Abstract
The transmission of information from DNA to RNA is a critical process. We compared RNA sequences from human B cells of 27 individuals to the corresponding DNA sequences from the same individuals and uncovered more than 10,000 exonic sites where the RNA sequences do not match that of the DNA. All 12 possible categories of discordances were observed. These differences were nonrandom as many sites were found in multiple individuals and in different cell types, including primary skin cells and brain tissues. Using mass spectrometry, we detected peptides that are translated from the discordant RNA sequences and thus do not correspond exactly to the DNA sequences. These widespread RNA-DNA differences in the human transcriptome provide a yet unexplored aspect of genome variation.
DNA carries genetic information that is passed onto mRNA and proteins that perform cellular functions, and it is assumed that the sequence of mRNA reflects that of the DNA. This assumed precision is important because mRNA serves as the template for protein synthesis. Hence, genetic studies have mostly focused on DNA sequence polymorphism as the basis of individual differences in disease susceptibility. Studies of mRNA and proteins analyze their expression and not sequence differences among individuals.
There are, however, known exceptions to the one-to-one relationship between DNA and mRNA sequences. These include errors in transcription (1, 2) and RNA-DNA differences that result from RNA editing (3–7). Errors are rare because proofreading and repair mechanisms ensure the fidelity of transcription (8–10). RNA editing is carried out by enzymes that target mRNA posttranscriptionally: ADARs (adenosine deaminases that act on RNA) that deaminate adenosine to inosine, which is then recognized by the translation machineries as a guanosine (A-to-G), and APOBECs (apolipoprotein B mRNA editing enzymes, catalytic polypeptide-like), which edit cytidine to uridine (C-to-U). Previously, sequence comparisons and computational predictions have identified many A-to-G editing sites (6, 7, 11–13). By contrast, C-to-U changes are rare; apolipoprotein B is one of the few known target genes of human APOBEC1 (14, 15).
We obtained sequences of DNA and RNA samples from immortalized B cells of 27 unrelated Centre d’Etude du Polymorphisme Humain (CEPH) (16) individuals, who are part of the International HapMap (17, 18) and the 1000 Genomes (19) projects. When we compared the DNA and RNA sequences of the same individuals, we found 28,766 events at over 10,000 exonic sites that differ between the RNA and the corresponding DNA sequences. Each of these differences was observed in at least two individuals; many of these were seen in B cells, as well as in primary skin cells and brain tissues from a separate set of individuals and in expressed sequence tags (ESTs) from cDNA libraries of various cell types. About 43% of the differences are transversions and therefore cannot be the result of typical deaminase-mediated RNA editing. By mass spectrometry, we also found peptide sequences that correspond to the RNA variant sequences, but not the DNA sequences, suggesting that the RNA forms are translated into proteins.
Samples
We compared the DNA and RNA sequences from B cells of 27 unrelated CEPH individuals (table S1). We chose these samples because much information is available on them, including dense DNA genotypes obtained using different technologies (20, 21). The genomes of B cells from the CEPH collection are stable as evidenced by Mendelian inheritance of genetic loci that allowed the construction of microsatellite- to single-nucleotide polymorphism (SNP)–based human genetic maps (20, 21). More recently, the International HapMap Consortium (17, 18) obtained millions of SNP genotypes, and the 1000 Genomes Project (19) sequenced the DNA of these individuals. Comparison of sequence data from these two projects showed high concordance (~99%). Here, we used the DNA genotypes and sequences from the two projects for our analyses. First, we considered sites that are monomorphic in the human genome. A monomorphic site is one where there is no evidence for sequence variation at that locus in dbSNP, the HapMap, and the 1000 Genomes Project. Different studies have analyzed these 27 and hundreds of additional individuals for DNA variants; thus, if a site has not been identified as polymorphic, most likely all individuals have the same sequences at these sites. But to be certain, for these sites in the 27 individuals, we compared their DNA sequences from the 1000 Genomes Project with the sequences of the human reference genome and carried out traditional Sanger sequencing (22). To be included in our analysis, we required that each site be covered by at least four reads in the 1000 Genomes Project and that the sequences from 1000 Genomes should be the same as those of the reference genome. To ensure the integrity of the aliquots of B cells that we used for analyses, we carried out Sanger sequencing of their DNA and found perfect concordance of sequences with data from the 1000 Genomes (thus also the reference genome sequences) (table S2). Second, we considered SNPs. For each individual, a SNP locus was included only if it was homozygous and the HapMap, as well as the 1000 Genomes Project, reported the same sequence. We have high confidence in those sequences because despite using different technologies (microarray-based genotyping in HapMap and high-throughput sequencing in 1000 Genomes), we obtained identical sequences in the two projects.
