Abstract
MicroRNAs (miRNAs) are short non-coding RNAs that regulate gene expression at the post-transcriptional level. While mostly intracellular, a portion of cellular miRNAs is released to the circulation and their level in the plasma is altered in certain pathological conditions such as cancer, and also during pregnancy. We examined the circulating levels of a set of trophoblastic miRNAs, which we recently found to be regulated by hypoxia, in the plasma of pregnant women with fetal growth restriction. As expected, pregnancy was associated with increased plasma levels of several placenta-specific miRNAs, compared to non-pregnant controls. Among pregnant women, the overall levels of miRNA species that we analyzed were increased by 1.84-fold (p ≤0.01) in plasma of women with pregnancies complicated by FGR, but decreased in FGR placentas by 24% (p ≤0.01) compared to values from uncomplicated pregnancies. Together, our results show that plasma concentration of miRNAs is regulated in pregnancy, and that FGR is associated with increased circulating miRNA levels, highlighting the need to explore plasma miRNAs as potential biomarkers for placental diseases.
Keywords: trophoblast, fetal growth restriction, hypoxia, plasma, microRNA
Introduction
Fetal growth restriction (FGR) is biologically characterized as failure of a fetus to reach its growth potential. Affecting 3–10% of births, FGR represents one of the leading causes of perinatal morbidity and mortality [1]. Growth-restricted newborns that survive the intrauterine period are at a greater risk of acute neonatal diseases, childhood neuro-developmental dysfunction, and the adult metabolic syndrome. FGR is a complex disease, resulting from an array of diverse etiologies. Its diagnosis remains difficult, and it has no effective therapy except for induced delivery, often required to avert additional harm in utero even at the cost of prematurity.
MicroRNAs (miRNAs) are small non-coding RNA molecules that regulate gene expression at the post-transcriptional level [2–6]. More than 650 miRNA species are known thus far, and each of them can potentially target more than a hundred mRNAs [7]. While a few miRNAs are tissue-specific, they are commonly expressed in different combinations across diverse organs. Consistent with their vast regulatory potential, miRNAs are involved in many cellular processes, including cell differentiation, proliferation, apoptosis, and metabolic homeostasis, as well as pathological processes such as viral infections [8], cardiovascular diseases [9], and cancer [10–14]. Unique organ- or disease-specific miRNA expression patterns constitute miRNA signatures that may be used as diagnostic or prognostic clinical tools [11,15,16].
Recent studies showed that miRNAs are released into the circulation where they are found in a surprisingly stable form [17–21]. The precise origin of circulating plasma miRNAs is not entirely clear. At least some miRNAs are released to the plasma within exosomes, which are circulating microparticles (50–100 nM in size) of endocytic origin that may support intercellular communication [21–23]. Exosomes from different cell types exhibit potent immunostimulatory and antitumorogenic effects [review in 24]. In contrast, exosomes released by tumor cells exhibit immunosuppressive capabilities and can promote tumor progression [25].
The human placenta also produces exosomes that may play a role in the establishment of maternal immune tolerance [26–28]. Discrete types of miRNA species are found in the plasma of pregnant women [18,19,29], with the plasma level of selected miRNA species altered in a gestational age-dependent manner. These observations imply a role for miRNAs as extracellular messengers. In this study we analyzed the plasma concentration of a set of miRNAs species that we recently identified to be regulated in human placental trophoblasts exposed to hypoxia [30]. We tested the hypothesis that the level of these miRNAs is altered in the plasma of women with pregnancies complicated by FGR.
Materials and methods
Participants
Plasma samples were collected from non-pregnant women as well as from pregnant women with a singleton healthy pregnancy, or pregnancy complicated by FGR. We used archived samples from the Preeclampsia Program Project (PEPP) study of pregnant women who delivered at Magee-Womens Hospital in Pittsburgh within the years 1995–2004. The study was approved by the University of Pittsburgh’s Institutional Review Board. All participants provided written informed consent for use of their samples and de-identified clinical data under the umbrella of the PEPP and related ancillary studies by the same investigators. All women aged 14–44 years were eligible to participate. Clinical diagnoses were established during monthly meetings of a jury of clinicians. FGR was defined as birth weight <10th percentile after exclusion of pregnancies complicated by systemic maternal diseases that might be associated with placental abnormalities such as preeclampsia, diabetes mellitus, fetal genetic anomalies or non-genetic congenital abnormalities. Clinical characteristics of these patients are listed in Table 1.
Table 1.
