Abstract
Multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates harboring New Delhi metallo-β-lactamase (NDM-1) were isolated from a patient who had returned to Canada from India. The NDM-1 gene was found on closely related incompatibility group A/C type plasmids. The occurrence of NDM-1 in North America is a major public health concern.
Keywords: Multidrug resistance, New Delhi metallo-β-lactamase, Klebsiella pneumoniae, Escherichia coli, carbapenemase, bacteria, expedited, Canada, dispatch
Carbapenem-hydrolyzing β-lactamases, i.e., carbapenemases, in bacterial clinical isolates are an increasing concern because they often also confer resistance to most other β-lactam antimicrobial agents. Among Enterobacteriaceae, carbapenemases are mainly found in the Ambler class A penicillinase or class B metallo-enzyme groups. VIM and IMP are the most frequently acquired class B enzymes and are commonly found in southern Europe and the Far East, rarely in North America. Klebsiella pneumoniae carbapenemase (KPC) class A has been found worldwide, although it emerged in the eastern United States in the mid 1990s and subsequently has successfully established itself in multiple states (1).
Recently, a new class B enzyme, New Delhi metallo-β-lactamase (NDM-1), was characterized from a K. pneumoniae isolate from Sweden; the bacteria seem to have been imported from India (2). Enterobacteriaceae isolates harboring NDM-1 have now been found in multiple areas of India and Pakistan and in the United Kingdom (3–5); such isolates were recently reported from 3 US states (6). In Canada, carbapenemase-harboring isolates are rare, reported only for a small outbreak of clonal Pseudomonas aeruginosa isolates harboring IMP-7, P. aeruginosa isolates harboring VIM-2, a Serratia marcescens isolate harboring a SME-2 class A β-lactamase, and 3 isolates of K. pneumoniae harboring KPC-3 (7–10). We characterized NDM-harboring clinical isolates from a patient who had recently traveled to India.
The Study
A 76-year-old woman returned to Vancouver, Canada, in early 2010 after spending 3.5 months in northern India. Before her trip, she had been in good health. In India, persistent nonbloody diarrhea developed, for which she did not seek medical attention. One month later, she was treated in the hospital for hypertension and congestive heart failure. She was discharged 3 days after admission but readmitted 3 days later because of ongoing diarrhea and decreased consciousness. Unspecified encephalitis and a urinary tract infection were diagnosed, but despite antibacterial drug therapy, her neurologic status did not improve over the next 3 weeks. She was discharged from the hospital in India and transferred to Canada.
When she arrived at the hospital in Vancouver on February 14, 2010, her vital signs reflected distributive shock: temperature 38.3°C, blood pressure 100/80 mm Hg, and heart rate 126 beats/minute. Sepsis was suspected and she was given imipenem and vancomycin. Within 24 hours, her level of consciousness had deteriorated, and she was admitted to the intensive care unit and intubated for airway protection. Blood cultures were negative, but urine culture (>1 × 108 CFU/mL) grew highly drug-resistant K. pneumoniae N10–0469 (February 15, 2010) with intermediate resistance to chloramphenicol and susceptibility to colistin. The urine was packed with leukocytes, and no other source for sepsis was found. A perirectal sample, screened for resistant gram-negative rods, grew K. pneumoniae N10–0506 and E. coli N10–0505 (February 16, 2010). A stool specimen was negative for ova and parasites but positive for Clostridium difficile toxin.
The patient was given vancomycin and metronidazole for the C. difficile infection and colistin for the K. pneumoniae infection. Treatment with colistin was discontinued shortly after its initiation because of the onset of renal complications; treatment with chloramphenicol was successful. Test results for infectious causes of encephalitis (e.g., malaria smears, cerebrospinal fluid culture for bacteria, and staining for acid-fast bacteria and fungi) were negative, as were test results for cryptococcal antigen, herpes simplex viruses 1 and 2, Japanese encephalitis virus, and rabies virus. Results of computed tomographic scans, magnetic resonance imaging of the head, and an electroencephalogram were suggestive of global metabolic encephalopathy. Neurologic symptoms did not improve, despite the successful treatment of the urinary tract infection. Subsequently, E. coli N10–0705 isolate with an extended-spectrum β-lactamase phenotype was obtained from urine on March 9, 2010, for which the patient was treated with imipenem.