We sequenced the RNA of B cells from the same 27 individuals using high-throughput sequencing technology from Illumina (23). The resulting RNA sequence reads were mapped to the Gencode genes (24) in the reference human genome. In total, we generated ~1.1 billion reads of 50 base pairs (bp) (~41 million reads and 2 Gb of sequence per individual), of which ~69% of the reads mapped uniquely to the transcriptome [see Methods in (25)]. To be confident of the base calls, for each individual, we focused our analysis on high-quality reads (quality score ≥25) and sites that were covered by at least 10 uniquely mapped reads. Another study (26) had carried out RNA sequencing of the same individuals but at a lower coverage; at these sites we compared our sequences with those from their study, and found that the concordance rate of the sequences is >99.5%. This is reassuring given that the samples were prepared and sequenced in different laboratories.
Differences between RNA and corresponding DNA sequences
For each of the 27 individuals, we compared the mRNA sequences from B cells with the corresponding DNA sequences (fig. S1). The comparison revealed many sites where the mRNA sequences differ from the corresponding DNA sequences of the same individual. To ensure that these are actual differences and to minimize the chance of sequencing errors, we required that at least 10% of the reads covering a site differ from the DNA sequence and at least two individuals show the same RNA-DNA difference at the site. We call each occurrence of a difference between RNA and DNA sequences an “event” and the chromosomal location where such a difference occurs a “site.” Each person can contribute an event to the site; thus, there could be multiple events at a site.
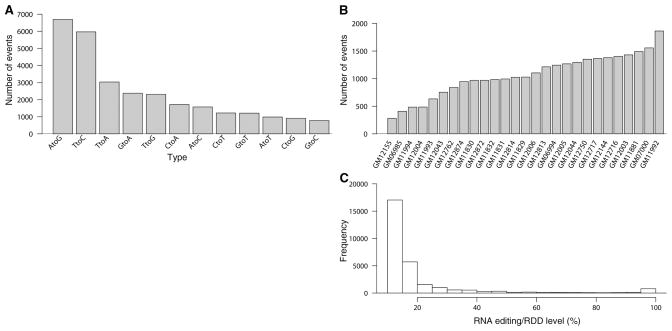
Among our 27 subjects, we identified 28,766 events where the RNA sequences do not match those of the corresponding DNA sequences. These events are found in 10,210 exonic sites (table S10) in the human genome and reside in 4741 known genes [36% of 13,214 genes that are covered by 10 or more RNA sequencing (RNA-Seq) reads in at least one part of the gene, in two or more individuals]. With gene orientation information in Gencode, we observed all 12 possible categories of base differences between RNA and its corresponding DNA (Fig. 1A). All 12 types of differences were found in each of the 27 samples; the relative proportion of each type is similar across individuals. There are 6698 A-to-G events, which can be the result of deamination by ADAR. There are 1220 C-to-T differences, which can also be mediated by a deaminase. However, it is notable that APOBEC1 and its complementation factor A1CF that deaminate cytidine are not expressed in our B cells [fragments per kilobase of exon per million fragments mapped (FPKM) (27) ~ 0 for both genes]; thus, it is likely that an unknown deaminase or other mechanism is involved. Even for relatively well-characterized proteins such as APOBEC1, a recent RNA-Seq study of Apobec1−/− mice uncovered many previously unknown targets (28). In addition, we found 12,507 transversions (43%), which cannot result from classic deaminase-mediated editing. Because we do not know the mechanism by which these differences between RNA and DNA sequences arise, we refer to them as RNA-DNA differences (RDD). An example of an RDD is a C-to-A difference on chromosome 12 (at position 54,841,626 bp) in the myosin light chain gene MYL6, where 16 of our subjects have C/C in their DNA but A/C in their RNA sequences. Another example is an A-to-C difference on chromosome 6 (at position 44,328,823 bp) in the gene HSP90AB1 that encodes a heat shock protein, where eight individuals have homozygous A/A DNA genotype but have A/C in their RNA. Additional examples are shown in Table 1. These sites where RNA sequences differ from the corresponding DNA sequences appear to be nonrandom because the identical differences were found in multiple individuals: 8163 (80.0%) of the sites were found in at least 50% of the informative individuals (i.e. with RNA-Seq coverage ≥10 and DNA-Seq coverage ≥4 at the site). Some sites were found in all or nearly all informative individuals. For example, the DNA sequences of all 19 informative individuals at position 49,369,615 bp of chromosome 3 in the GPX1 gene are G/G, whereas their RNA sequences are G/A. (The remaining individuals were not included because available data did not meet our inclusion criteria although the data suggest the same RDD in all remaining individuals: G/G in DNA, and G/A in RNA.)
Fig. 1.
Characteristics of RDD sites. (A) Frequency of the 12 categories of RNA-DNA differences identified in B cells of 27 normal individuals. (B) Number of RNA editing or RDD events in 27 normal individuals. (C) Bimodal distribution of the levels of 28,766 RNA editing or RDD events.
Table 1.