Maternal and neonatal characteristics
| Control (n=14, mean ± SD) |
FGR (n=14, mean ± SD) |
p value | |
|---|---|---|---|
| Maternal age (yrs) | 25.2 ± 5.8 | 25.4 ± 5.4 | NS |
| Pre-pregnancy weight (kg) | 63.15 ± 10.82 | 61.72 ± 11.5 | NS |
| Gestational age at delivery (wks) | 39.7 ± 1.8 | 39.5 ± 1.1 | NS |
| Neonatal weight (g) | 3239.0 ± 335 | 2539.4 ± 185 | p <0.001 |
| APGAR score < 7 at 5 min | 0 | 0 | NS |
Specimen processing and total RNA isolation
Maternal non-fasting blood samples were collected at the usual times for clinical indications into 10 ml EDTA-containing tubes (BD Vacutainer, Franklin Lakes, NJ). Plasma was collected by centrifugation and aliquoted at −80°C. Total RNA was extracted from plasma samples using miRNeasy mini columns (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. Briefly, 300 µl of plasma were thawed on ice and mixed with 700 µl of QIAzol reagent. After mixing, 140 µl of chloroform were added, and the sample was centrifuged at 12,000 g for 15 min. The upper phase was collected and transferred to a new collection tube containing 1.5 v/v ethanol. The mixture was applied to an RNeasy Mini spin column and centrifuged at 8000 g for 15 sec. After washings the RNA was eluted from the column using RNase-free water.
Quantitative RT-PCR (RT-qPCR) assays
Plasma circulating miRNAs were determined by RT-qPCR. RNA (100 ng) was reverse transcribed using the miScript Reverse Transcription kit (Qiagen) according to the manufacturer instructions. RT product was diluted by a factor of four, and 2 µl of RT product was combined with 10 µl of 2× QuantiTect SYBR Green PCR Master Mix, 2 µl of 10× miScript Universal Primer, 2 µl of 10× miScript Primer Assay in a total volume of 20 µl. PCR as carried out using a 7900HT Fast RT-PCR system (Applied Biosystems Foster City, CA, USA). Data were analyzed with SDS Relative Quantification Software version 2.3 (Applied Biosystems), with automatic setting for adapting baseline and threshold for Ct determination.
Statistical analysis
All RT-qPCR measurements were performed in duplicates. Because of unequal sample sizes for different experimental groups, as well as wide range of miRNA levels, we elected to analyze these unbalanced datasets using linear mixed effect models, with the random effect representing the sample-to-sample variation. We fitted the models with and without the treatment factor as the fixed effect, and performed analysis of variance to compare the null model (without the treatment factor as the fixed effect) and the alternative model (with the treatment factor as the fixed effect). Using a log likelihood ratio test, we calculated the p values against the null hypotheses for each miRNA, assuming that the “exposure” (e.g., FGR) had no effect on its level. The p values were corrected by Benjamini and Hochberg’s method to control for false discovery rate (FDR). All analyses were done using the R statistical computing software (http://www.R-project.org, 2009). The linear mixed effect model analysis was done using the lme4: Linear mixed-effects models lme4 packages of R (http://www.R-project.org/package=lme4, 2009)
Results
We recently defined a set of miRNAs (miR-93, miR-205, miR-224, miR-335, miR-424, miR-451 and miR-491) that are regulated in hypoxic trophoblasts in vitro [30]. We initially sought to assess the basal level of these miRNA species in plasma of a subset of healthy non-pregnant (n=11) and pregnant women (n=10), to which we added four ubiquitous species that are abundant in the placenta (miR-27a-1, miR30d, miR-141, and miR-200c). We found that while the level of the entire group of miRNAs was unchanged between pregnant and non-pregnant individuals, plasma levels of miR-141 and miR-424 was significantly higher during pregnancy. For control, we measured the levels of a subset of miRNA species that represents a miRNA cluster from chromosome 19 (C19MC), known to be almost exclusively expressed by the placenta [31,32]. As expected, the circulating levels of four members of this family (miR-517a, −518b, −518e and −524) was markedly increased in plasma samples of pregnant women (Fig. 1). Together, these data indicate that when compared to non-pregnant women, the level of plasma miRNAs during pregnancy is miRNA species-specific.
Fig. 1. The levels of miRNAs in plasma samples from non-pregnant and pregnant women.
Box plots of the log-transformed relative values by RT-qPCR, normalized to the median of the levels in the control (non-pregnant women). RNA samples were extracted from plasma samples, obtained from non-pregnant women (n=10) or uncomplicated pregnancies (n=11). Base 2 logarithms of expression values are plotted. Each box shows the variance of relative values of miRNAs with the lower boundary indicating the 25th percentile, the line within the box indicating the median, and the upper edge marking the 75th percentile. Upper and lower whisker caps indicate the 95th and 5th percentiles. Outliers are indicated by circles. * denotes miRNA species that are significantly different in the plasma of non-pregnant women and pregnant women (adjusted p value ≤0.05).
