Abstract
Recent advances have provided substantial insight into the maintenance of mucosal immunity and the pathogenesis of inflammatory bowel disease. Cellular programs responsible for intestinal homeostasis use diverse intracellular and intercellular networks to promote immune tolerance, inflammation or epithelial restitution. Complex interfaces integrate local host and microbial signals to activate appropriate effector programs selectively and even drive plasticity between these programs. In addition, genetic studies and mouse models have emphasized the role of genetic predispositions and how they affect interactions with microbial and environmental factors, leading to pro-colitogenic perturbations of the host–commensal relationship.
Inflammatory bowel disease (IBD) comprises the chronic relapsing inflammatory disorders Crohn’s disease and ulcerative colitis. Family history is a risk factor for developing IBD, with a peak incidence in early adult life, although individuals of any age can be affected. IBD is thought to result from an inappropriate and continuing inflammatory response to commensal microbes in a genetically susceptible host. Recent progress in understanding IBD pathobiology offers insight into relevant disease mechanisms in mucosal immunity, including how genetic factors interact with microbial and environmental cues within tissue-specific contexts, the biological checkpoints involved, the selective decisions made during the course of disease and how plasticity of the biological response results in the capacity for different phenotypes.
Ulcerative colitis is characterized by inflammation that is limited to the colon: it begins in the rectum, spreads proximally in a continuous fashion and frequently involves the periappendiceal region. By contrast, Crohn’s disease involves any part of the gastrointestinal tract — most commonly the terminal ileum or the perianal region — in a non-continuous fashion and, unlike ulcerative colitis, is commonly associated with complications such as strictures, abscesses and fistulas. Histologically, ulcerative colitis shows superficial inflammatory changes limited to the mucosa and submucosa with cryptitis and crypt abscesses. The microscopic features of Crohn’s disease include thickened submucosa, transmural inflammation, fissuring ulceration and non-caseating granulomas.
Among complex diseases, genome-wide association studies (GWAS) have been successful in IBD, identifying 99 non-overlapping genetic risk loci, including 28 that are shared between Crohn’s disease and ulcerative colitis1,2 (Fig. 1). The genes implicated in childhood-onset and adult-onset IBD overlap, suggesting similar contributory genetic predispositions and pathophysiological pathways. Adding to the complexity of understanding disease mechanisms, a susceptibility allele often requires other genetic and non-genetic cues to manifest disease. The concordance rate in monozygotic twins of 10–15% in ulcerative colitis compared with 30–35% in Crohn’s disease suggests that non-genetic factors may have an even more important role in ulcerative colitis than in Crohn’s disease3. Furthermore, the higher penetrance of common Crohn’s-disease-associated polymorphisms in genetic case-control studies than in population-based studies of cohorts of the same ethnicity is probably due to the concomitant aggregation of both genetic and environmental factors in the case-control studies4. Smoking is an example of a disease-specific modifier that seems to exacerbate Crohn’s disease while being protective against ulcerative colitis. Evidence suggests that smoking impairs autophagy, a process thought to be involved especially in Crohn’s disease, demonstrating how exposure to a disease modifier in a genetically predisposed individual may mechanistically affect IBD development5.
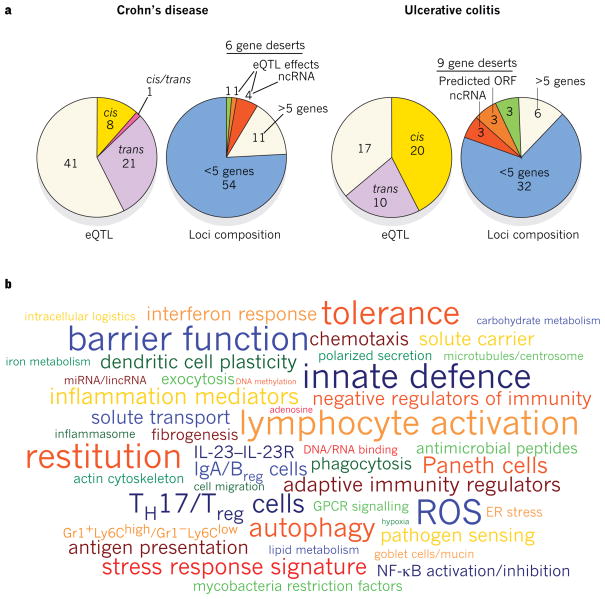
Figure 1. Genetic architecture of IBD-linked susceptibility loci.