Selected examples of sites that show RNA-DNA differences in B cells and EST clones.
| Gene | Chr. | Position (bp)* | Type | No. of informative individuals†‡ | No. of individuals with RDD‡ | Average level‡§ [range] | EST |
|---|---|---|---|---|---|---|---|
| HSP90AB1 | 6 | 44,328,823 | A-to-C | 11 | 8 | 0.39 [0.15, 0.79] | BQ355193 (head, neck), BX413896 (B cell) |
| AZIN1 | 8 | 103,910,812 | A-to-G | 17 | 10 | 0.22 [0.12, 0.37] | CD359333 (testis), BF475970 (prostate) |
| CNBP | 3 | 130,372,812 | A-to-T | 18 | 16 | 0.13 [0.10, 0.21] | EL955109 (eye), BJ995106 (hepatoblastoma) |
| MYL6 | 12 | 54,841,626 | C-to-A | 16 | 16 | 0.35 [0.12, 0.60] | EC496428 (prostate), BG030232 (breast adenocarcinoma) |
| RBM23 | 14 | 22,440,217 | C-to-G | 11 | 5 | 0.18 [0.11, 0.35] | BQ232763 (testis, embryonic) |
| RPL23 | 17 | 34,263,515 | C-to-T | 12 | 8 | 0.16 [0.10, 0.22] | BP206252 (smooth muscle), CK128791 (embryonic stem cell) |
| BLNK | 10 | 97,957,645 | G-to-A | 14 | 7 | 0.14 [0.11, 0.17] | BF972964 (leiomyosarcoma), BE881159 (lung carcinoma) |
| C17orf70 | 17 | 77,117,583 | G-to-C | 2 | 2 | 0.26 [0.24, 0.28] | AA625546 (melanocyte), AA564879 (prostate) |
| HMGN2 | 1 | 26,674,349 | G-to-T | 7 | 4 | 0.22 [0.14, 0.43] | BX388386 (neuroblastoma), BE091398 (breast) |
| CANX | 5 | 179,090,533 | T-to-A | 9 | 8 | 0.20 [0.13, 0,30] | EL950052, DB558106 |
| EIF3K | 19 | 43,819,430 | T-to-C | 19 | 14 | 0.16 [0.10, 0.27] | AI250201 (ovarian carcinoma), AI345393 (lung carcinoma) |
| RPL37 | 5 | 40,871,072 | T-to-G | 6 | 6 | 0.27 [0.16, 0.45] | CF124792 (T cell), DW459229 (liver) |
hg18 build of the human genome.
RNA-Seq ≥ 10 reads, DNA-Seq ≥ 4 reads.
B cells.
Calculated by tallying RNA-Seq reads that contain RDDs and those that do not.
RDD in expressed sequence tags
Computational and experimental validations also upheld these observed RNA-DNA differences. First, for 120 sites (10 sites per RDD type; randomly selected and all examples cited in this paper; see Table 1 and table S3), we looked for evidence of RDD in the human EST database by BLAST alignment (29) and manual inspection of each result. For 81 of the 120 sites, we found ESTclones that contain the RDD alleles. The numbers of sites found in human ESTs are similar across different RDD types (average 67.5%; range: 60 to 90%). Second, we examined previously identified A-to-G editing sites (6). Fourteen of the A-to-G sites that we identified were found in their data even though different cell types were studied. Even the levels of editing at these sites are similar between the two studies (fig. S2). Twelve additional sites were found in both studies but were filtered out because they did not meet our selection criteria.
Sanger sequencing of B cells, skin, and brain
Next, we validated our findings experimentally by Sanger sequencing of both DNA and RNA at 12 randomly selected sites in B cells (2 to 9 individuals per site), primary skin (foreskin; 8 to 10 individuals per site), and brain cortex (6 to 10 individuals per site). We regrew the B cells from our subjects and extracted DNA and mRNA from the same aliquots of cells. By sequencing the paired DNA and RNA samples and analysis of each chromatogram by two individuals independently, we confirmed 57 events in 11 sites (Table 2 and fig. S3). In EIF2AK2, in all of the eight individuals whose samples were sequenced, three sites were found within 10 nucleotides (nt) (see below). RDD was not found in one site in NDUFC2. Sanger sequencing is not very sensitive or quantitative; thus, we do not expect to validate all sites, especially those with low levels of RDD.
Table 2.