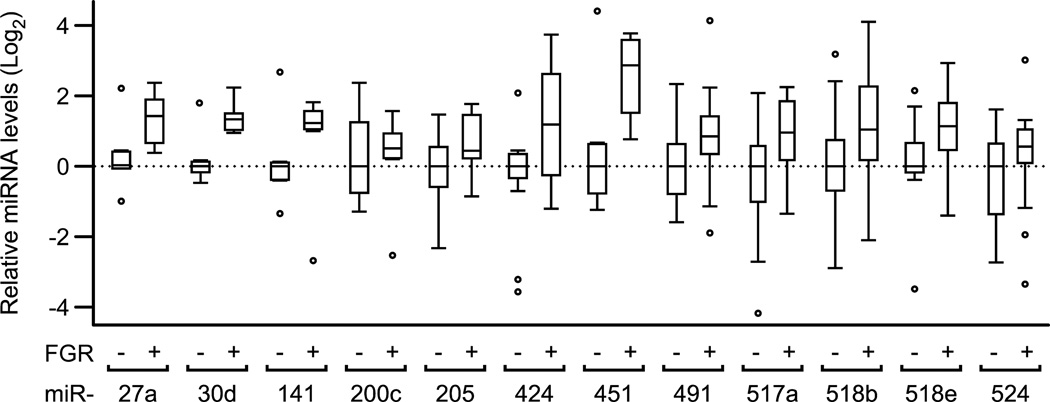
We next compared the plasma levels of a subset of the selected miRNAs from uncomplicated pregnancies and from women with pregnancies complicated with term FGR (n=14 in each group). We found that none of the differences in the level of individual miRNA was significant between the two experimental groups (Fig. 2). In contrast, when considered as a group, the level of all tested miRNA species was elevated by 1.8-fold (p ≤0.01, two sided Wilcoxon test) in plasma samples from women with FGR, compared to healthy pregnant controls. We also assessed the levels of miRNAs in RNA previously extracted from placental biopsies of women with term FGR vs. controls [33]. As shown in Fig. 3, we found no difference in the level of any of the miRNAs between the two experimental groups. Notably, as a group the level of all miRNA species was reduced by 24% (p ≤0.01, two sided Wilcoxon test) in placental samples from women with FGR, compared to controls. To assess a possible correlation between the change in plasma miRNAs and placental miRNAs in pregnancies complicated by FGR vs controls, we plotted miRNA ratios from the two groups. We uncovered a significant negative correlation of −0.7 (p ≤0.01, two sided t test) between plasma and placental miRNAs (Fig. 4), supporting a miRNA-specific relationship between the levels of miRNAs in the plasma and the placenta.
Fig. 2. Relative miRNA levels in plasma from normal vs FGR pregnancies.
Box plots of the base-2 log-transformed relative values by RT-qPCR, analyzed and plotted as described in the legend to Fig. 1. RNA samples were extracted from plasma samples obtained from uncomplicated pregnancies (n=14) or FGR pregnancies (n=14). Note that the levels of all miRNA species, considered as a single group, was higher by 1.8-fold in FGR than in control plasma samples (p ≤0.01).
Fig. 3. Relative miRNA levels in the normal vs FGR placentas.
Box plots of the base-2 log-transformed relative values by RT-qPCR, analyzed and plotted as described in the legend to Fig. 1, and normalized to RNU6B levels. RNA samples were extracted from placental biopsies, obtained from uncomplicated pregnancies (n=4) or FGR pregnancies (n=5). Data are presented as described in Fig. 1. Note that the levels of all miRNA species, considered as a single group, was lower by 24% in FGR than in control plasma samples (p ≤0.01).
Fig. 4. The relative change of miRNA levels from plasma vs placental samples in pregnancies complicated by FGR.
Plot of the log-transformed ratio of miRNA levels in plasma vs placental samples in FGR/control, measured by RT-qPCR. Base 2 logarithms of ratio are plotted. The dotted line is the regression line for the entire set of miRNAs (r = −0.7; p ≤0.01).