a, GWAS have identified 71 risk loci in Crohn’s disease and 47 risk loci in ulcerative colitis (P value of association < 5×10−8). Of these, 28 risk loci exhibit shared associations (defined as P < 5×10−8 for either Crohn’s disease or ulcerative colitis, and P < 1×10−4 for the other form of IBD). Approximately half of the loci implicated in Crohn’s disease and ulcerative colitis are associated with cis- and/or trans-expression quantitative trait loci (eQTL) effects (left panels). Genes whose expression are affected by these variants could also be involved in IBD pathogenesis. The loci composition (right panels) shows the number of genes that either lie within or segregate in linkage disequilibrium with IBD-implicated loci (coefficient of correlation r2 > 0.8). These loci are structurally heterogeneous, and are associated with widely ranging numbers of genes. Loci not associated with any genes, known as gene deserts, frequently contain non-coding transcripts or predicted open reading frames (ORFs), and can be associated with trans-eQTL effects. b, Recurring terms illustrating biological processes implicated by at least three genes represented in IBD loci; font sizes are proportional to the number of genes associated with each respective process. Breg cells, B regulatory cells; ER, endoplasmic reticulum; GPCR, G-protein-coupled receptor; IL, interleukin; lincRNA, large intervening non-coding RNA; miRNA, microRNA; ncRNA, non-coding RNA; NF-κB, nuclear factor-κB; ROS, reactive oxygen species; TH17 cells, T helper 17 cells; Treg cells, T regulatory cells.
In this Review, we provide an overview of genes and susceptibility loci implicated in IBD by GWAS and other genetic studies. Candidate genes are discussed in the context of IBD-relevant pathways, as well as how these molecular pathways interact with environmental factors to modulate intestinal homeostasis.
Genes and pathways in IBD
International collaborative research groups focusing on an unbiased appraisal of the human genome have been particularly successful in identifying genes and genetic loci that contribute to IBD susceptibility1,2. Despite distinct clinical features, approximately 30% of IBD-related genetic loci are shared between ulcerative colitis and Crohn’s disease, indicating that these diseases engage common pathways and may be part of a mechanistic continuum (Fig. 1).
Analyses of the genes and genetic loci implicated in IBD show several pathways that are crucial for intestinal homeostasis, including barrier function, epithelial restitution, microbial defence, innate immune regulation, reactive oxygen species (ROS) generation, autophagy, regulation of adaptive immunity, endoplasmic reticulum (ER) stress and metabolic pathways associated with cellular homeostasis (Fig. 2). Early studies have suggested the existence of both protective and predisposing alleles6. Disease-relevant biological pathways are further highlighted when several components are implicated as risk factors together (Fig. 3).
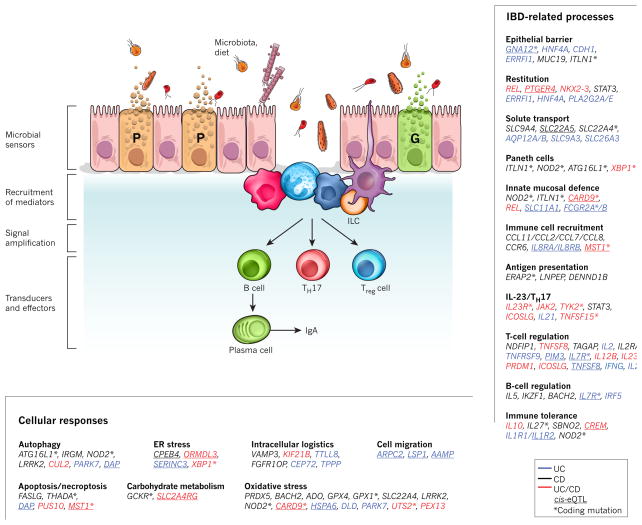
Figure 2. A model for IBD pathways based on GWAS.
