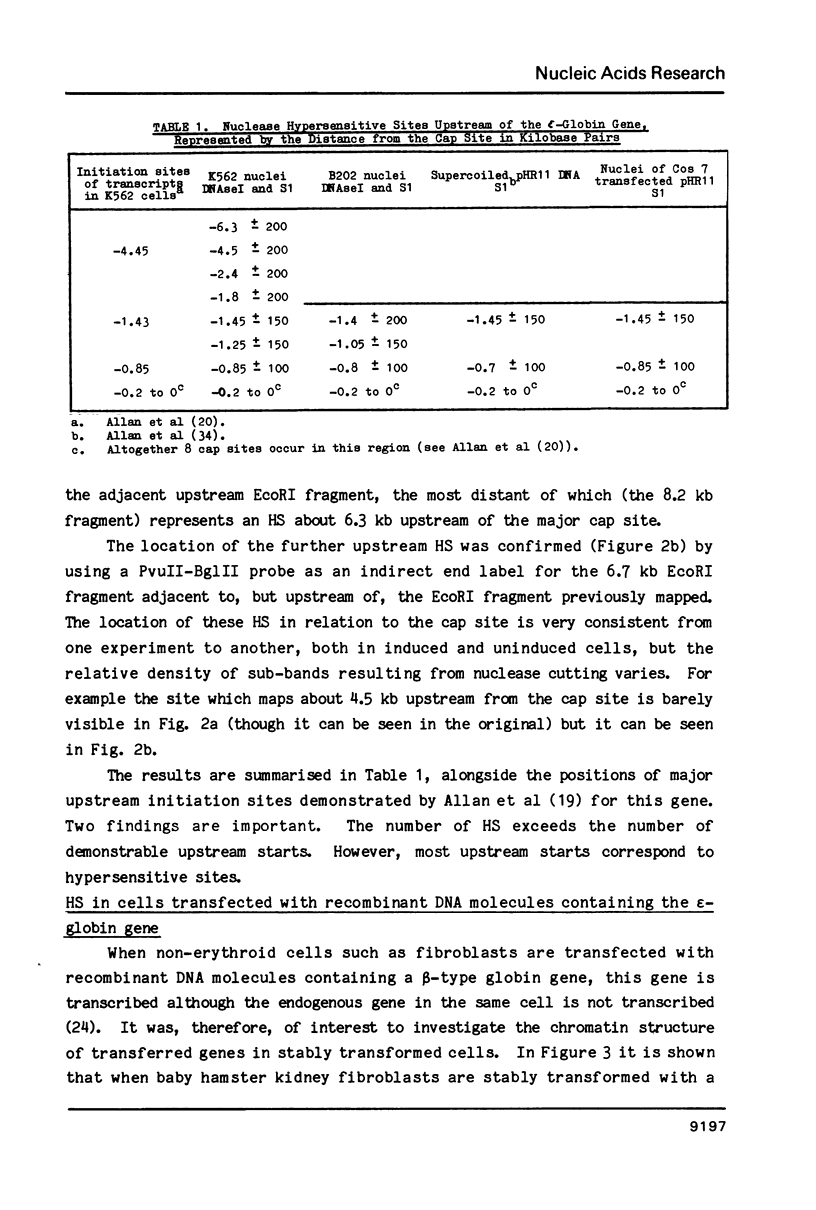
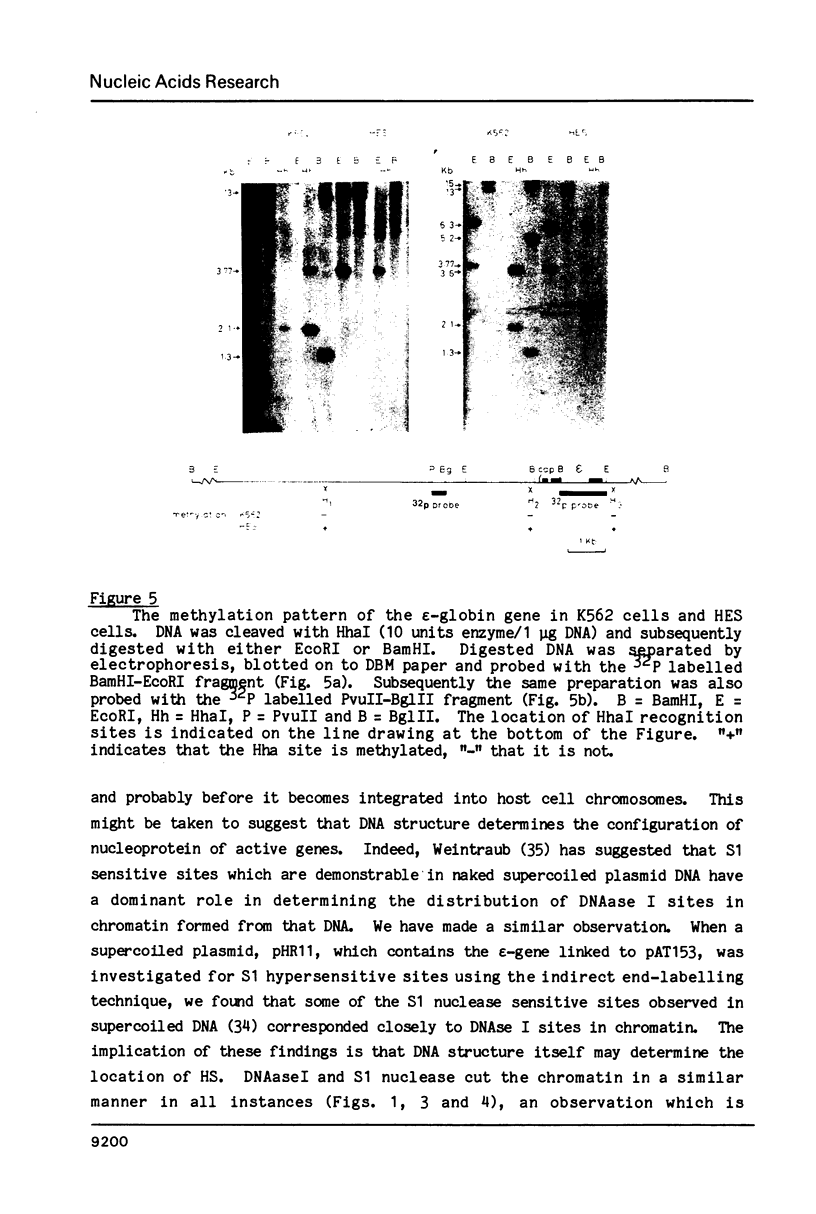
Abstract
We have mapped sites in chromatin flanking the epsilon-globin gene in the K562 cell which are hypersensitive to digestion with DNAseI, micrococcal nuclease and S1 nuclease. Many of those in the 5' flanking region correspond to minor upstream transcriptional starts. However, one prominent site occurs upstream of the boundary of transcription; it maps to a region with an unusual DNA sequence. In baby hamster kidney cells stably transformed with recombinant DNA containing the human epsilon-globin gene and in Cos 7 cells transiently transfected with DNA containing the epsilon-globin gene, hypersensitive sites can be demonstrated.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alan M., Grindlay G. J., Stefani L., Paul J. Epsilon globin gene transcripts originating upstream of the mRNA cap site in K562 cells and normal human embryos. Nucleic Acids Res. 1982 Sep 11;10(17):5133–5147. doi: 10.1093/nar/10.17.5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan M., Lanyon W. G., Paul J. Multiple origins of transcription in the 4.5 Kb upstream of the epsilon-globin gene. Cell. 1983 Nov;35(1):187–197. doi: 10.1016/0092-8674(83)90221-0. [DOI] [PubMed] [Google Scholar]
- Allan M., Zhu J. D., Montague P., Paul J. Differential response of multiple epsilon-globin cap sites to cis- and trans-acting controls. Cell. 1984 Sep;38(2):399–407. doi: 10.1016/0092-8674(84)90495-1. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Olah J., Birnstiel M. L. Major changes in the 5' and 3' chromatin structure of sea urchin histone genes accompany their activation and inactivation in development. Cell. 1983 Jul;33(3):843–848. doi: 10.1016/0092-8674(83)90026-0. [DOI] [PubMed] [Google Scholar]
- Burch J. B., Weintraub H. Temporal order of chromatin structural changes associated with activation of the major chicken vitellogenin gene. Cell. 1983 May;33(1):65–76. doi: 10.1016/0092-8674(83)90335-5. [DOI] [PubMed] [Google Scholar]
- Carlson D. P., Ross J. Human beta-globin promoter and coding sequences transcribed by RNA polymerase III. Cell. 1983 Oct;34(3):857–864. doi: 10.1016/0092-8674(83)90543-3. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation and gene activity. Annu Rev Biochem. 1983;52:93–124. doi: 10.1146/annurev.bi.52.070183.000521. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour R. S., Spandidos D. A., Vass J. K., Gow J. W., Paul J. A negative regulatory sequence near the mouse beta-maj globin gene associated with a region of potential Z-DNA. EMBO J. 1984 Jun;3(6):1263–1272. doi: 10.1002/j.1460-2075.1984.tb01961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindlay G. J., Lanyon W. G., Allan M., Paul J. Alternative sites of transcription initiation upstream of the canonical cap site in human gamma-globin and beta-globin genes. Nucleic Acids Res. 1984 Feb 24;12(4):1811–1820. doi: 10.1093/nar/12.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M., Eisenman R., Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981 Jul 23;292(5821):311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- Groudine M., Weintraub H. Propagation of globin DNAase I-hypersensitive sites in absence of factors required for induction: a possible mechanism for determination. Cell. 1982 Aug;30(1):131–139. doi: 10.1016/0092-8674(82)90019-8. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Kruczek I., Doerfler W. The unmethylated state of the promoter/leader and 5'-regions of integrated adenovirus genes correlates with gene expression. EMBO J. 1982;1(4):409–414. doi: 10.1002/j.1460-2075.1982.tb01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M. T., Mandel J. L., Chambon P. DNA methylation: correlation with DNase I sensitivity of chicken ovalbumin and conalbumin chromatin. Nucleic Acids Res. 1979 Dec 20;7(8):2105–2113. doi: 10.1093/nar/7.8.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Ley T. J., Nienhuis A. W. A weak upstream promoter gives rise to long human beta-globin RNA molecules. Biochem Biophys Res Commun. 1983 May 16;112(3):1041–1048. doi: 10.1016/0006-291x(83)91723-0. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D., Oudet P., Chambon P. Structure of transcribing chromatin. Prog Nucleic Acid Res Mol Biol. 1980;24:1–55. doi: 10.1016/s0079-6603(08)60670-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- McGinnis W., Shermoen A. W., Heemskerk J., Beckendorf S. K. DNA sequence changes in an upstream DNase I-hypersensitive region are correlated with reduced gene expression. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1063–1067. doi: 10.1073/pnas.80.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D. M., Turner P., Nienhuis A. W., Axelrod D. E., Gopalakrishnan T. V. Active conformation of the globin genes in uninduced and induced mouse erythroleukemia cells. Cell. 1978 Jul;14(3):511–521. doi: 10.1016/0092-8674(78)90237-4. [DOI] [PubMed] [Google Scholar]
