical density at 280 nm; mAu, milli-absorbance units.
B. Determination of V. cholerae FadR solution structure according to elution patterns of a series of standard proteins (Pharmacia). The standard proteins were ribonuclease (~13.7 kDa), chymotrypsinogen (~25 kDa), bovine serum albumin, 67 kDa), aldolase (153 kDa), and ferritin (~440 kDa). The elution position of V. cholerae FadR is indicated with an arrow. KAV, partition coefficient; M, molecular weight.
C. Chemical cross-linking assay of V. cholerae FadR solution structure.
D. MS identification of V. cholerae FadR. The matched amino acid residues are given bold and underlined type.
V. cholerae plsB gene is a Functional Homologue of the E. coli protein
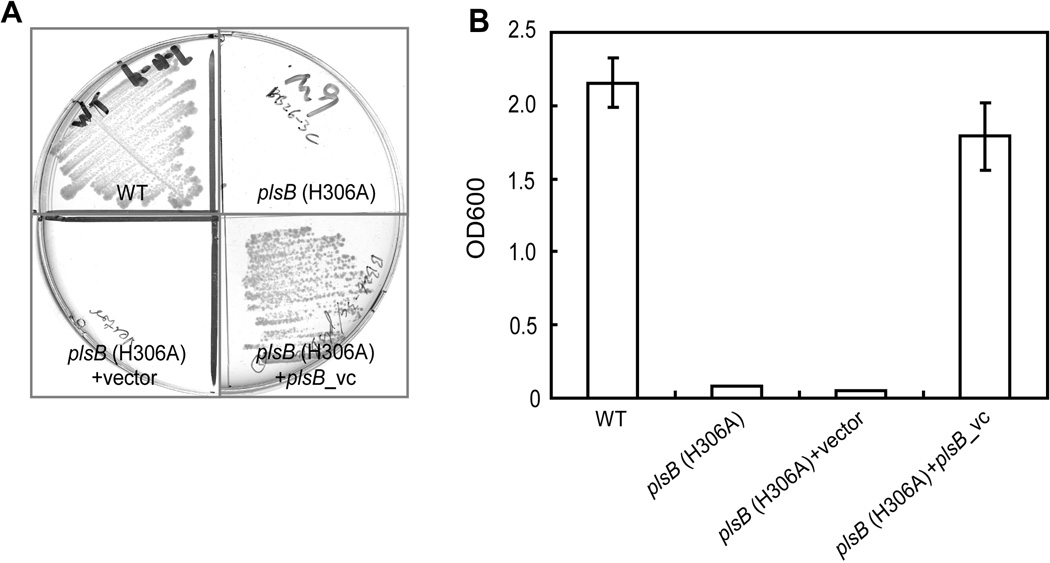
The V. cholerae plsB gene product (811 residues) aligned well with E. coli PlsB protein (807 residues) with 56.3% identity. To test the function of this plsB orthologue we assayed the ability to complement growth of an E. coli plsB mutant. Due to a point mutation that result in a H306A PlsB E. coli strain BB26-36 requires an exogenous supply of G3P to overcome a Km defect in the mutant enzyme (Cronan & Bell, 1974, Bell, 1975). The V. cholerae plsB gene carried by vector pCR2.1 allowed growth of strain BB26-36 on minimal medium M9 agar plates lacking G3P, whereas the strain carrying the empty vector failed to grow (Fig. 3A). A similar result was obtained in liquid medium (Fig. 3B). Thus, V. cholerae plsB gene encodes a functional G3P acyltransferase.
Fig. 3. Functional identification of V. cholerae plsB.
A & B. Growth of E. coli plsB mutant strain BB26-36 (which encodes a mutant H306A PlsB acyltransferase) on minimal medium in the presence or absence of V. cholerae plsB gene expression on either solid (A) or liquid medium (B). The vector was pCR2.1.
The V. cholerae plsB promoter
The genetic organization of the plsB gene on the V. cholerae chromosome I differs somewhat from that of the E. coli plsB locus. In both cases the ubiA gene (4-hydroxybenzoate polyprenyltransferase) is encoded downstream on the other DNA strand. In E. coli the the dgkA (diacylglycerol kinase) gene is upstream of plsB whereas in V. cholerae, dgkA is replaced by lexA which encodes the repressor of the bacterial SOS regulon. In E. coli it was recently reported that plsB and dgkA are inversely regulated by several stress responses at the transcriptional level (Wahl et al., 2011).
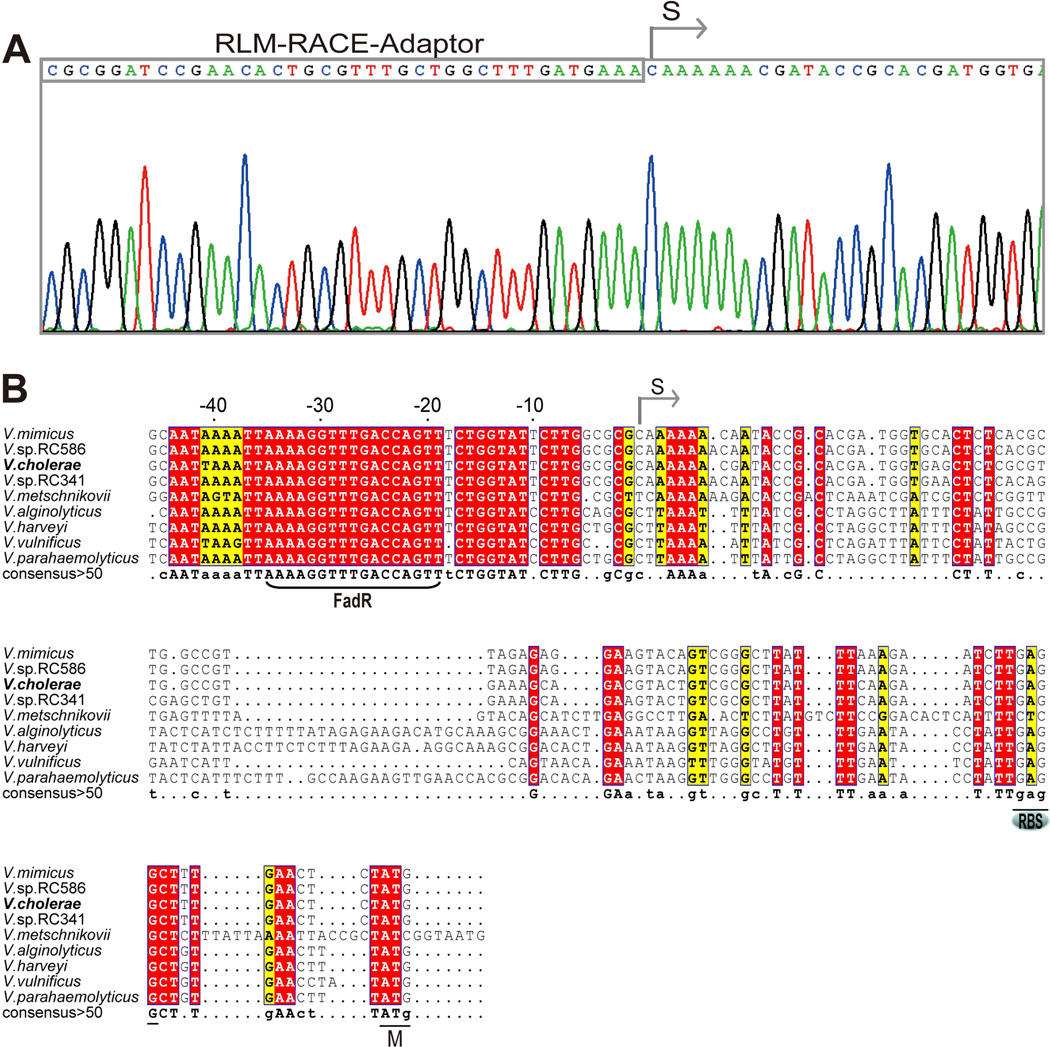
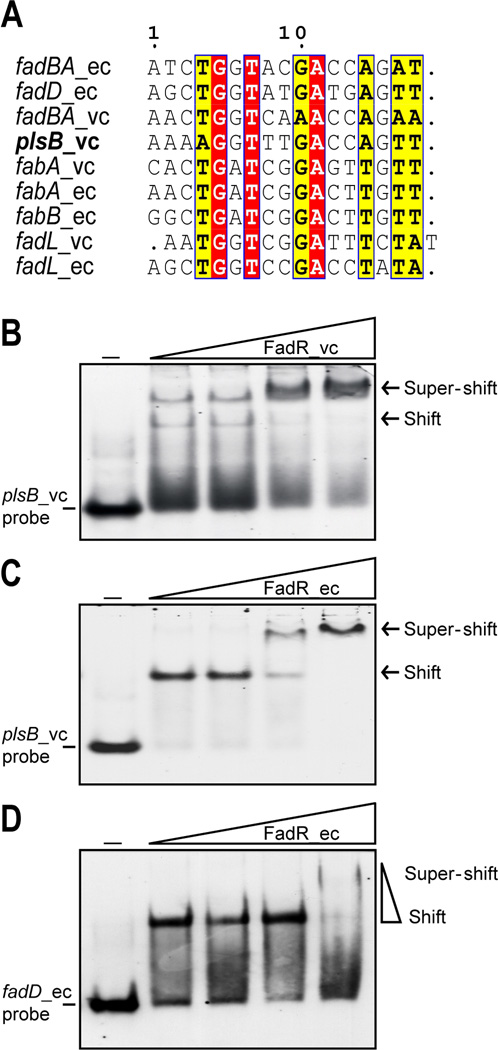
To determine the transcriptional start site of V. cholera plsB, RLM-RACE, an improved version of 5’-RACE using tobacco acid pyrophosphatase was applied as described (Feng & Cronan, 2011). Although several truncated plsB transcripts were encountered we isolated two full-length 5’-RACE PCR products. DNA sequence analyses of several cloned 5’-RACE products showed the same sequence beginning with a C nucleotide adjacent to the RLM-RACE adaptor (Fig. 4B). This located the 5’-end of the transcript 96 nucleotides upstream of the V. cholerae plsB initiation codon (Fig. 4B) and defined the promoter (Fig. 4C). Surprisingly, although in most Vibrio species plsB_vc orthologues are found within the same genetic context, the sequences of plsB promoter regions differ very markedly (e.g. V. harveyi, V. parahaemolyticus and V. alginolyticus). BLAST searches of all the Vibrionales of known genome sequences plus whole genome shotgun sequences showed that the only Vibrio species having plsB promoter sequences highly similar to that of V. cholerae were four strains of V. mimicus (VM603, VM223, VM573 and MB451and two Vibrio sp. isolates (RC586 and RC341, recently named V. metecus and V. parilis, respectively) (Haley et al., 2010). The conservation of the V. cholerae plsB promoter sequences in these bacteria is not surprising since these bacteria were formerly all considered non-toxigenic environmental