Intestinal homeostasis involves the coordinated actions of epithelial, innate and adaptive immune cells. Barrier permeability permits microbial incursion, which is detected by the innate immune system, which then orchestrates appropriate tolerogenic, inflammatory and restitutive responses in part by releasing extracellular mediators that recruit other cellular components, including adaptive immune cells. Genetic variants, the microbiota and immune factors affect the balance of these signals. Genes in linkage disequilibrium (r2 > 0.8) with IBD-associated single nucleotide polymorphisms (SNPs) were manually curated and classified according to their function(s) in the context of intestinal homeostasis and immunity. Text colour indicates whether the genes are linked to risk loci associated with Crohn’s disease (CD; black), ulcerative colitis (UC; blue) or both (red). Asterisk denotes corresponding coding mutations; cis-eQTL effects are underlined. G, goblet cell; P, Paneth cell.
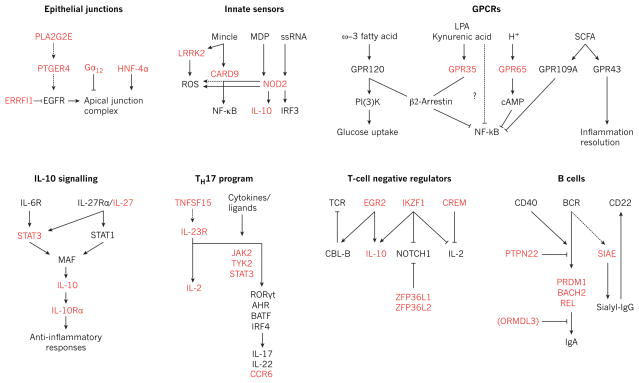
Figure 3. Genetic variants in IBD signalling modules.
Schematic of selected signalling pathways involved in the maintenance of intestinal homeostasis, including epithelial junctional complex assembly, innate immune recognition of pathogen-associated motifs, GPCRs and immune defence, anti-inflammatory interleukin-10 (IL-10) signalling, TH17-cell differentiation, inhibitory pathways in lymphocyte signalling, and B-cell activation and IgA antibody responses. Proteins encoded by genes identified as being in linkage disequilibrium with IBD-risk SNPs (r2 > 0.8) are highlighted in red. BCR, B-cell receptor; cAMP, cyclic AMP; EGFR, epidermal growth factor receptor; ERRFI1, ERBB receptor feedback inhibitor 1; Gα12, G protein subunit α12; GPCR, G-protein-coupled receptor; HNF-4α, hepatocyte nuclear factor-4α; LPA, lysophosphatidic acid; MDP, muramyl dipeptide; NF-κB, nuclear factor-κB; PI(3)K, phosphatidylinositol-3-OH kinase; PLA2G2E, phospholipase A2, group IIE; PTGER4, prostaglandin E receptor 4; SCFA, short-chain fatty acid; SIAE, sialic acid acetylesterase; ssRNA, single-stranded RNA; TCR, T-cell receptor.
Multidisease comparative analysis can uncover common disease-causing genes and pathways. More than 50% of IBD susceptibility loci have also been associated with other inflammatory and autoimmune diseases. These overlapping genes can have contrasting effects in different diseases. For example, the same coding variant of PTPN22 (R620W) is a strong risk factor for type 1 diabetes and rheumatoid arthritis, but is protective against Crohn’s disease7. These data suggest that crucial clues to disease biology may reside in understanding the function of these shared genes. Several loci containing genes such as MST1, IL2, CARD9 and REL are shared between ulcerative colitis and the associated complication primary sclerosing cholangitis (PSC)8. This overlap may help to identify subsets of patients with ulcerative colitis who are at risk of PSC. Risk loci for Crohn’s disease present an unexpected overlap with susceptibility regions for Mycobacterium leprae infection, including genes such as NOD2, C13orf31 and LRRK2 (ref. 9). Although absent from the leprosy GWAS, other Crohn’s-disease-associated genes are also implicated in host responses to mycobacterial infection, including CARD9, LTA, ITLN1 and IRGM1. Thus, studies to delineate immune responses to antigens from, and infection by, mycobacteria, or other microbes that elicit similar host cell responses, may also be pertinent to Crohn’s disease.