Ventilator and vasopressor support were eventually removed, and the patient was transferred to the general medicine ward. Several days later, her condition worsened and she died; the final diagnosis was toxic metabolic leukoencephalopathy, probably related to sepsis. The patient’s family refused to allow an autopsy.
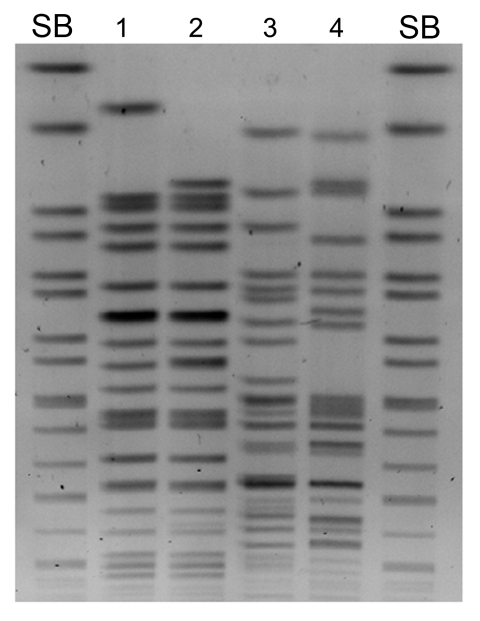
Macrorestriction analysis of the 4 isolates showed that K. pneumoniae N10–0469 and N10–0506 were closely related, although E. coli N10–0505 and N10–0705 were not (Figure 1). All 4 isolates contained multiple plasmids (data not shown). β-lactamase PCR and sequencing were conducted (primers listed in Table 1). K. pneumoniae N10–0469 and N10–0506 harbored the genes for SHV-1, CTX-M-15, OXA-1, CMY-6, and NDM-1; E. coli N10–0505 harbored TEM-1, CTX-M-15, CMY-6, and NDM-1; and E. coli N10–0705 harbored TEM-1, CTX-M-15, OXA-1, and CMY-42. The first characterized isolate from Sweden that harbored blaNDM-1 also harbored CMY-4, a Trp221Arg variant of the widespread CMY-2 β-lactamase (3). CMY-6 is a Trp221Leu variant of CMY-2 (GenBank accession no. AJ011293). CMY-42 is a Val231Ser variant of CMY-2.
Figure 1.
Macrorestriction analysis (XbaI) by pulsed-field gel electrophoresis of Klebsiella pneumoniae N100469 (lane 1), K. pneumoniae N10–0506 (lane 2), Escherichia coli N10–0505 (lane 3), E. coli N10–0705 (lane 4), Salmonella enterica serovar Branderup molecular mass marker (XbaI) (lanes SB)..
Table 1. Primers used in study of New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada, 2010.
| Name | Sequence, 5′ → 3′* | Product size, bp |
|---|---|---|
| SHV-UP | CGCCGGGTTATTCTTATTTGTCGC | 1,016 |
| SHV-LO | TCTTTCCGATGCCGCCGCCAGTCA | |
| TEM-G | TTGCTCACCCAGAAACGCTGGTG | 708 |
| TEM-H | TACGATACGGGAGGGCTTACC | |
| CTX-U1 | ATGTGCAGYACCAGTAARGTKATGGC | 593 |
| CTX-U2 | TGGGTRAARTARGTSACCAGAAYCAGCGG | |
| OXA1-F | CGCAAATGGCACCAGATTCAAC | 464 |
| OXA1-R | TCCTGCACCAGTTTTCCCATACAG | |
| CMY2-A | TGATGCAGGAGCAGGCTATTCC | 323 |
| CMY2-B | CTAACGTCATCGGGGATCTGC | |
| KPC1 | ATGTCACTGTATCGCCGTC | 863 |
| KPC2 | AATCCCTCGAGCGCGAGT | |
| VIM1 | GTTTGGTCGCATATCGCAAC | 382 |
| VIM2 | AATGCGCAGCACCAGGATAGAA | |
| IMP1 | CCWGATTTAAAAATYGARAAGCTTG | 522 |
| IMP2 | TGGCCAHGCTTCWAHATTTGCRTC | |
| NDM-F | GGTGCATGCCCGGTGAAATC | 660 |
| NDM-R | ATGCTGGCCTTGGGGAACG | |
| NDM-A | CACCTCATGTTTGAATTCGCC | 984 |
| NDM-B | CTCTGTCACATCGAAATCGC |
*H is A, C, or T; K is G or T; R is A or G; S is G or C; and W is A or T.