Sanger sequencing of RDD sites.
| Gene | Chr. | Position (bp)* | Type | Location | Amino acid change | B cells† |
Primary skin fibroblast† |
Brain (cortex)† |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of informative individuals | No. of individuals showing RDD | No. of informative individuals | No. of individuals showing RDD | No. of informative individuals | No. of individuals showing RDD | ||||||
| EIF2AK2 | 2 | 37,181,512 | A-to-G | 3′ UTR | Not applicable | 8 | 8 | 8 | 0 | 10 | 10 |
| 2 | 37,181,517 | A-to-G | 3′ UTR | Not applicable | 8 | 8 | 8 | 3 | 10 | 10 | |
| 2 | 37,181,520 | A-to-G | 3′ UTR | Not applicable | 8 | 8 | 8 | 3 | 10 | 10 | |
| 2 | 37,181,538 | A-to-G | 3′ UTR | Not applicable | 8 | 8 | 8 | 6 | 10 | 10 | |
| AZIN1‡ | 8 | 103,910,812 | A-to-G | Coding, exonic | S to G | 2 | 2 | 10 | 0 | 9 | 8 |
| DPP7 | 9 | 139,128,755 | C-to-T | Coding, exonic | Synonymous (P) | 9 | 2 | 8 | 1 | 10 | 0 |
| PPWD1 | 5 | 64,894,960 | G-to-A | Coding, exonic | E to K | 2 | 2 | 8 | 8 | 8 | 8 |
| HLA-DQB2 | 6 | 32,833,537 | G-to-A | Coding exonic | G to S | 2 | 2 | 10 | 10 | NE§ | NE |
| 6 | 32,833,545 | G-to-A | Coding, exonic | R to H | 2 | 2 | 10 | 10 | NE | NE | |
| 6 | 32,833,550 | C-to-T | Coding, exonic | Synonymous (I) | 2 | 2 | 10 | 10 | NE | NE | |
| BLCAP# | 20 | 35,580,977 | A-to-G | Coding, exonic | Q to R | 6 | 4 | 10 | 4 | 6 | 6 |
| NDUFC2 | 11 | 77,468,303 | C-to-G | Coding, exonic | L to V | 10 | 0 | 10 | 0 | 10 | 0 |
hg18 build of the human genome.
In all cases, matched DNA and RNA samples from the same individuals were sequenced.
Also reported by Li et al. (6).
NE, not expressed.
Known site that we used as a positive control.
To assess whether RDD shows cell type specificity, we looked for evidence of RNA-DNA sequence differences using primary human cells. We studied the same sites as above by Sanger sequencing of DNA and RNA samples from primary skin fibroblasts and brain (cortex) of a separate set of normal individuals (for each site, we examined the DNA and RNA of 6 to 10 samples per cell type). We identified 55 RDD events in primary skin cells and 62 events in brain cortex (Table 2). The results suggest that most sites are shared across cell types (Table 2), although there are exceptions, for example, an A-to-G difference in EIF2AK2 (chromosome 2: 37,181,512), which was only found in B cells and brain cortex but not in primary skin cells. We also queried the EST database for evidence of RDD (Table 1 and table S3). The RNA alleles are seen in a wide range of tissues from embryonic stem cells to brain and testis; they are also found in tumors such as lung carcinoma and neuroblastoma.
Proteomic evidence for RDD
Validation at the sequence level is important but does not address concerns such as the difficulty in aligning sequences that are highly similar and errors introduced by enzymes in reverse transcription steps. We believe that such artifacts are unlikely considering the consistent patterns across sequencing methods and that we observed all 12 types of nucleotide differences. An alternative and independent validation would be to ask whether the RNA variants in RDD sites are translated to proteins. To do so, first we searched mass spectrometry data from human ovarian cancer cells (30) and leukemic cells for putative RDD sites. Because the levels of most RDDs are less than 100%, both DNA and the RDD forms of the mRNAs should be available to be translated (hereafter, we refer to mRNAs that correspond identically to the DNA sequences as DNA forms and those that contain an RDD as RNA forms). In the ovarian cancer and leukemic cells, we indeed found examples of proteins with peptides encoded by both DNA and RNA forms of mRNA (table S4). Encouraged by the search results and cognizant of possible genome instability and thus DNA mutations in cancer cells, we carried out mass spectrometry analysis of our B cells.
We analyzed the proteome of our B cells using liquid chromatography–tandem mass spectrometry and detected peptides for 3217 proteins. Despite advances in mass spectrometry, far less than 50% of peptides can be detected in most studies (31, 32). We identified 327 peptides that cover RDD sites: 299 of them are encoded by the DNA forms and 28 by RNA forms of RDD-containing mRNAs [false discovery rate (FDR) <1%; tables S5 and S9]. For 17 RDD sites, peptides that correspond to both DNA and RNA forms were identified (Table 3). By BLAST alignment, we ensured that these 28 peptides were unique to the genes that contain the RDD sites. In addition, we sequenced the DNA of the B cells used for mass spectrometry and validated that the DNA sequences were the same as those of the reference genome but differed from the RNA sequences and thus did not encode the RNA forms of the peptides (table S2). It is easier to detect more abundant proteins by mass spectrometry; for most RDD sites, the unaltered DNA forms are more abundant than variant RNA forms of mRNA (see below). Thus, it is not surprising to find more peptides that correspond to the DNA than to the RNA sequences. However, the counts of peptides corresponding to the DNA and RNA forms of RDD sites should not be taken as a measure of the proportions of DNA versus RNA forms of mRNA that are translated because differences in the amino acid sequences of the DNA and RNA forms of the peptides affect the ability of mass spectrometry to detect them. In addition, when a peptide is not detected, it does not mean that it is absent from the proteome; it could be a result of sampling.