variants of V. cholerae and are regarded as closely related sister species of common ancestry (Haley et al., 2010, Thompson et al., 2008, Thompson et al., 2009). The sequences of these plsB promoter regions showed a conserved site upstream of the mapped transcriptional start site, which is the proposed FadR binding site (AAAAGGTTTGACCAGT) of Kazanov and coworkers (Kazakov et al., 2009). The site is centered at 27 bp upstream of the transcriptional start site (Fig. 4B) and thus FadR binding would seem likely to hinder RNA polymerase binding or action (Balleza et al., 2009) and therefore function as a repressor as in the case of the fad regulon genes (rather than the activation seem with the fabA, fabB and iclR genes). However, since FadR binding sites contain only three strictly conserved base pairs (Fig. 5A), these sites must be experimentally validated.
Fig. 4. Mapping of the V. cholerae plsB promoter.
A. 5’-RACE-based mapping of the V. cholerae plsB transcriptional start site
B. Multiple sequence alignments of V. cholerae plsB promoter region with those of some other Vibrio species. The predicted FadR-binding site is bracketed. Designations: S, transcriptional start site; RBS, ribosome binding site; M: initiator methionine. Identical residues are in given as white letters with red background, similar residues are in black letters with yellow background, varied residues are in grey letters and dots denote gaps. The plsB promoter region sequences are from V. mimicus (ACYU01000137.1, only one of four isolates with available contig sequences is given), V. vulnificus YJ016 (NC_005139.1), V. parahaemolyticus (BA000031), V. alginolyticus 40B (ACZB01000122.1), V. harveyi (CP000789), V. metschnikovii CIP (ACZO01000002.1), V. sp. RC586 (ADBD01000014.1), V. sp. RC341 (ACZT01000013.1), and V. cholerae ATCC14547 (results of direct sequencing in this study), respectively.
Fig. 5. The V. cholerae plsB promoter region binds both V. cholerae FadR and E. coli FadR.
A. Sequence alignments of the putative plsB FadR-binding site with known sites. Identical residues are black letters with red background, similar residues are in black letters in yellow background and differing residues are in grey letters.
B & C. Electrophoretic mobility shift assays of the binding of V. cholera FadR (B) or E. coli FadR (C) to the V. cholera plsB promoter region.
D. Binding of E. coli FadD promoter region to FadR protein.
The minus sign denotes no addition of FabR protein whereas the DIG-labeled probe shifted by FadR protein is indicated with an arrow. The FadR levels in the right hand four lanes of each panel were (from left to right) 5, 10, 20 and 50 pmol. The protein samples were incubated with 1 pmol of DIG-labeled probe in a total volume of 20 µl. Designations ec and vc denote E. coli and V. cholerae, respectively. FadR_ec and FadR_vc denote the FadRs of E. coli and V. cholerae, respectively. Representative gels (8% native PAGE) from at least three independent gel shift assays are given.
Binding of V. cholerae FadR to the plsB promoter region and its reversal by long chain acyl-CoAs
The predicted FadR-binding site of the plsB promoter region was tested by electrophoretic mobility shift assays using a 39 bp probe containing the predicted 17 bp FadR-binding site (Fig. 4A and 5A) and either V. cholerae FadR or E. coli FadR. Gel shift assays showed that binding of the DNA probe to either FadR_ec or FadR_vc was dose-dependent, as previously observed with the fadM (Feng & Cronan, 2009b) and fadH (Feng & Cronan, 2010) promoter regions. As before super-shifted bands were frequently observed at high FadR concentrations (Fig. 5 B&C). Similar super-shifted bands were seen our prior analyses (Feng & Cronan, 2010). We believe that the super-shifted bands are due to a portion of the protein forming soluble aggregates (perhaps tetramers) in the buffer used in the mobility shift experiments.