Discussion
We recently defined a set of miRNAs that are regulated in primary human trophoblasts exposed to hypoxia [30]. When we assessed the level of these miRNAs in plasma of healthy pregnant women, we found that the level of most of these miRNAs was unchanged during pregnancy, except for miR-424, which is increased. In agreement with previous work, we also found that plasma miR-141, which was not changed by hypoxia, is elevated in the plasma of pregnant women [18,19]. As control, we found that the level of placenta-specific miRNAs from the C19MC cluster was elevated during pregnancy, as expected. These results suggest that the placenta is an important contributor of circulating miRNAs during pregnancy. Although plasma volume is increased during pregnancy, this physiological change is relatively small (1.5-fold) and therefore unlikely to significantly influence miRNA levels. We also doubt that sample handling might have altered our results, as miRNAs are extremely stable even when stored in room temperature up to 24 h, or when undergone up to eight freeze-thaw cycles [20].
The change in miRNA species that were quantified in our study suggests an increase in total miRNA levels in the plasma of women with pregnancies complicated by FGR. This increase of circulating miRNAs in FGR was accompanied by a small diminution of miRNAs in placentas from FGR. The negative correlation between the positive fold-change of plasma miRNAs in FGR vs. the negative fold-change of placenta miRNAs in FGR is intriguing. While this relationship may represent two independent mechanisms, it may reflect related process where placental injury in FGR attenuates miRNA biogenesis and increases exosome-dependent or independent release of miRNA to the plasma. Lastly, our results do not rule out an extra-placental source of the elevated plasma miRNAs.
Elevated plasma levels of placental DNA and RNA are associated with clinical conditions related to placental dysfunction, such as preeclampsia [34–38]. A fraction of these nucleic acids, released by the placenta, originates within exosome microparticles, which are secreted from the late endosomal compartment via membrane blebbing [24]. Placental exosomes are implicated in the suppression of T-cell signaling and the promotion of immune tolerance to the developing fetus [26–28,39,40]. Exosomes from diverse tissues, including the placenta, were recently found to contain miRNAs [22,23,28,29,41]. The encapsulation of miRNAs within membrane-limited exosomes is likely to contribute to the remarkable stability of circulating miRNAs in plasma.
Although we found that the expression of individual miRNAs is altered by hypoxia in trophoblasts [30], the plasma level of these individual miRNA species was not significantly altered in women with FGR. This can be attributed to the following factors: (a) the inherent difference between cellular levels of miRNAs in injured trophoblasts in vitro and release to the circulation in FGR in vivo, (b) a lack of confirmation of trophoblast injury in placentas of women with clinical FGR, which might have led to inclusion of women without placental dysfunction, and therefore increased data variability, (c) the relatively high inter-individual variability in plasma miRNA levels, and (d) our use of a relatively conservative statistical approach, where we adjusted the p-values using Benjamini and Hochberg’s FDR method for multiple comparisons instead of more permissive approaches [42]. Notably, the use of equal amounts of plasma miRNA template in our RT-qPCR reactions was based on total extracted plasma RNA, and not on internal controls, which are not standardized at the present time. In order to normalize our results to a constant miRNA we attempted to assess the levels of two small plasma RNAs species, RNU6B and RNU5A, but detected excessive variability, which prohibited us from performing this normalization procedure. As most miRNA species trended toward a higher level in plasma samples from FGR pregnancies, a larger number of participants might have yielded significant differences.
We note that our results thus far do not support the use of the miRNA species targeted by our study as biomarkers for pregnancies complicated by placental dysfunction. Larger, high-throughput analyses of plasma miRNAs are needed prior to application of these and similar discoveries to clinical care.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jean-Francois Mouillet, Email: mouilletj@mwri.magee.edu.
Tianjiao Chu, Email: tchu@mwri.magee.edu.
Carl A. Hubel, Email: chubel@mwri.magee.edu.
D. Michael Nelson, Email: nelsondm@wudosis.wustl.edu.
W. Anthony Parks, Email: parkswa@upmc.edu.
Yoel Sadovsky, Email: ysadovsky@mwri.magee.edu.