- Pays E., Gilmour R. S. Specificity of chromatin transcription in vitro. Asymmetric transcription of the globin gene by Escherichia coli RNA polymerase. Biochim Biophys Acta. 1981 May 29;653(3):356–367. doi: 10.1016/0005-2787(81)90192-1. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Rutherford T., Clegg J. B., Higgs D. R., Jones R. W., Thompson J., Weatherall D. J. Embryonic erythroid differentiation in the human leukemic cell line K562. Proc Natl Acad Sci U S A. 1981 Jan;78(1):348–352. doi: 10.1073/pnas.78.1.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal B., Worcel A., Louis C., Schedl P. Chromatin structure of the histone genes of D. melanogaster. Cell. 1981 Feb;23(2):401–409. doi: 10.1016/0092-8674(81)90135-5. [DOI] [PubMed] [Google Scholar]
- Saragosti S., Moyne G., Yaniv M. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell. 1980 May;20(1):65–73. doi: 10.1016/0092-8674(80)90235-4. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Rifkind R. A., Marks P. A. Murine erythroleukemia cell differentiation: DNase I hypersensitivity and DNA methylation near the globin genes. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1180–1184. doi: 10.1073/pnas.79.4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spandidos D. A., Paul J. Transfer of human globin genes to erythroleukemic mouse cells. EMBO J. 1982;1(1):15–20. doi: 10.1002/j.1460-2075.1982.tb01117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder J., Groudine M., Dodgson J. B., Engel J. D., Weintraub H. Hb switching in chickens. Cell. 1980 Apr;19(4):973–980. doi: 10.1016/0092-8674(80)90088-4. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Tuan D., London I. M. Mapping of DNase I-hypersensitive sites in the upstream DNA of human embryonic epsilon-globin gene in K562 leukemia cells. Proc Natl Acad Sci U S A. 1984 May;81(9):2718–2722. doi: 10.1073/pnas.81.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H. A dominant role for DNA secondary structure in forming hypersensitive structures in chromatin. Cell. 1983 Apr;32(4):1191–1203. doi: 10.1016/0092-8674(83)90302-1. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wigler M. H. The inheritance of methylation patterns in vertebrates. Cell. 1981 May;24(2):285–286. doi: 10.1016/0092-8674(81)90317-2. [DOI] [PubMed] [Google Scholar]
- Wigler M., Sweet R., Sim G. K., Wold B., Pellicer A., Lacy E., Maniatis T., Silverstein S., Axel R. Transformation of mammalian cells with genes from procaryotes and eucaryotes. Cell. 1979 Apr;16(4):777–785. doi: 10.1016/0092-8674(79)90093-x. [DOI] [PubMed] [Google Scholar]
- Wu C., Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]