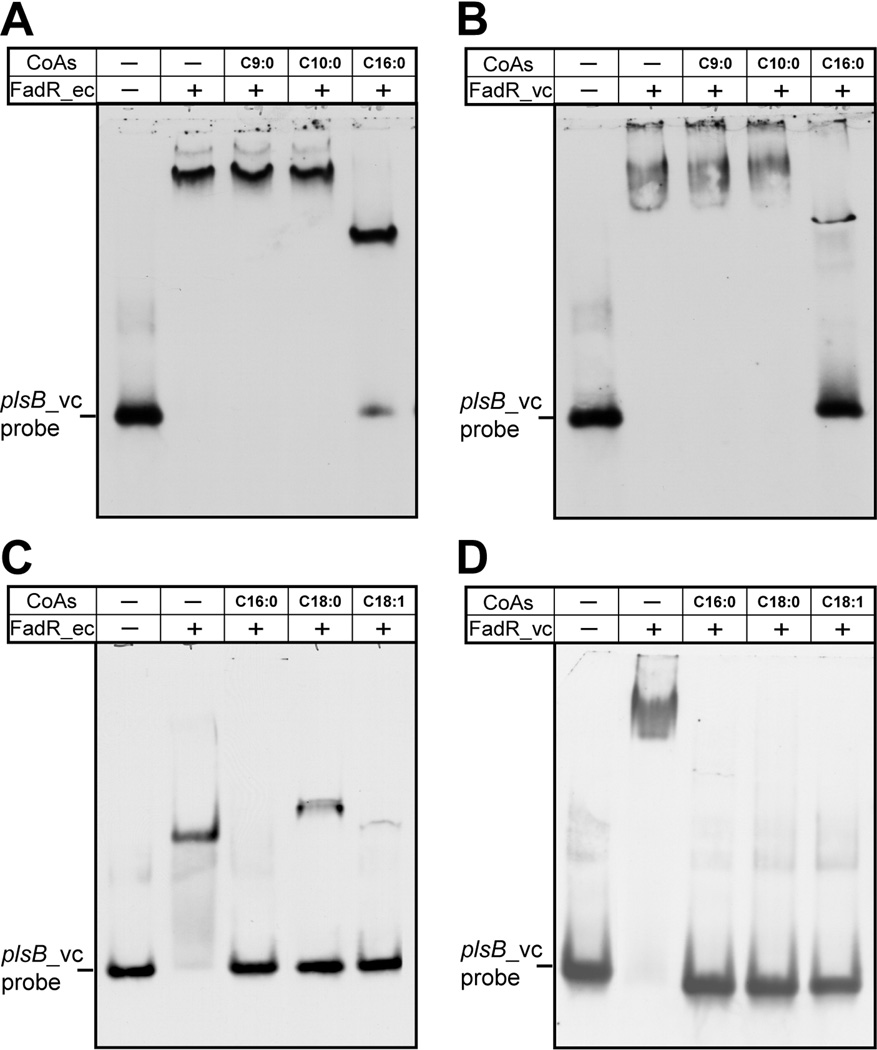
We also tested the effects of fatty acyl-CoA addition on FadR-promoter binding. As we expected, long chain (but not short chain) acyl-CoAs such as oleoyl-CoA (C18:1) antagonized DNA binding by both FadRs (Fig. 6). Addition of long chain acyl-CoA thioesters to the probe-FadR_vc complex resulted in liberation of the probe (Fig. 6B and 6D) (Note, due to an unknown reason traces of shifted bands remained after addition of stearoyl-CoA, Fig. 6C). The super-shifted bands caused by FadR_vc multimers behaved similarly to the bands shifted by FadR_ec dimers in that they disappeared upon an addition of a long chain acyl-CoA (palmitoyl-CoA, stearoyl-CoA or oleoyl-CoA) (Figs. 6C & D). Our in vitro data therefore indicate that the V. cholerae plsB palindromic site predicted by (Kazakov et al., 2009) is specifically recognized by FadR and the interaction is eliminated by physiologically relevant small molecule ligands.
Fig. 6. Acyl-CoA species having long chain but not medium chain length acyl chains impair interaction between FadR and the plsB_vc promoter region.
Panels A & B. Effects of medium and a long chain acyl-CoA species on binding of E. coli FadR (A) or V. cholerae FadR (B) to the V. cholerae plsB probe. Panels C & D. Effects of different long chain acyl-CoA species on binding of E. coli FadR (C) or V. cholerae FadR (D) to the V. cholerae plsB probe. Designations ec and vc denote E. coli and V. cholerae, respectively. Minus sign denotes no addition, whereas plus sign denotes addition. Designations C9:0, nonanoyl-CoA; C10:0, decanoyl-CoA; C16:0, palmitoyl-CoA; C18:0, stearoyl-CoA; C18:1, oleoyl-CoA. Note the quantitative discrepancy between the right lane of A and the center lane of C (the C16:0 lanes). The discrepancy correlates with the puzzling tendency of FadR binding to occasionally give super-shifted bands that seem more refractory to dissociation by long chain acyl-CoAs. In the binding reaction mixtures (20 µl total), the FadR (~20 pmol) was incubated with 1 pmol of DIG-labeled plsB_vc probe. When required, acyl-CoA (~50 pmol) was added as we recently described (Feng & Cronan, 2011). The gel shift assays were conducted for more than three times using 8% native PAGE, and the representative result is given.
Repression of V. cholerae plsB expression by FadR and its induction by oleate in vivo
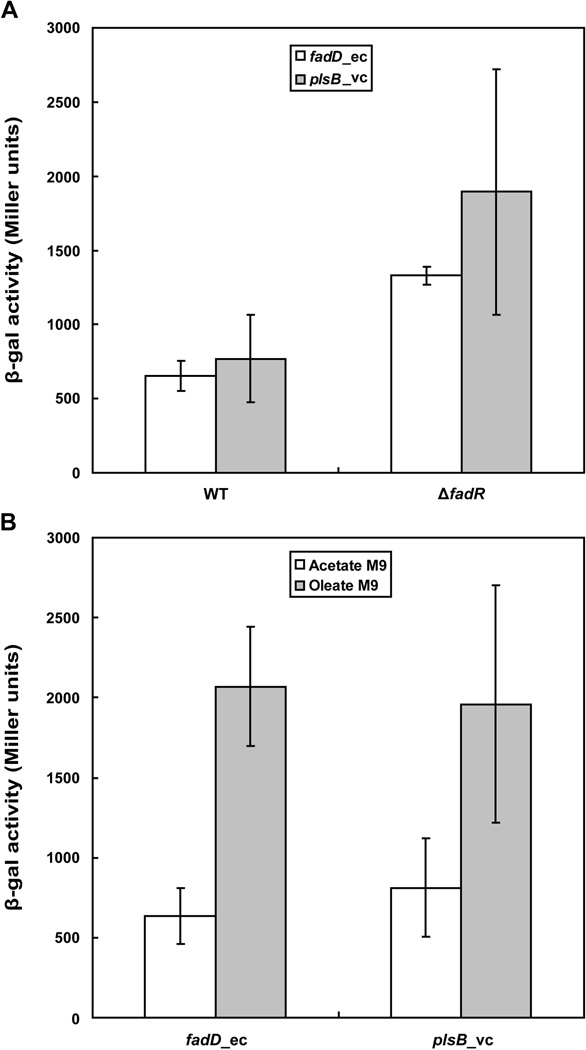
We first tested the physiological role of FadR binding to the V. cholerae plsB promoter in E. coli because it is more amenable to genetic manipulation than V. cholerae. A plsB_vc-lacZ transcriptional fusion was constructed and inserted into the chromosome to give E. coli strain FYJ135. A fadR null mutant derivative of this strain was then constructed and called FYJ136. Assays of LacZ activity showed that the level of plsB_vc transcription was about 2.5 fold higher in the strain that lacked FadR. This extent of FadR regulation was similar to that of E. coli fadD which encodes acyl-CoA synthetase (Fig. 7A). Repression by FadR was largely reversed by addition of oleic acid to the medium which upon conversion to oleoyl-CoA provides a potent antagonist of FadR binding to its operator sites (Fig. 7B).
Fig. 7. Transcription of plsB_vc is repressed by FadR in E. coli and induced upon oleic acid supplementation.
A. The LacZ (β-gal) activity of plsB_vc-lacZ transcriptional fusion integrated onto the E. coli chromosome was assayed. Strains FYJ135 (wild type fadR) and FYJ136 (ΔfadR) were assayed for the levels of plsB_vc transcription whereas strains FYJ159 (wild type fadR) and FYJ161 (ΔfadR) were assayed for the levels of fadD_ec expression.
B. Effects of oleate supplementation on transcription of plsB_vc-lacZ and fadD_ec-lacZ in E. coli. The lacZ-fusion constructs were fadD_ec-lacZ (FYJ159) and plsB_vc-lacZ (FYJ135), respectively. The data are expressed in average ± standard deviation and were derived from more than six independent experiments.