Table 3.
Peptides encoded by both DNA and RNA forms of mRNA at RDD sites.
| Protein | Position (bp)* | RDD | Amino acid change | DNA form† | RNA form† |
|---|---|---|---|---|---|
| AP2A2 | Chr 11: 976,858 | T-to-G | Y-to-D | YLALESMCTLASSEFSHEAVK | DLALESMCTLASSEFSHEAVK |
| DFNA5‡ | Chr 7: 24,705,225 | T-to-A | L-to-Q | VFPLLLCITLNGLCALGR | VFPQLLCITLNGLCALGR |
| ENO1 | Chr 1: 8,848,125 | T-to-C | L-to-P | EGLELLK | EGPELLK |
| ENO3 | Chr 17: 4,800,624 | T-to-G | V-to-G | LAQSNGWGVMVSHR | LAQSNGWGGMVSHR |
| FABP3 | Chr 1: 31,618,424 | T-to-A | W-to-R | MVDAFLGTWK | MVDAFLGTR |
| FH‡ | Chr 1: 239,747,217 | T-to-A | I-to-K | IEYDTFGELK | KEYDTFGELK |
| HMGB1 | Chr 13: 29,935,772 | T-to-A | Y-to-N | MSSYAFFVQTCR | MSSNAFFVQTCR |
| NACA | Chr 12: 55,392,932 | G-to-A | D-to-N | DIELVMSQANVSR | NIELVMSQANVSR |
| NSF | Chr 17: 42,161,411 | T-to-C | V-to-A | LLDYVPIGPR | LLDYAPIGPR |
| POLR2B | Chr 4: 57,567,852 | T-to-A | L-to-Q | IISDGLK | IISDGQK |
| RAD50‡ | Chr 5: 131,979,610 | T-to-G | L-to-R | WLQDNLTLR | WRQDNLTLR |
| RPL12 | Chr 9: 129,250,509 | A-to-G | N-to-D | HSGNITFDEIVNIAR | HSGDITFDEIVNIAR |
| RPL32‡ | Chr 3: 12,852,658 | G-to-T | A-to-S | AAQLAIR | SAQLAIR |
| RPS3AP47‡ | Chr 4: 152,243,651 | C-to-A | T-to-K | EVQTNDLK | EVQKNDLK |
| SLC25A17 | Chr 22: 39,520,485 | A-to-G | E-to-G | TTHMVLLEIIK | TTHMVLLGIIK |
| TUBA1‡ | Chr 2: 219,823,379 | A-to-G | E-to-G | EDMAALEK | EDMAALGK |
| TUBB2C | Chr 9: 139,257,297 | G-to-A | G-to-D | LHFFMPGFAPLTSR | LHFFMPDFAPLTSR |
hg 18 build of the human genome.
For each peptide, the amino acid that differs between the DNA and RNA forms are underlined.
Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
DNA sequences of these and other proteins were verified by Sanger sequencing (table S2).
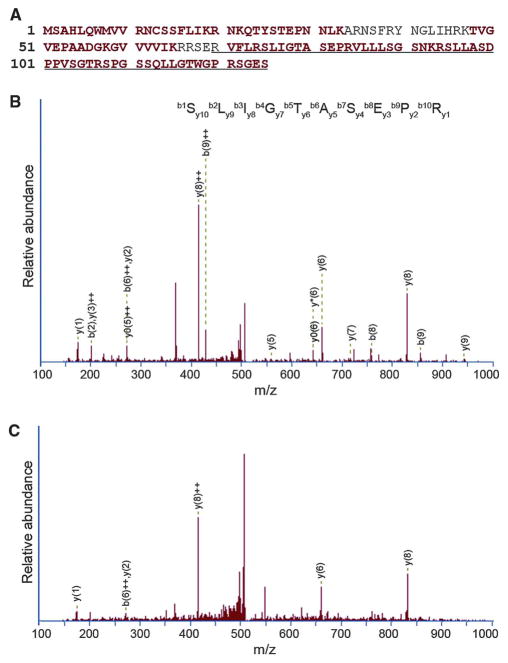
The proteomic data provide an independent validation that mRNA sequences are not always identical to DNA sequences and demonstrate that RNA forms of genes are translated to proteins. They also show that there are peptides in human cells that are not exactly encoded by the DNA sequences. An example of a protein variant that results from RDD is RPL28 (T-to-A; chromosome 19: 60,590,467). The RDD led to a loss of a stop codon. We identified peptides corresponding to the 55–amino acid extension of RPL28 protein in the ovarian cancer cells and in our B cells (Fig. 2). Previously identified cases of RNA editing leading to proteins not encoded by genomic DNA, such as apolipoprotein B (3, 4), serotonin and glutamate receptors (33–35) in humans, and plant ribosomal protein S12 (36), also support our hypothesis that RDD leads to protein isoforms that do not correspond to the DNA sequences of the encoding genes.