Antimicrobial drug susceptibility testing by Sensititer panels ESB1F and CMV1AGNF (Trek Diagnostic Systems, Cleveland, OH, USA) found that all 4 isolates were multidrug resistant and that 3 of the 4 isolates, not E. coli N10–0705, were nonsusceptible to carbapenems (Table 2). Nonsusceptibility to carbapenems was also shown by Etest (bioMérieux, St. Laurent, Quebec, Canada) and disk diffusion (Table 2). K. pneumoniae N10–0469 and E. coli N10–0505 were susceptible to colistin, polymixin B, and tigecycline.
Table 2. Antimicrobial drug susceptibilities of the isolates from study of New Delhi metallo-β-lactamase in Klebsiella pneumoniae and Escherichia coli, Canada, 2010*.
| Test/antimicrobial agent | K. pneumoniae N10–0469 | E. coli DH10B (pNDM-Kp10469) | E. coli N10–0505 | E. coli DH10B (pNDM-Ec10505) | E. coli DH10B |
|---|---|---|---|---|---|
| Broth microdilution (MIC, μg/mL) | |||||
| Amoxicillin/clavulanic acid | >32 | >32 | 32 | >32 | 4 |
| Piercillin/tazobactam | >64 | >64 | >64 | >64 | <4 |
| Ampicillin | >16 | >16 | >16 | >16 | 2 |
| Cefazolin | >16 | >16 | >16 | >16 | <8 |
| Cefotaxime | >64 | >64 | >64 | >64 | <0.25 |
| Cefotaxime/clavulanic acid | >64 | >64 | >64 | >64 | <0.12 |
| Ceftazidime | >128 | >128 | >128 | >128 | <0.25 |
| Ceftazidime/clavulanic acid | >128 | >128 | >128 | >128 | 0.25 |
| Cefepime | >16 | 16 | >16 | 16 | <1 |
| Cefoxitin | >64 | >64 | >64 | >64 | 8 |
| Imipenem | >16 | 4 | 2 | 2 | <0.5 |
| Meropenem | >8 | 2 | 8 | 2 | <1 |
| Chloramphenicol | 16 | 4 | 32 | 4 | <2 |
| Tetracycline | >32 | ≤4 | 32 | ≤4 | <4 |
| Amikacin | >64 | >64 | >64 | >64 | 2 |
| Gentamicin | >16 | >16 | >16 | >16 | <0.25 |
| Ciprofloxacin | >2 | <0.015 | >2 | <0.015 | <0.015 |
| Trimethoprim/sulfamethoxazole |
>4 |
<0.12 |
>4 |
<0.12 |
<0.12 |
| Etest (MIC, μg/mL) | |||||
| Aztreonam | >256 | 4 | >256 | 4 | 0.094 |
| Imipenem | >32 | 4 | >32 | 4 | 0.19 |
| Meropenem | >32 | 8 | 32 | 6 | 0.032 |
| Ertapenem | >32 | 16 | >32 | 16 | 0.008 |
| Colistin | 0.38 | ND | 0.38 | ND | ND |
| Polymyxin B | 0.19 | ND | 0.25 | ND | ND |
| Tigecycline |
1.5 |
ND |
0.125 |
ND |
ND |
| Disk diffusion (zone diameter, mm) | |||||
| Imipenem | 6 | 14 | 11 | 13 | 33 |
| Meropenem | 6 | 15 | 10 | 14 | 35 |
| Ertapenem | 6 | 13 | 6 | 13 | 35 |
*NDM, New Delhi metallo-β-lactamase; ND, not done.