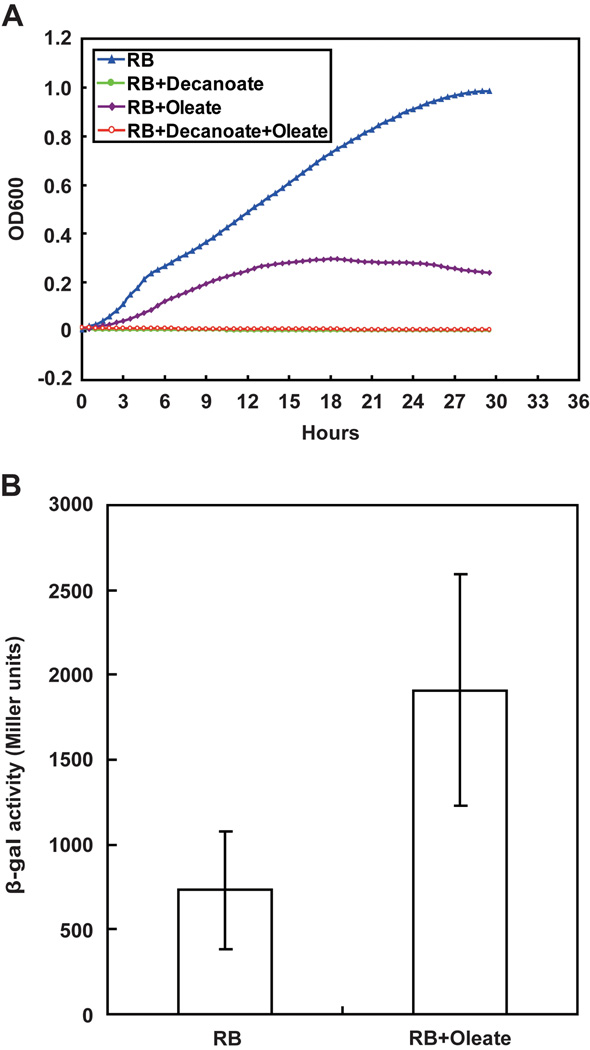
Given these data we had hoped to isolate V. cholerae strains that lack fadR function by their ability to grow on decanoate, the method used to isolate fadR mutants of E. coli and Salmonella enterica (in those bacteria decanoate is a carbon source, but cannot induce β-oxidation) (Iram & Cronan, 2005). However, decanoic acid was found to completely block V. cholerae growth which precluded this approach (Fig. 8 A and B). We subsequently found that other workers have reported inhibition of the growth of Vibrio species by medium chain length fatty acids (Immanuel et al., 2011). We also attempted to delete the fadR gene of the V. cholerae chromosome by homologous recombination, but repeatedly failed presumably due to the inefficiency and strong strain-dependence of genetic manipulation of Vibrio species which can probably be attributed to the known high genome plasticity of these bacteria (Thompson et al., 2010). Finally, we constructed a plsB_vc-lacZ transcriptional fusion in the V. cholerae plasmid pTL61T and after many attempts managed to transform this plasmid into V. cholerae to give strain FYJ176. This allowed us to test the effects of fatty acid supplementation on plsB_vc transcription in its biological context and we found that plsB_vc transcription level increased significantly upon supplementation of RB medium with oleic acid (Fig. 8C), although the fatty acid inhibited growth. Thus, we conclude that V. cholerae FadR negatively regulates plsB expression with a regulatory strength similar to that exerted by FadR on E. coli fadD expression.
Fig. 8. Oleate addition slows growth of V. cholerae and induces plsB gene expression.
A. Growth curve of V. cholerae in RB medium with or without fatty acid supplementation (decanoate and/or oleate). The growth curve was recorded automatically with a BioScreen C instrument in which each time point was measured in five parallel wells. A representative result of three independent trails is shown.
B. Induction of V. cholerae plsB expression by oleate.
The LacZ (β-gal) data are expressed as the mean ± standard deviation and are from more than nine independent experiments. Strain FYJ176 (V. cholerae carrying pTL61T-PplsB_vc, a plasmid encoding a plsB-lacZ transcriptional fusion) was assayed.
DISCUSSION
Our data show that the V. cholerae PlsB homologue is functional and its expression is regulated by FadR thereby validating the bioinformatics prediction of Kazakov et al. (Kazakov et al., 2009). However, transcriptional regulation of plsB expression by FadR seems to be an uncommon mechanism among the proteobacteria. We extended the prior search for this regulatory mechanism (Kazakov et al., 2009) to Vibrionales genome sequences (and finished contigs) that became available since the Kazakov report. From these data FadR regulation of plsB seems to occur in eight Vibrionales species (Fig. 4C). The V. cholerae plsB FadR-binding site (AAAAGGTTTGACCAGTT) is completely conserved in these species, although their promoter sequences vary greatly. An exception is V. fischeri which seems to contain a derivative FadR binding site of uncertain function (Fig. 4C). Among these Vibrionales species, the FadR orthologs are 76.6%–85.7% identical whereas the putative PlsB proteins are 76.7%–94.9% identical (data not shown) and thus it seems very likely that FadR regulates plsB expression in these bacteria. It should be noted that it is “notoriously difficult” to define a Vibrio species due to mobile genetic elements and horizontal gene transfer (Thompson et al., 2010). However, all of the Vibrio FadR orthologs contain the 40-residue insertion relative to E. coli, the presence of which may be useful in defining Vibrio species especially since no obvious relics of gene horizontal transfer are seen (the GC content of Vibrio fadR is essentially the same as that of V. cholerae chromosome I). A prior attempt to remove this Vibrio FadR insertion by mutagenesis resulted in loss of detectable protein expression presumably due to proteolytic destruction of a misfolded protein (Iram & Cronan, 2005). In that work the V. cholerae orthologue was found to have the greatest dynamic regulatory range of the five FadR proteins examined suggesting that the insertion provides the protein with a gain of function (Iram & Cronan, 2005). The distribution of FadR sites in lipid synthetic genes is asymmetric in V. cholerae and E. coli. In both bacteria FadR regulates fabA expression and FadR also regulates fabB expression in E. coli but not in V. cholerae whereas the opposite is true of plsB (Cronan & Subrahmanyam, 1998, Feng & Cronan, 2009a, Feng & Cronan, 2011).
Up-regulation of plsB expression seems likely to be of advantage for acquiring fatty acids from a dilute environment such as seawater, the usual habitat of most Vibrio species. Long chain fatty acid uptake in E. coli requires three known proteins, the first of which is the outer membrane protein FadL (Clark, 2005). The available complete genome sequences indicate that each of the Vibrionales encodes three homologues of FadL rather than the single fadL found in E. coli. However, FadL does not perform active transport, it functions only a ligand-gated channel for substrate diffusion (Lepore et al., 2011). Thus in order to obtain vectorial uptake, the fatty acids must be converted to their acyl-CoA esters by the FadD acyl-CoA synthetase (VC_1985 in V. cholerae El Tor N16961) which in turn would pass the acyl chains to PlsB (which readily accepts acyl-CoAs as acyl donors). Based on transposon mutagenesis studies, it seems probable that utilization of exogenous fatty acids for phospholipid synthesis may be the primary physiological role of V. cholerae PlsB. In V. cholerae a transposon insertion early in plsB is not a lethal mutation (Cameron et al., 2008) whereas in E. coli plsB is an essential gene synthesis (Yoshimura et al., 2007) (Baba et al., 2006). In contrast plsX, which encodes a component of the other known bacterial G3P acyltransferase system (PlsX/PlsY), is essential in V. cholerae (Cameron et al., 2008), but not in E. coli (Yoshimura et al., 2007) (Baba et al., 2006). Therefore it may be that V. cholerae PlsX, together with its membrane bound partner PlsY, catalyzes acylation of G3P with de novo synthesized acyl chains whereas PlsB performs this task in E. coli. Since in vitro E. coli PlsX prefers acyl-ACPs rather than the CoA substrates formed upon uptake of exogenous fatty acids (Lu et al., 2006), V. cholerae PlsX seems unlikely to play a major role in fatty acid uptake.