Fig. 2.
Identification of peptides coded by both RNA and DNA sequences. (A) The RNA form of a RDD leads to loss of a stop codon in RPL28 and extension of 55 amino acids. Peptides detected by mass spectrometry are shown in red. Extended protein sequence due to RDD is underlined. (B and C) Tandem mass spectrometry (MS-MS) data confirm the detection of peptides encoded by the RDD-containing RPL28 mRNA. The representative spectra of one peptide (SLIGTASEPR) from ovarian cancer cells (B) and cultured B cells (C) are shown.
Individual variation in abundance of RDD
Using our selection criteria, we found that in each person among the Gencode genes, there are on average 1065 exonic events that differ in the RNA and DNA sequences. But the number of events varied among individuals (range: 282 to 1863) by up to sixfold across our 27 subjects (Fig. 1B). The degree of sequence coverage and sequencing errors in DNA or RNA samples do not explain these individual differences (25). Thus, there is likely a biological basis for the individual variation in the number of editing and RDD events. We found no significant correlation between ADAR expression and the number of RDDs or the numbers of A-to-G events (P > 0.5). Thus, either ADAR expression does not affect the number of editing or RDD events, or our sample size is not sufficient to detect the correlation.
Characteristics of RDD sites
The 10,210 sites that showed RNA and DNA sequence differences are not evenly distributed across the ge-nome: Chromosome 19 has the most sites, whereas chromosome 13 has the fewest. This pattern is observed after correction for differences in size and gene density among chromosomes. RDD sites are significantly (P < 10−10) enriched in genes that play a role in helicase activity, and in protein and nucleotide binding (table S6).
The 10,210 sites that showed RNA and DNA sequence differences are not evenly distributed within genes. About 44% (4453 sites) of them are located in coding exons (10% were found in the last exons), 4% (386 sites) are in the 5′ untranslated regions (UTRs), and 39% (3977 sites) are in the 3′ UTRs (table S7; those remaining cannot be classified because of differences in gene structures across isoforms). The results suggest that there are more sites in the 3′ ends than in the 5′ ends of genes; a pattern that was also observed in deamination-mediated RNA editing (28, 37). Seventy-one percent of the coding sites result in nonsynonymous amino acid changes, including 2.1% that lead to the gain or loss of a stop codon if translated into proteins. Relative to other structural features in genes, we found that 4% of RDD sites are within 2 nt of exon borders and 5% are within 30 nt of polyadenylate [poly(A)] signals (table S7). Among RDD types, the numbers of sites near splice junctions are quite similar, but the numbers near poly(A) sites are more different. C-to-A and G-to-A differences are found more often near poly(A) sites.
Sites also tended to cluster; for example, 2613 sites (26%) are within 25 bp, and 1059 sites (10%) are adjacent to each other. Statistical analysis using a runs test supports the nonrandom locations of the sites (median P= 0.22). We did not find obvious patterns or associations with motifs shared across the sites, except for the A-to-G and A-to-C differences that show a preference for a cytidine 5′ to the adenosine, as previously observed in ADAR-mediated A-to-G changes (7, 35).
RDD levels
We examined the percentage of mRNAs that differ in sequence from the corresponding DNA. For each site, to determine the RDD level, we counted the number of reads with a different nucleotide from that in the corresponding DNA sequence. The distribution of the level is bimodal (Fig. 1C); the average level is 20% (median = 13%). However, for some sites, RDD was detected in nearly 100% of the RNA sequences such as the A-to-C difference in the gene that encodes an mRNA decapping enzyme, DCP1A (chromosome 3: 53,297,343). This level is correlated with the frequency and types of RNA-DNA differences. Sites found in more than 50% of the informative individuals tend to have higher levels of RNA editing or RDD than other sites (P < 10−5; fig. S5). The levels also differ across individuals. For example, at a G-to-A site in the gene RHOT1, which encodes a RAS protein that plays a role in mitochondrial trafficking (chromosome 17: 27,526,465), in one person, the level was 90% while in another person, it was only 18%. We identified 437 sites with 10 or more informative individuals where the individuals with the highest levels and the lowest levels differed by twofold or more (range: 2- to 8.6-fold).