Multilocus sequence typing of K. pneumoniae N10–0469 showed that it was sequence type (ST) 16 (11). The first characterized strain of K. pneumoniae that harbored NDM-1 was ST14, unrelated to ST16 (3). Mutilocus sequence typing of E. coli N10–0505 showed it to be ST405 (12). The worldwide spread of blaCTX-M-15 has been partly attributed to 2 epidemic strains, ST131 and ST405 (13).
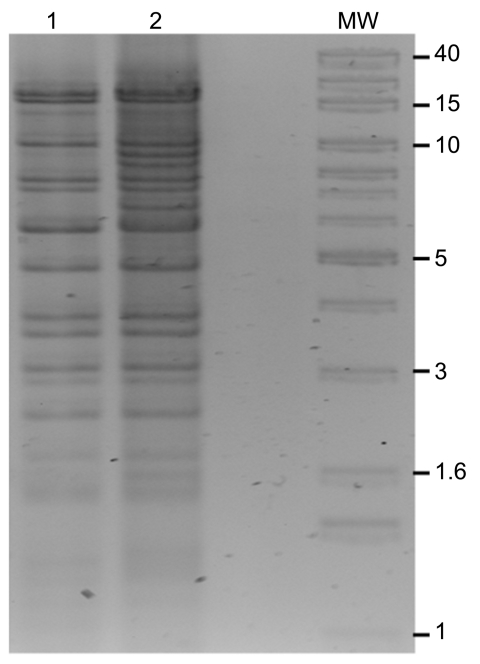
E. coli DH10B harboring NDM-1 plasmids was obtained by electrotransformation with whole plasmid DNA with selection on 0.25 μg/mL meropenem. Agarose gel analysis of transformants showed that they carried only a single, large plasmid. PCR and sequencing showed that CMY-6 and NDM-1 were located on an ≈102-kb plasmid (pNDM-Kp10469), originating from K. pneumoniae N10–0469, and on an ≈129-kb plasmid (pNDM-Ec10505), originating from E. coli N10–0505 (Figure 2). Incompatibility (Inc) group PCR (14) and fingerprinting (15) showed that both plasmids were related Inc A/C types, indicating possible horizontal transfer in vivo (Figure 2).
Figure 2.
BglII restriction digests of New Delhi metallo-β-lactamase (pNDM)–Kp10469 (lane 1) and pNDM-Ec10505 (lane 2). MW, 1-kb extension ladder molecular mass marker (Invitrogen, Carlsbad, CA, USA). Sizes (kb) are indicated for some bands.
Transformant susceptibility to aztreonam is consistent with the inability of class B enzymes to efficiently hydrolyze this drug. Transformants exhibited lower carbapenem MICs than did clinical isolates, likely reflecting total β-lactamase content and additional resistance mechanisms such as porin mutations in the clinical isolates (Table 2). The recent emergence of blaNDM-1 in India has been linked to its spread on the Inc A/C-type, IncF1/FII-type, or unknown plasmid types (3). Analysis for class 1 integron cassettes found that both plasmids contained a 0.8-kb cassette that coded for aac(6′)-Ib–type aminoglycoside 6′-N-acetyl transferase. PCR for the ISCR1 element associated with some class 1 integrons was negative. Plasmids harboring NDM-1 plasmids were successfully transferred by conjugation from K. pneumoniae N10–0469 and E. coli N10–0505 to recipient strain E. coli RG192 (rifampin resistant).