References
- 1.Bernstein IM, Horbar JD, Badger GJ, Ohlsson A, Golan A. Morbidity and mortality among very-low-birth-weight neonates with intrauterine growth restriction. The Vermont Oxford Network. Am J Obstet Gynecol. 2000;182:198–206. doi: 10.1016/s0002-9378(00)70513-8. [DOI] [PubMed] [Google Scholar]
- 2.Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 5.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mello CC, Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 10.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Garzon R, Cimmino A, Fabbri M, Croce CM. MicroRNAs and leukemias: how strong is the connection? Leuk Res. 2006;30:653–655. doi: 10.1016/j.leukres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 12.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2007;(96 Suppl):R40–R44. [PubMed] [Google Scholar]
- 14.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 16.Fabbri M, Garzon R, Andreeff M, Kantarjian HM, Garcia-Manero G, Calin GA. MicroRNAs and noncoding RNAs in hematological malignancies: molecular, clinical and therapeutic implications. Leukemia. 2008;22:1095–1105. doi: 10.1038/leu.2008.30. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 18.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–490. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 19.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 23.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 25.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 26.Frangsmyr L, Baranov V, Nagaeva O, Stendahl U, Kjellberg L, Mincheva-Nilsson L. Cytoplasmic microvesicular form of Fas ligand in human early placenta: switching the tissue immune privilege hypothesis from cellular to vesicular level. Mol Hum Reprod. 2005;11:35–41. doi: 10.1093/molehr/gah129. [DOI] [PubMed] [Google Scholar]
- 27.Taylor DD, Akyol S, Gercel-Taylor C. Pregnancy-associated exosomes and their modulation of T cell signaling. J Immunol. 2006;176:1534–1542. doi: 10.4049/jimmunol.176.3.1534. [DOI] [PubMed] [Google Scholar]
- 28.Mincheva-Nilsson L, Baranov V. The Role of Placental Exosomes in Reproduction. Am J Reprod Immunol. 2010;63:520–533. doi: 10.1111/j.1600-0897.2010.00822.x. [DOI] [PubMed] [Google Scholar]
- 29.Luo SS, Ishibashi O, Ishikawa G, Ishikawa T, Katayama A, Mishima T, et al. Human villous trophoblasts express and secrete placenta-specific microRNAs into maternal circulation via exosomes. Biol Reprod. 2009;81:717–729. doi: 10.1095/biolreprod.108.075481. [DOI] [PubMed] [Google Scholar]
- 30.Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Chen S, Zhang W, Liu X, Shi H, Che H, et al. Human differentiation-related gene NDRG1 is a Myc downstream-regulated gene that is repressed by Myc on the core promoter region. Gene. 2008;417:5–12. doi: 10.1016/j.gene.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Biron-Shental T, Schaiff WT, Rimon E, Shim TL, Nelson DM, Sadovsky Y. Hypoxia enhances the expression of follistatin-like 3 in term human trophoblasts. Placenta. 2008;29:51–57. doi: 10.1016/j.placenta.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong XY, Laivuori H, Livingston JC, Ylikorkala O, Sibai BM, Holzgreve W, et al. Elevation of both maternal and fetal extracellular circulating deoxyribonucleic acid concentrations in the plasma of pregnant women with preeclampsia. Am J Obstet Gynecol. 2001;184:414–419. doi: 10.1067/mob.2001.109594. [DOI] [PubMed] [Google Scholar]
- 35.Levine RJ, Qian C, Leshane ES, Yu KF, England LJ, Schisterman EF, et al. Two-stage elevation of cell-free fetal DNA in maternal sera before onset of preeclampsia. Am J Obstet Gynecol. 2004;190:707–713. doi: 10.1016/j.ajog.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 36.Swinkels DW, de Kok JB, Hendriks JC, Wiegerinck E, Zusterzeel PL, Steegers EA. Hemolysis, elevated liver enzymes, and low platelet count (HELLP) syndrome as a complication of preeclampsia in pregnant women increases the amount of cell-free fetal and maternal DNA in maternal plasma and serum. Clin Chem. 2002;48:650–653. [PubMed] [Google Scholar]
- 37.Alberry MS, Maddocks DG, Hadi MA, Metawi H, Hunt LP, Abdel-Fattah SA, et al. Quantification of cell free fetal DNA in maternal plasma in normal pregnancies and in pregnancies with placental dysfunction. Am J Obstet Gynecol. 2009;200(98):e1–e6. doi: 10.1016/j.ajog.2008.07.063. [DOI] [PubMed] [Google Scholar]
- 38.Pang WW, Tsui MH, Sahota D, Leung TY, Lau TK, Lo YM, et al. A strategy for identifying circulating placental RNA markers for fetal growth assessment. Prenat Diagn. 2009;29:495–504. doi: 10.1002/pd.2230. [DOI] [PubMed] [Google Scholar]
- 39.Sabapatha A, Gercel-Taylor C, Taylor DD. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am J Reprod Immunol. 2006;56:345–355. doi: 10.1111/j.1600-0897.2006.00435.x. [DOI] [PubMed] [Google Scholar]
- 40.Hedlund M, Stenqvist AC, Nagaeva O, Kjellberg L, Wulff M, Baranov V, et al. Human placenta expresses and secretes NKG2D ligands via exosomes that down-modulate the cognate receptor expression: evidence for immunosuppressive function. J Immunol. 2009;183:340–351. doi: 10.4049/jimmunol.0803477. [DOI] [PubMed] [Google Scholar]
- 41.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]