Genetic variants associated with IBD can vary in frequency depending on the cohort ethnicity, raising the possibility that some such variants may have emerged in the context of historical selective pressures. Although this notion remains to be demonstrated in IBD, lessons from other autoimmune and infectious contexts lend support. For example, variants of apolipoprotein L1 and the inhibitory Fc receptor FcγRIIb that confer protection against trypanosomiasis and malaria, respectively, are more common in populations endemically exposed to these pathogens, but these variants also confer increased susceptibility to focal segmental glomerulosclerosis and systemic lupus erythematosus (SLE), respectively10,11.
Current GWAS are typically powered to characterize variants of >1% frequency and do not include the contributions from rare variants (<1% frequency). Exome sequencing can be useful for identifying rare variants, whereas whole-genome sequencing is of value in elucidating modifier loci. If pedigrees are available, rare variant discovery can be further targeted by fine mapping, as shown by the identification of IL10RA polymorphisms associated with the development of early-onset IBD12. Other interleukin-10 receptor (IL-10R) signalling components have also been implicated by GWAS, including STAT3, TYK2, JAK2 and IL10 itself, in concordance with the notion that both rare and common variants may highlight the same pathway. Although these components can also function in other contexts — for example, the transcription factor STAT3 and the kinase proteins TYK2 and JAK2 are involved in the signalling of the interleukins IL-6, IL-22 and IL-23 — these results illustrate the value of genetic studies in determining not just single genes, but also disease-relevant pathways. Recent resequencing studies in IBD recovered both known and new variants of CARD9, NOD2 and IL23R, with independent effects on disease risk. The IL23R variants were protective, supporting previous findings of a common protective IL23R allele and illustrating how studies of rare variants can reinforce GWAS findings6. Furthermore, T helper 17 (TH17) cells generated ex vivo from subjects with a variant IL23R allele (R381Q) show decreased production of the pro-inflammatory cytokine IL-17A in response to IL-23 stimulation, emphasizing the importance of IL-23-related pathways in human IBD13.
Early functional studies attempting to determine causality have largely focused on coding variants, although non-coding single nucleotide polymorphisms (SNPs) can be associated with qualitative and quantitative changes. Alternative splicing exemplifies a qualitative change affected by non-coding modifications. In the context of regulating immune responses, IL23R and NOD2 can encode truncated variants that inhibit their signalling pathways14,15. Furthermore, genetic changes may affect transcription-factor-binding sequences, locus accessibility, translational efficiency and trans-regulators such as non-coding RNAs and microRNAs (miRNAs). In this regard, a Crohn’s-disease-associated synonymous variant in IRGM (c.313C>T) perturbs regulation by miR-196A and miR-196B, and is associated with altered IRGM expression in patients with Crohn’s disease who bear this SNP16. Cis- or trans-expression quantitative trait loci (eQTL) are detected for approximately half of the IBD risk regions, indicating that allele-specific gene-expression changes contribute to disease risk (Fig. 1). Furthermore, IBD-implicated loci contain more than 10 miRNA-encoding sequences and 39 large intervening non-coding RNAs (lincRNAs), 5 of which interacted with the histone methyltransferase polycomb repressive complex 2 (PRC2), supporting the notion that regulation of gene expression by miRNAs and lincRNAs may be mechanistically relevant in IBD17.
So far, GWAS account for 23% and 16% of the heritability in Crohn’s disease and ulcerative colitis, respectively1,2. Although these may be underestimates owing to the net effect of common variants that are individually too small to calculate accurately; the missing heritability may further comprise genetic, epigenetic and non-genetic (including environmental) components. Genetic factors such as rare variants, private mutations, structural variants and interactions between genes are not well captured by GWAS. Nevertheless, a key success of GWAS in IBD has been the ability to provide insight into disease pathobiology by highlighting key molecular pathways.