Vibrio species have diverse sources of fatty acids. The luminescent bacterium V. fischeri, incorporates unusual fatty acids derived from the lipids of its squid symbiont host (Wier et al., 2010). Moreover, Giles and coworkers (Giles et al., 2011) reported that five Vibrio species including V. cholerae efficiently incorporate exogenous fatty acids from both bile and from marine sediments into their membrane phospholipids. It might be argued that for efficient fatty acid utilization plsC, the gene that encodes the acyltransferase that attaches the second acyl chain, should be regulated in concert with plsB. However, in V. cholerae PlsC seems to be in functional excess over PlsB because no singly acylated G3P (lysophosphatidic acid) species accumulate (Giles et al., 2011).
Experimental procedures
Bacterial strains and growth conditions
Bacterial strains used here were E. coli K-12 derivatives (Table 1) and ATCC 14547, an avirulent strain of V. cholerae (Feng & Cronan, 2011, Massengo-Tiasse & Cronan, 2008). The media used for E. coli included Luria-Bertani (LB) medium (tryptone, 10g/L; yeast extract, 5 g/L; NaCl, 10 g/L; pH 7.5), rich broth (RB medium; 10 g of tryptone, 1 g of yeast extract, and 5 g of NaCl per liter) and the minimal medium M9 supplemented with 0.4% glucose or another carbon source, 0.1% Vitamin-Free Casamino Acids and 0.001% thiamine. Although LB and RB media both can support V. cholerae growth, Mueller-Hinton broth (Difico) (MH medium, 300 g of beef infusion, 17.5 g of Casamino Acids, and 1.5 g of starch per liter) was required to obtain electroporation competent V. cholerae cells (Hamashima et al., 1995). Fatty acids were neutralized with potassium hydroxide, solubilized with Tergitol NP-40 and used for induction experiments at the final concentrations of 5 mM (E. coli) or 2 mM (V. cholerae) (Feng & Cronan, 2009b, Feng & Cronan, 2010). For assays of β-galactosidase activities, either 5 mM acetate or 5 mM fatty acid were the carbon sources used (Feng & Cronan, 2009b, Feng & Cronan, 2010, Feng & Cronan, 2011). When necessary, antibiotics were used as at the following concentrations (in mg/liter): sodium ampicillin, 100; kanamycin sulfate, 50; and tetracycline HCl, 15.
Table 1.
Bacteria and plasmids in this study
|
E. coli strain or plasmid |
Relevant characteristics | Sources/References |
|---|---|---|
| Strains | ||
| MG1655 | Wild type stain of E. coli K-12 | Lab stock |
| JW1176-1 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) ΔfadR776::kan, rph-1 Δ(rhaD-rhaB)568, hsdR514 | CGSCa, (Baba et al., 2006) |
| Topo10 | ΔlacX74, cloning strain | Invitrogen |
| DH5α | Cloning strain | Lab stock |
| BL21(DE3) | E. coli protein expression strain | Lab stock |
| MFH8 | UB1005, fadR::Tn10 | (Henry & Cronan, 1992, Feng & Cronan, 2010, Feng & Cronan, 2011) |
| BB26-36 |
plsB26 plsX50 fadL701 glpR2(glpc) glpD3 glpK14(fbR) Encodes a defective G3Pacyltransferase, G3P auxotroph |
(Cronan & Bell, 1974) |
| MC4100 | araD139, Δ(argF-lac)169 | (Feng & Cronan, 2009b) |
| DH5α (λ-pir) | Δlac host for pAH125 and its derivatives | (Feng & Cronan, 2009a) |
| FYJ1 | DH5α (λ-pir) carrying pAH125 | (Feng & Cronan, 2009a) |
| FYJ57 | JW1176-1, ΔfadR | (Feng & Cronan, 2010, Feng & Cronan, 2011) |
| FYJ135 | MC4100 with a plsB_vc-lacZ fusion integrated into the chromosomal attλ site | This work |
| FYJ136 | plsB_vc-lacZ fusion, ΔfadR::Tn10 | P1vir(MFH8)×FYJ135 |
| FYJ158 | DH5α (λ-pir) carrying pAH125-PfadD_ec | This work |
| FYJ159 | MC4100 with a fadD_ec-lacZ fusion integrated into the chromosomal attλ site | This work |
| FYJ161 | FYJ57, ΔfadR, fadD-lacZ transcriptional fusion | P1vir(FYJ159)×FYJ57 |
| FYJ168 | Topo 10 carrying pTL61T-PplsB_vc | This work |
| FYJ176 | Vibrio cholerae carrying pTL61T-PplsB_vc | This work |
| FYJ187 | MC4100 pINT-ts | This work |
| Plasmids | ||
| pCR2.1-TOPO | Topo-cloning vector, AmpR, KanR, | Invitrogen |
| pET28a | T7 promoter-driven expression vector, KanR | Novagen |
| pAH125 | A promoter-less lacZ reporter plasmid used in E. coli, KanR | (Feng & Cronan, 2009a, Haldimann & Wanner, 2001) |
| pCR-plsBvc | pCR2.1 carrying V. cholerae plsB gene, AmpR, KanR | This work |
| pAH-PplsBvc | pAH125 carrying the promoter region of V. cholerae plsB gene, KanR | This work |
| pINT-ts | Temperature sensitive plasmid expressing phage λ Int. | (Haldimann & Wanner, 2001) |
| pTL61T | A promoter-less lacZ fusion plasmid used for V. cholerae | (Bellair & Withey, 2008, Linn & St Pierre, 1990, Withey & Dirita, 2005) |
| pTL61T-PplsBvc | pTL61T carrying the promoter region of the V. cholerae plsB gene. | This work |
| pET28-fadRec | Recombinant plasmid carrying the E. coli fadR gene, KanR | (Feng & Cronan, 2009b, Feng & Cronan, 2009a, Iram & Cronan, 2005, Feng & Cronan, 2010) |
| pET16-fadRvc | Recombinant plasmid carrying the V. cholerae fadR gene | Lab stock |
CGSC denotes Coli Genetic Stock Center, Yale University.
Plasmids and DNA manipulations
The pCR2.1-TOPO vector (Invitrogen) was applied for PCR cloning, and Topo 10 strain is the corresponding recipient host (Table 1). Recombinant plasmid pCR-plsBvc that carries V. cholerae plsB gene along with its promoter region was introduced into strain BB26-36, which is an E. coli G3P auxotroph encoding a defective PlsB that results in an elevated Km for G3P, to (Table 2). Strain BL21 (DE3) carrying either pET28-fadRec or pET16-fadRvc plasmids was used to prepare FadR_ec or FadR_vc proteins (Feng & Cronan, 2009b, Feng & Cronan, 2010, Iram & Cronan, 2005).
Table 2.