Conclusions
We have uncovered thousands of exonic sites where the RNA sequences do not match those of the DNA sequences, including transitions and transversions. These findings challenge the long-standing belief that in the same individuals, DNA and RNA sequences are nearly identical. To increase the confidence in our results, we obtained the DNA, RNA, and protein sequences from different individuals and cell types using a range of technologies (fig. S1B). The samples included cell lines and primary cells from healthy individuals and tumors. We used data from public resources such as EST databases, the HapMap, and 1000 Genomes Project, as well as those that we generated with traditional Sanger sequencing, high-throughput sequencing technologies, and mass spectrometry. Table 4 shows the DNA, RNA, and peptide sequences at 15 confirmed sites, which illustrate that the RNA and peptide sequences are the same but differ from the corresponding DNA sequences. The results support our observation that in an individual, DNA and RNA sequences from the same cells are not always identical and some of the variant RNA sequences are translated into proteins. The consistent pattern of the observations suggests that the RDDs have biological significance and are not just “noise.” At nearly all RDD sites, we observed only one RDD type across cell types and in different individuals. If the DNA sequence is A/A, and the RNA is A/C in one sample, in other samples, we see the same A-to-C difference, but not other types of differences. These results suggest that there are unknown aspects of transcription and/or posttranscriptional processing of RNA. These differences may now be studied along with those in other genomes and organisms such as the mitochondrial genomes of trypanosomes and chloroplasts of plants, where RNA editing and modifications are relatively common (36, 37).
Table 4.
Corresponding DNA, RNA, and peptide sequences at selected sites.
| RDD | Gene | Location | DNA*† | RNA† | Peptide (DNA form, LC-MS/MS)‡ | Peptide (RNA form, LC-MS/MS) |
|---|---|---|---|---|---|---|
| T-to-G | CD22 | Chr 19: 40,514,815 | CTG | CGG | ND§ | MHLLGPWLLLR |
| T-to-A | DFNA5 | Chr 7: 24,705,225 | CTG | CAG | VFPLLLCITLNGLCALGR | VFPQLLCITLNGLCALGR |
| T-to-C | ENO1 | Chr 1: 8,848,125 | CTG | CCG | EGLELLK | EGPELLK |
| T-to-A | FH | Chr 1: 239,747,217 | ATA | AAA | IEYDTFGELK | KEYDTFGELK |
| T-to-A | HMGB1 | Chr 13: 29,935,772 | TAT | AAT | MSSYAFFVQTCR | MSSNAFFVQTCR |
| A-to-C | HMGB1 | Chr 13: 29,935,469 | AAA | AAC | ND | TMSAKEN |
| A-to-C | ITPR3 | Chr 6: 33,755,773 | GAC | GCC | ND | DGVEDHSPLMYHISLVALLAACAEGK |
| T-to-G | RAD50 | Chr 5: 131,979,610 | CTA | CGA | WLQDNLTLR | WRQDNLTLR |
| G-to-T | ROD1 | Chr 9: 114,026,264 | GGA | GTA | ND | NLFIEAVCSVK |
| G-to-T | RPL32 | Chr 3: 12,852,658 | GCT | TCT | AAQLAIR | SAQLAIR |
| A-to-G | RPS25P8 | Chr 11: 118,393,375 | AAC | GAC | ND | EVPDYK |
| C-to-A | RPS3AP47 | Chr 4: 152,243,651 | ACA | AAA | EVQTNDLK | EVQKNDLK |
| G-to-T | SUPT5H | Chr 19: 44,655,806 | CAG | CAT | ND | TPMYGSQTPLHDGSR |
| T-to-C | TOR1AIP1 | Chr 1: 178,144,365 | TCA | CCA | ND | QPSVLSPGYQK |
| A-to-G | TUBA1 | Chr 2: 219,823,379 | GAG | GGG | EDMAALEK | EDMAALGK |
DNA sequences are monomorphic according to dbSNP, 1000 Genomes, and HapMap projects; all individuals should have the reference allele. We verified this by Sanger sequencing of the B cells used for mass spectrometry.
RDD sites are underlined.
LC-MS/MS: liquid chromatography and tandem mass spectrometry.
ND: not detected by mass spectrometry; however, this does not mean that the peptides are absent from the B cell proteome. It is likely a result of sampling.
The underlying mechanisms for these events are largely unknown. For most of the cases, we do not know yet whether a different base was incorporated into the RNA during transcription or if these events occur posttranscriptionally. About 23% of the sites are A-to-G differences; some of these are likely mediated by ADAR, but other, currently unknown, mechanisms can be involved. If it is a cotranscriptional process, then the signal can be in the DNA or the RNA such as secondary structures or modified nucleotides. In addition, as some of the RDDs are found near splice and poly(A) sites, it is possible that this may be a facet of systematic RNA processing steps such as splicing and cleavage (38, 39).
Our findings supplement previous studies demonstrating RNA-DNA differences in the human genome and show that these differences go beyond A-to-G transition. These findings affect our understanding of genetic variation; in addition to DNA sequence variation, we identify individual variation in RNA sequences. For monomorphic DNA sequences that show RDD, there is an overall increase in genetic variation. Thus, this variation not only contributes to individual variation in gene expression, but also diversifies the proteome because some identified sites lead to nonsynonymous amino acid changes. We speculate that this RNA sequence variation likely affects disease susceptibility and manifestations. To date, mapping studies have focused on identifying DNA variants as disease susceptibility alleles. Our results suggest that the search may need to include RNA sequence variants that are not in the DNA sequences.