Conclusions
North America has thus far escaped the widespread establishment of metallo-β-lactamase–harboring organisms. Therefore, the emergence of blaNDM-1-harboring Enterobacteriaceae in North America is of concern because such isolates exhibit resistance to drugs commonly used to treat gram-negative infections (β-lactams, fluoroquinolones, and aminoglycosides) and have shown a propensity to spread rapidly (4–6)).
Biography
Dr Mulvey is chief of the Antimicrobial Resistance and Nosocomial Infections section of the National Microbiology Laboratory of the Public Health Agency of Canada. His research interests include molecular epidemiology of antimicrobial drug resistant pathogens.
Footnotes
Suggested citation for this article: Mulvey MR, Grant JM, Plewes K, Roscoe D, Boyd DA. New Delhi metallo-β-lactamase in Klebsiella pnemoniae and Escherichia coli, Canada. Emerg Infect Dis [serial on the Internet]. 2011 Jan [date cited]. http://dx.doi.org/10.3201/eid1701.101358
References
- 1.Kitchel B, Rasheed J, Patel J, Srinivasan A, Navon-Venzia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. 10.1128/AAC.00126-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, blaNDM-1, and a novel esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;50:5046–54. 10.1128/AAC.00774-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumarasamy K, Toleman M, Walsh T, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597–602. 10.1016/S1473-3099(10)70143-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumarasamy K, Thirunarayan MA, Krishnan P. Coexistence of blaOXA-23 with blaNDM-1 and armA in clinical isolates of Acinetobacter baumanii from India. J Antimicrob Chemother. 2010;65:2253–4 . 10.1093/jac/dkq273 [DOI] [PubMed] [Google Scholar]
- 5.Health Protection Agency. Multi-resistant hospital bacteria linked to India and Pakistan. Health Protection Report 2009;3:3–4 [cited 2010 Jul 14]. http://www.hpa.org.uk/hpr/archives/2009/hpr2609.pdf
- 6.Centers for Disease Control and Prevention. Detection of Enterobacteriaceae isolates carrying metallo-beta-lactamase—United States, 2010. MMWR Morb Mortal Wkly Rep. 2010;59:750. [PubMed] [Google Scholar]
- 7.Gibb PA, Tribuddharat C, Moore RA, Louie TJ, Krulicki W, Livermore DM, et al. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new blaIMP allele, blaIMP7. Antimicrob Agents Chemother. 2010;202:255–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pitout J, Chow BL, Gregson DB, Laupland KB, Elsayed S, Church DL. Molecular epidemiology of metallo-beta-lactamase–producing Pseudomonas aeruginosa in the Calgary Health Region: emergence of VIM-2-producing isolates. J Clin Microbiol. 2007;45:294–8. 10.1128/JCM.01694-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrer A, Poirel L, Pitout JD, Church D. Occurrence of an SME-2-producing Serratia marcescens isolate in Canada. Int J Antimicrob Agents. 2008;31:175–87. 10.1016/j.ijantimicag.2007.10.007 [DOI] [PubMed] [Google Scholar]
- 10.Goldfarb D, Harvey S-B, Jessamine K, Jessamine P, Toye B, Desjardins M. Detection of plasmid-mediated KPC-producing Klebsiella pneumoniae in Ottawa, Canada: evidence of intrahospital transmission. J Clin Microbiol. 2009;47:1920–2. 10.1128/JCM.00098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diancourt L, Passet V, Verhoef J, Grimont P, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43:4178–82. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–51. 10.1111/j.1365-2958.2006.05172.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coque T, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, et al. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis. 2008;14:195–200. 10.3201/eid1402.070350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caratolli A, Bertini A, Villa L, Falbo V, Hopkins K, Threlfall E. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–28. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- 15.Mulvey MR, Susky E, McCracken M, Morck DW, Read RR. Similar cefoxitin-resistance plasmids circulating in Escherichia coli from human and animal sources. Vet Microbiol. 2009;134:279–87. 10.1016/j.vetmic.2008.08.018 [DOI] [PubMed] [Google Scholar]