Primers used in this study
| Primers | Primer sequences |
|---|---|
| plsBvc-Fa | 5'-CG GGATCC CTG GAT TGC GTC CTC CAT AAT G-3’ |
| plsBvc-Ra | 5'-CCG CTCGAG GTT TGG TAA TCC CAG CTT AAG GC-3’ |
| plsBvc_FadR_BS-Fb | 5’-TTA AAT TAA AAG GTT TGA CCA GTT TCT GGT ATT CTT GGC-3’ |
| plsBvc_FadR_BS-Rb | 5'-GCC AAG AAT ACC AGA AAC TGG TCA AAC CTT TTA ATT TAA-3' |
| fadDec_FadR_F2 | 5’-GTA ATT ATC AAG CTG GTA TGA TGA GTT AAT ATT ATG-3’ |
| fadDec_FadR_R2 | 5’-CAT AAT ATT AAC TCA TCA TAC CAG CTT GAT AAT TAC-3’ |
| plsBvc_P-F | 5'-GTC CTC CAT AAT GGC TTC AAA G-3' |
| plsBvc_P-R | 5'-GTT CAA AGC CTC AAG ATT CTT G-3' |
| PplsBvc-F1a | 5'-CCG GTCGAC CAG TTG ACT GTG AGT ATA TCC AG-3' |
| PplsBvc-R1a | 5'-AACC GAATTC AGA GTT CAA AGC CTC AAG ATT C-3' |
| PplsBvc-F2a | 5'-CCG CTCGAGCAG TTG ACT GTG AGT ATA TCC AG-3' |
| PplsBvc-R2a | 5'-CG GGATCC AGA GTT CAA AGC CTC AAG ATT C-3' |
| plsBvc-Nested (212–235) | 5'-CAT TCA GCA TCA AAG GCT CTA AAG-3' |
| plsBvc-GSP (212–235) | 5'-GAA TCA CCT GCA CAT CCA ACT C-3' |
| RLM-RACE Adaptor | 5’-GCU GAU GGC GAU GAA UGA ACA CUG CGU UUG CUG GCU UUG AUG AAA-3’ |
| RLM-RACE Outer | 5’-GCT GAT GGC GAT GAA TGA ACA CTG-3’ |
| RLM-RACE Inner | 5’-CGC GGA TCC GAA CAC TGC GTT TGC TGG CTT TGA TGA AA-3’ |
| fadD-promoter-Fa | 5'-CCG GTCGAC GTT GCG GTA CAA AAC CAG CA-3' |
| fadD-promoter-Ra | 5'-AACC GAATTC CTC TAA AAT GCG TGT TCG TCG-3' |
| fadD-check2 | 5'-GTC TGA CGA CTG ACT TAA CGC-3' |
| lacZ-R | 5’-GAC CAT GAT TAC GGA TTC ACT G-3’ |
| 16Svc-F | 5’-GGA AAC GAT GGC TAA TAC CGC A-3’ |
| 16Svc-R | 5’-AGT GTG GCT GAT CAT CCT CTC A-3’ |
The underlined italic sequences are the introduced restriction sites.
The bold letters are the predicted core FadR binding palindromes.
To quantify V. cholerae plsB transcription two versions of plsB_vc-lacZ fusion constructs were made using two pairs of specific primers, PplsBvc-F1 plus PplsBvc-R1 and PplsBvc-F2 plus PplsBvc-R2 (Table 2) to amplify the V. cholerae promoter region. These products were directly inserted into the promoter-less lacZ reporter plasmids pAH125 and pTL61T, respectively (Table 1). The pAH-PplsB_vc plasmid requires pir protein for replication and replicates in DH5α λ-pir, but to impart antibiotic resistance the plasmid must specifically integrate into the attλ site of the chromosome of E. coli MC4100 (a lacZ strain lacking pir) in a reaction catalyzed by the pINT-ts helper plasmid.
Plasmid pTL61T-PplsB_vc, replicates in V. cholerae species (Withey & Dirita, 2005) (Table 1). Using a similar strategy, E. coli strain FYJ159 which has a fadD_ec-lacZ transcriptional fusion specifically integrated into the attλ site of the E. coli MC4100 chromosome was made (Table 1). PCR assays with specific primers plsBvc_P-F (fadD-check2) plus lacZ-R (Table 2) were conducted to determine the plsB_vc-lacZ (or fadD_ec-lacZ) junction. X-Gal plates were used to test function of the plsB_vc-lacZ transcriptional fusion.
Transformation of V. cholera
Electroporation was utilized to transform the lacZ reporter plasmid pTL61T-PplsB_vc into V. cholerae. Electro-competent cells were prepared according to Hamashima et al. (Hamashima et al., 1995). In brief, a single colony of V. cholerae was cultivated overnight in 3 ml of MH medium and then 1 ml of overnight culture was transferred into 100 ml of MH medium in a 500 ml of flask with vigorous shaking at 37 °C. Once the OD600 reached about 0.6, bacterial cultures were chilled on ice for 30 min and then collected by spinning at 2,000×g for 15 min at 4 °C. The collected pellets in each tube were resuspended in 30 ml of iced-chilled EP washing buffer (272 mM sucrose, 1 mM MgCl2, 7 mM KH2PO4-Na2HPO4 buffer, pH 7.4) and then centrifuged at 2,000×g for 15 min at 4 °C. Following two more washes the cells were suspended in 1 ml of ice-cold EP buffer and this was divided into small aliquots (200 µl each tube) which were frozen on dry ice and kept at −80 °C until use. Prior to electroporation, cells (200 µl) were thawed quickly at 37 °C, mixed with about 1 µg of plasmid DNA and then placed on ice for about 20 min prior to transfer into a chilled 2 cm electrode gap cuvette. Electroporation was a done with a time constant of 25 ms (25 µF capacitance, 1000 Ω) at 1.6 kV (8.0 kV/cm). The pulsed cells were quickly diluted with 600 µl of pre-warmed MH medium. The cultures were cultivated at 37 °C for 60 min and the cells were plated on MH agar plates containing ampicillin at 50 µg/ml. The plates were incubated at 37 °C for about 36 h. Colonies containing the reporter plasmid were confirmed by colony PCR, assay of β-galactosidase activity plus DNA sequencing of the isolated plasmid.
P1vir phage transduction
P1vir transductions were performed as described by Miller (Miller, 1992) with little modifications. Strain FYJ136 (plsBvc-lacZ, ΔfadR) was generated by P1vir transduction of strain FYJ135 (plsBvc-lacZ) using a lysate grown on strain MFH8 (fadR::Tn10) with selection for tetracycline resistance. Strain FYJ161 was obtained by P1vir transduction of strain FYJ57 (ΔfadR) with a lysate grown on strain FYJ159 (fadDec-lacZ) and selection for kanamycin resistance (Table 1).
β-Galactosidase assays
Mid-log phase cultures grown in either RB or minimal medium were collected for assay of β-galactosidase (Feng & Cronan, 2009b, Feng & Cronan, 2009a, Miller, 1992). The data were recorded in triplicate in more than three independent experiments.
RNA isolation and 5’-RACE
As documented previously (Feng & Cronan, 2009b, Feng & Cronan, 2009a, Feng & Cronan, 2010, Feng & Cronan, 2011) bacterial RNAs were isolated from cells of V. cholerae ATCC14547 grown to mid-log phase in (either LB or rich broth medium) using the RNeasy bacterial RNA isolation kit (Qiagen). RNA quality was determined by agarose gel electrophoresis, and PCR detection (using total RNA as template with primers 16Svc-F plus 16Svc-R, in Table 2) to test possible DNA contamination of the RNA samples (Feng & Cronan, 2009b, Feng & Cronan, 2009a, Feng & Cronan, 2010). The qualified RNA preparations then were used for subsequent 5’-RACE experiments.
RLM-RACE (Ambicon), an improved version of the customary 5’-RACE methods was used to determine the putative transcription start site of V. cholerae plsB gene as recently described (Feng & Cronan, 2009b, Feng & Cronan, 2009a, Feng & Cronan, 2010). Following three steps of customary treatments (calf intestinal alkaline phosphatase digestion, tobacco acid pyrophosphatase treatment and ligation of a 5’-RACE adaptor to the pyrophosphatase-treated mRNAs), the reverse-transcription reaction was performed. Subsequently, a set of nested PCR reactions were performed in which combinations of Outer Primer plus plsBvc-GSP and Inner Primer plus plsBvc-Nested (Table 2) were used. The PCR program was a denaturing cycle at 95 °C for 5 min followed by 35 cycles comprising 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The purified PCR products were cloned into the pCR2.1 TOPO vector (Invitrogen) for direct DNA sequencing. The transcriptional start site was taken to be the first nucleotide adjacent to the RLM-RACE adaptor (Feng & Cronan, 2009a, Feng & Cronan, 2010, Feng & Cronan, 2011)}.