Supplementary Material
Acknowledgments
Dedicated to the memory of Dr. Tom Kadesch who gave us important suggestions, taught us salient and subtle points on gene expression, and inspired us with his enthusiasm. Dr. Kadesch died during the preparation of this manuscript. We thank D. Epstein, H. Kazazian, D. Puppione, and L. Simpson for suggestions and discussions. We thank C. Gunter, R. Nussbaum, and J. Puck for comments on the manuscript, M. Morley for help with data analysis, W. Ankener for sample processing, and J. Devlin and CHOP NAP core for results on Sanger sequencing. The mass spectrometry analysis was carried out at the Wistar Proteomic Facility; we thank K. Speicher for help and suggestions. Funded by grants from the National Institutes of Health (to V.G.C. and M.L.) and support from the Howard Hughes Medical Institute. The RNA-Seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under the accession no. GSE25840.
Footnotes
References and Notes
- 1.Libby RT, Gallant JA. Mol Microbiol. 1991;5:999. doi: 10.1111/j.1365-2958.1991.tb01872.x. [DOI] [PubMed] [Google Scholar]
- 2.Sydow JF, Cramer P. Curr Opin Struct Biol. 2009;19:732. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Chen SH, et al. Science. 1987;238:363. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 4.Powell LM, et al. Cell. 1987;50:831. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 5.Bass BL, Weintraub H. Cell. 1988;55:1089. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 6.Li JB, et al. Science. 2009;324:1210. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 7.Athanasiadis A, Rich A, Maas S. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas MJ, Platas AA, Hawley DK. Cell. 1998;93:627. doi: 10.1016/s0092-8674(00)81191-5. [DOI] [PubMed] [Google Scholar]
- 9.Wang D, et al. Science. 2009;324:1203. doi: 10.1126/science.1168729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zenkin N, Yuzenkova Y, Severinov K. Science. 2006;313:518. doi: 10.1126/science.1127422. [DOI] [PubMed] [Google Scholar]
- 11.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Nat Chem Biol. 2010;6:733. doi: 10.1038/nchembio.434. [DOI] [PubMed] [Google Scholar]
- 12.Nishikura K. Annu Rev Biochem. 2010;79:321. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levanon EY, et al. Nat Biotechnol. 2004;22:1001. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 14.Conticello SG. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chester A, Scott J, Anant S, Navaratnam N. Biochim Biophys Acta-Gene Struct Expression. 2000;1494:1. doi: 10.1016/s0167-4781(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 16.Dausset J, et al. Genomics. 1990;6:575. doi: 10.1016/0888-7543(90)90491-c. [DOI] [PubMed] [Google Scholar]
- 17.International HapMap Consortium. Nature. 2003;426:789. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.International HapMap Consortium. Nature. 2005;437:1299. [Google Scholar]
- 19.The 1000 Genomes Project Consortium. Nature. 2010;467:1061. [Google Scholar]
- 20.Cann HM. Curr Opin Genet Dev. 1992;2:393. doi: 10.1016/s0959-437x(05)80148-0. [DOI] [PubMed] [Google Scholar]
- 21.Matise TC, et al. Am J Hum Genet. 2003;73:271. doi: 10.1086/377137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson AR. Proc Natl Acad Sci USA. 1977;74:5463. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bentley DR, et al. Nature. 2008;456:53. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrow J, et al. Genome Biol. 2006;7(suppl 1):S4. [Google Scholar]
- 25.Supporting material is available on Science Online.
- 26.Montgomery SB, et al. Nature. 2010;464:773. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trapnell C, et al. Nat Biotechnol. 2010;28:511. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Nat Struct Mol Biol. 2011;18:230. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Sodek KL, et al. Mol Biosyst. 2008;4:762. doi: 10.1039/b717542f. [DOI] [PubMed] [Google Scholar]
- 31.Michalski A, Cox J, Mann M. J Proteome Res. 2011;10:1785. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 32.de Godoy LM, et al. Genome Biol. 2006;7:R50. doi: 10.1186/gb-2006-7-6-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burns CM, et al. Nature. 1997;387:303. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 34.Lomeli H, et al. Science. 1994;266:1709. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 35.Maas S, Patt S, Schrey M, Rich A. Proc Natl Acad Sci USA. 2001;98:14687. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phreaner CG, Williams MA, Mulligan RM. Plant Cell. 1996;8:107. doi: 10.1105/tpc.8.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hundley HA, Krauchuk AA, Bass BL. RNA. 2008;14:2050. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rueter SM, Burns CM, Coode SA, Mookherjee P, Emeson RB. Science. 1995;267:1491. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- 39.Rueter SM, Dawson TR, Emeson RB. Nature. 1999;399:75. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.