Expression and purification of V. cholerae FadR
Recombinant hexahistidine-tagged FadR_vc protein was expressed in E. coli BL21 (DE3) carrying the pET16-fadRvc plasmid (Table 1). Induction with 0.3 mM isopropyl β-D-1-thiogalactopyranoside at 30 °C for 3 h gave soluble FadR_vc protein. The clarified bacterial supernatant obtained by lysis in a French pressure cell and removal of bacterial debris by centrifugation was loaded onto a nickel chelate column (Qiagen). The column was washed with buffer containing 20 mM imidazole and then the protein was eluted in elution buffer containing 150 mM imidazole. The protein was concentrated by ultrafiltration (30 kDa cutoff) and exchanged into 20 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl (Feng & Cronan, 2010)
Liquid chromatography quadrupole time-of-flight mass spectrometry
A Waters Q-Tof API-US Quad-ToF mass spectrometer connected to a Waters nano Acquity UPLC) was applied to determine identify the in vitro prepared FadR_vc protein (Feng & Cronan, 2011). The protein band of interest was cut from SDS-PAGE gel. The gel slices were de-stained and the proteins digested with 25 µl of Sequencing Grade Trypsin (G-Biosciences St. Louis, MO, 12.5 ng/µl in 25 mM ammonium bicarbonate) using a CEM Discover Microwave Digestor (Mathews, NC) for 15 min at 55 °C and 50W. Subsequently, the resulting peptides were extracted using 50% acetonitrile containing 5% formic acid, dried using a Savant SpeedVac and suspended in 13 µl of 5% acetonitrile containing 0.1% formic acid. Samples (10 µl) were loaded on a Waters Atlantis C-18 column (0.03 mm particle, 0.075 mm × 150 mm) and eluted at a flow rate of 250 nl per min using a linear gradient of water/acetonitrile containing 0.1% formic acid 0–60% B in 60 min. The mass spectrometer was set for data dependent acquisition and ms/ms analysis was carried out on the most abundant four peaks at any given time. Data collected were processed using the Waters Protein Lynx Global Server 2.2.5, Mascot (Matrix Sciences) and BLAST against NCBI nr database (Feng & Cronan, 2011).
Size exclusion chromatography
Relative to FadR_ec, FadR_vc contains a 40 residue insertion into the center of the protein sequence (Iram & Cronan, 2005). To test if this insertion affected the solution structure of the protein the purified FadR_vc protein was subjected to gel filtration analyses using a Superdex 200 column (Pharmacia) run on an Äkta fast protein liquid chromatography system (GE Healthcare) essentially as recently described (Feng & Cronan, 2010, Feng & Cronan, 2011). The column effluent was monitored at a flow rate of 0.5 ml/min in running buffer (20 mM Tris-HCl,100 mM NaCl, pH 8.0). When necessary, samples of interest were separated by 15% SDS-PAGE and stained with Coomassie Brilliant Blue R250 (Sigma, St. Louis, MO).
Chemical cross-linking assays
To further test the solution structure of V. cholerae FadR, chemical cross-linking with ethylene glycol bis-succinimidylsuccinate (Pierce) was performed (Feng & Cronan, 2010). In each chemical cross-linking reaction (20 µl in total), the purified FadR protein (~10 mg/ml) was separately mixed with cross-linker at different concentrations (0, 1.0, 2.5, 5.0, 7.5 and 10 µM), and kept 30 min at room temperature before analysis. All the reaction products were assayed using SDS-PAGE as above.
Electrophoretic mobility shift assays
To address whether FadR can bind to the V. cholerae plsB promoter region, gel shift assays were conducted as we recently reported (Feng & Cronan, 2009b, Feng & Cronan, 2010, Feng & Cronan, 2011) with minor modifications. Two DNA probes were used, one of which is about 100 bp of a PCR product obtained by amplification with specific primers plsBvc_P-F plus plsBvc_P-R (Table 2). The second DNA probe containing the predicted FadR binding palindrome was generated by annealing two complementary oligonucleotides (plsBvc_FadR_BS-F plus plsBvc_FadR_BS-R, Table 2) by incubated at 95 °C in TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl, pH 8.0) for 5 min followed by slow cooling to 25 °C. The probes were labeled with digoxigenin by terminal transferase incorporation of digoxigenin-ddUTP (Roche). The digoxigenin-labeled DNA probes (~1 pmol) were mixed with purified FadR (in appropriate concentrations) in the binding buffer (Roche) and incubated 15–20 min at room temperature. The DNA/protein mixtures were then analyzed by native PAGE (6% PAGE for the ~100 bp PCR probe, and 8% PAGE for the 39 bp synthetic probe). Contact blotting-aided gel transfer was conducted,followed by UV cross-linking (120 mJ for 180 s), 1 h of blocking of the nylon membrane in 50 ml blocking buffer, and 1 h of incubation with an anti-digoxigenin antibody solution (1:10,000) at room temperature. Subsequently the nylon membrane was washed 5 times (15 min each) and equilibrated with detection buffer for 15 min before 1 h of development of the luminescent reaction in CSPD working solution (Roche) at 37 °C. Finally the membrane was exposed to ECL film (Amersham) for signal capture.
Bioinformatic analyses
The known DNA binding sites by FadR_ec were all from literature compiled in the E. coli Regulon Data Base (http://regulondb.cs.purdue.edu/), whereas those putative DNA binding sites recognized by FadR_vc were searched by BLAST using known FadR_ec binding sites as probes.
Acknowledgements
This work was supported by National Institutes of Health Grant AI15650 from the National Institute of Allergy and Infectious Diseases. We thank Prof. Jeffrey H. Withey of Wayne State University School of Medicine for kindly providing pTL61T.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 20060008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balleza E, Lopez-Bojorquez LN, Martinez-Antonio A, Resendis-Antonio O, Lozada-Chavez I, Balderas-Martinez YI, Encarnacion S, Collado-Vides J. Regulation by transcription factors in bacteria: beyond description. FEMS Microbiol Rev. 2009;33:133–151. doi: 10.1111/j.1574-6976.2008.00145.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis. Properties of wild type and Km defective sn-glycerol-3-phosphate acyltransferase activities. J Biol Chem. 1975;250:7147–7152. [PubMed] [Google Scholar]
- Bellair M, Withey JH. Flexibility of Vibrio cholerae ToxT in transcription activation of genes having altered promoter spacing. J Bacteriol. 2008;190:7925–7931. doi: 10.1128/JB.00512-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron DE, Urbach JM, Mekalanos JJ. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A. 2008;105:8736–8741. doi: 10.1073/pnas.0803281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Cronan JE., Jr Escherichia coli FadR positively regulates transcription of the fabB fatty acid biosynthetic gene. J Bacteriol. 2001;183:5982–5990. doi: 10.1128/JB.183.20.5982-5990.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JW, Cronan JE., Jr The enigmatic Escherichia coli fadE gene is yafH. J Bacteriol. 2002;184:3759–3764. doi: 10.1128/JB.184.13.3759-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D, Cronan J. Two-carbon compounds and fatty acids as carbon sources. In: Bock A, Curtiss R III, Karper J, Karp P, Neidhardt F, Nystrom T, Slauch J, Squiress C, Ussery D, editors. EcoSal--Escherichia coli and Salmonella: cellular and molecular biology. Washington, DC: ASM Press; 2005. http://www.ecosal.org. [Google Scholar]
- Cronan JE., Jr In vivo evidence that acyl coenzyme A regulates DNA binding by the Escherichia coli FadR global transcription factor. J Bacteriol. 1997;179:1819–1823. doi: 10.1128/jb.179.5.1819-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Jr, Bell RM. Mutants of Escherichia coli defective in membrane phospholipid synthesis: mapping of sn-glycerol 3-phosphate acyltransferase Km mutants. J Bacteriol. 1974;120:227–233. doi: 10.1128/jb.120.1.227-233.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Jr, Subrahmanyam S. FadR, transcriptional co-ordination of metabolic expediency. Mol Microbiol. 1998;29:937–943. doi: 10.1046/j.1365-2958.1998.00917.x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Escherichia coli unsaturated fatty acid synthesis: complex transcription of the fabA gene and in vivo identification of the essential reaction catalyzed by FabB. J Biol Chem. 2009a;284:29526–29535. doi: 10.1074/jbc.M109.023440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. A new member of the Escherichia coli fad regulon: transcriptional regulation of fadM (ybaW) J Bacteriol. 2009b;191:6320–6328. doi: 10.1128/JB.00835-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Overlapping repressor binding sites result in additive regulation of Escherichia coli FadH by FadR and ArcA. J Bacteriol. 2010;192:4289–4299. doi: 10.1128/JB.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Cronan JE. Complex binding of the FabR repressor of bacterial unsaturated fatty acid biosynthesis to its cognate promoters. Mol Microbiol. 2011 doi: 10.1111/j.1365-2958.2011.07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles DK, Hankins JV, Guan Z, Trent MS. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol. 2011;79:716–728. doi: 10.1111/j.1365-2958.2010.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui L, Sunnarborg A, LaPorte DC. Regulated expression of a repressor protein: FadR activates iclR. J Bacteriol. 1996;178:4704–4709. doi: 10.1128/jb.178.15.4704-4709.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley BJ, Grim CJ, Hasan NA, Choi SY, Chun J, Brettin TS, Bruce DC, Challacombe JF, Detter JC, Han CS, Huq A, Colwell RR. Comparative genomic analysis reveals evidence of two novel Vibrio species closely related to V. cholerae. BMC Microbiol. 2010;10:154. doi: 10.1186/1471-2180-10-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamashima H, Iwasaki M, Arai T. A simple and rapid method for transformation of Vibrio species by electroporation. Methods Mol Biol. 1995;47:155–160. doi: 10.1385/0-89603-310-4:155. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry MF, Cronan JE., Jr Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J Mol Biol. 1991;222:843–849. doi: 10.1016/0022-2836(91)90574-p. [DOI] [PubMed] [Google Scholar]
- Henry MF, Cronan JE., Jr A new mechanism of transcriptional regulation: release of an activator triggered by small molecule binding. Cell. 1992;70:671–679. doi: 10.1016/0092-8674(92)90435-f. [DOI] [PubMed] [Google Scholar]
- Immanuel G, Sivagnanavelmurugan M, Palavesam A. Antibacterial effect of medium-chain fatty acid: caprylic acid on gnotobiotic Artemia franciscana nauplii against shrimp pathogens Vibrio harveyi and V. parahaemolyticus. Aquaculture International. 2011;19:91–101. [Google Scholar]
- Iram SH, Cronan JE. Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. J Biol Chem. 2005;280:32148–32156. doi: 10.1074/jbc.M504054200. [DOI] [PubMed] [Google Scholar]
- Kazakov AE, Rodionov DA, Alm E, Arkin AP, Dubchak I, Gelfand MS. Comparative genomics of regulation of fatty acid and branched-chain amino acid utilization in proteobacteria. J Bacteriol. 2009;191:52–64. doi: 10.1128/JB.01175-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson TJ, Lightner VA, Green PR, Modrich P, Bell RM. Membrane phospholipid synthesis in Escherichia coli. Identification of the sn-glycerol-3-phosphate acyltransferase polypeptide as the plsB gene product. J Biol Chem. 1980;255:9421–9426. [PubMed] [Google Scholar]
- Lepore BW, Indic M, Pham H, Hearn EM, Patel DR, van den Berg B. Ligand-gated diffusion across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1018532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightner VA, Bell RM, Modrich P. The DNA sequences encoding plsB and dgk loci of Escherichia coli. J Biol Chem. 1983;258:10856–10861. [PubMed] [Google Scholar]
- Lightner VA, Larson TJ, Tailleur P, Kantor GD, Raetz CR, Bell RM, Modrich P. Membrane phospholipid synthesis in Escherichia coli. Cloning of a structural gene (plsB) of the sn-glycerol-3-phosphate acyl/transferase. J Biol Chem. 1980;255:9413–9420. [PubMed] [Google Scholar]
- Linn T, St Pierre R. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Zhang YM, Grimes KD, Qi J, Lee RE, Rock CO. Acyl-phosphates initiate membrane phospholipid synthesis in Gram-positive pathogens. Mol Cell. 2006;23:765–772. doi: 10.1016/j.molcel.2006.06.030. [DOI] [PubMed] [Google Scholar]
- Massengo-Tiasse RP, Cronan JE. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem. 2008;283:1308–1316. doi: 10.1074/jbc.M708171200. [DOI] [PubMed] [Google Scholar]
- Miller JH. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- Paoletti L, Lu YJ, Schujman GE, de Mendoza D, Rock CO. Coupling of fatty acid and phospholipid synthesis in Bacillus subtilis. J Bacteriol. 2007;189:5816–5824. doi: 10.1128/JB.00602-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, Thompson FL, Vicente AC. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA) Int J Syst Evol Microbiol. 2008;58:617–621. doi: 10.1099/ijs.0.65461-0. [DOI] [PubMed] [Google Scholar]
- Thompson CC, Vicente AC, Souza RC, Vasconcelos AT, Vesth T, Alves N, Jr, Ussery DW, Iida T, Thompson FL. Genomic taxonomy of Vibrios. BMC Evol Biol. 2009;9:258. doi: 10.1186/1471-2148-9-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson FL, Thompson CC, Vicente AC, Klose KE. Vibrio2009: the third international conference on the biology of Vibrios. Mol Microbiol. 2010;77:1065–1071. doi: 10.1111/j.1365-2958.2010.07286.x. [DOI] [PubMed] [Google Scholar]
- van Aalten DM, DiRusso CC, Knudsen J. The structural basis of acyl coenzyme A-dependent regulation of the transcription factor FadR. EMBO J. 2001;20:2041–2050. doi: 10.1093/emboj/20.8.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Aalten DM, DiRusso CC, Knudsen J, Wierenga RK. Crystal structure of FadR, a fatty acid-responsive transcription factor with a novel acyl coenzyme A-binding fold. EMBO J. 2000;19:5167–5177. doi: 10.1093/emboj/19.19.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl A, My L, Dumoulin R, Sturgis JN, Bouveret E. Antagonistic regulation of dgkA and plsB genes of phospholipid synthesis by multiple stress responses in Escherichia coli. Mol Microbiol. 2011;80:1260–1275. doi: 10.1111/j.1365-2958.2011.07641.x. [DOI] [PubMed] [Google Scholar]
- Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, Koroleva I, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107:2259–2264. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey JH, Dirita VJ. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J Bacteriol. 2005;187:7890–7900. doi: 10.1128/JB.187.23.7890-7900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Heath RJ, Li Z, Rock CO, White SW. The FadR.DNA complex. Transcriptional control of fatty acid metabolism in Escherichia coli. J Biol Chem. 2001;276:17373–17379. doi: 10.1074/jbc.M100195200. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Oshima T, Ogasawara N. Involvement of the YneS/YgiH and PlsX proteins in phospholipid biosynthesis in both Bacillus subtilis and Escherichia coli. BMC Microbiol. 2007;7:69. doi: 10.1186/1471-2180-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Rock CO. Thematic review series: Glycerolipids. Acyltransferases in bacterial glycerophospholipid synthesis. J Lipid Res. 2008;49:1867–1874. doi: 10.1194/jlr.R800005-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]