Epithelial encounters and pathogenicity
The intestinal mucosa exists in a functional equilibrium with the complex luminal milieu, which is dominated by a spectrum of microbial species and their products. Maintaining this functional balance is central to preserving normal mucosal physiology, with perturbations contributing to the pathophysiology of many gastrointestinal disorders, including IBD. In addition to nutrient absorption, intestinal epithelial cells (IECs) perform both barrier and signal-transduction functions, with the capacity to sense luminal contents through surface receptors and, in return, secrete regulatory products that can orchestrate an appropriate response in the underlying lamina propria.
Molecular details of the epithelial barrier and the structure of tight junctions, which are crucial to its integrity, have been characterized. Abnormal intestinal permeability has been observed in IBD patients and in some of their first-degree relatives. Genes within several IBD-associated loci indicate a role for barrier integrity in disease predisposition, implicating candidate genes such as CDH1, GNA12 and PTPN2. Genetic studies have shown that truncated forms of the adherens junction protein E-cadherin (encoded by CDH1) are associated with Crohn’s disease, and intestinal biopsies from patients with Crohn’s disease carrying these mutant alleles show inappropriate protein localization and cytosolic accumulation18. Activation of the G protein Gα12 (encoded by GNA12) leads to phosphorylation of the tight junction proteins ZO-1 and ZO-2, resulting in destabilization of cell junctions in epithelial cell lines19. In vitro studies show that the protein tyrosine phosphatase family member PTPN2 protects against interferon-γ (IFN-γ)-induced epithelial permeability; concordantly, Ptpn2-deficient mice show increased susceptibility to experimental colitis20,21.
Genetic studies have associated IBD with several transcription factors involved in epithelial regeneration, such as HNF4A and NKX2-3, which control crypt cell proliferation and IEC differentiation, respectively22–24. Spontaneous colitis did not occur in all animal models with IEC-specific deletion of Hnf4a, suggesting that further environmental triggers are required for disease22,23. STAT3, the gene encoding which lies within an IBD-implicated locus, is activated in epithelial cells from patients with IBD, and IEC-specific Stat3 deletion affects epithelial repair25.
The intestinal barrier is enhanced by the presence of a pre-epithelial layer formed primarily of mucus glycoproteins, trefoil peptides, IgA and antimicrobial peptides (AMPs). Goblet cells generate the mucus layer, a protective polysaccharide bilayer rich in cationic proteins, the inner layer of which is essentially devoid of microbes. Patients with IBD frequently have a compromised mucus layer and increased mucolytic bacteria; mucus layer defects are also observed in Muc2−/− and IEC-specific C1galt1−/− mice, which develop spontaneous colitis26. Interestingly, some patients with ulcerative colitis show defective intestinal O-glycosylation resembling that seen in C1galt1−/− mice26. Paneth cells are located in the crypts of the small intestine. In addition to the role of these cells in crypt homeostasis and maintenance of the intestinal stem-cell niche, they also secrete antimicrobial effectors that prevent microbial invasion and control the composition of the gut microflora. These effectors include lysozyme, RegIIIγ, secreted phospholipase A2 (which degrades bacterial membrane phospholipids) and defensins HD5 and HD6 (pore-forming hydrophobic peptides that can integrate into bacterial membranes, resulting in lysis). Production of AMPs is regulated by Toll-like receptor (TLR) and NOD2 signals triggered by commensal flora. Paneth cell defects and susceptibility to intestinal inflammation have been uncovered in mice deficient in several Crohn’s-disease-associated genes, including Nod2, Atg16l1 and Xbp1 (refs 27–29). These results highlight pathways important to Paneth cell biology, such as the regulation of AMP production (Nod2), granule exocytosis (Atg16l1) and the ER stress response (Xbp1). Similar phenotypes have been observed in human disease, such that patients with Crohn’s disease carrying the ATG16L1 (T300A) mutation show Paneth cell granule abnormalities. These findings suggest that defects in Paneth cell biology may define a subset of patients with Crohn’s disease.
Cells with high synthetic capacity and secretory activity, such as Paneth cells and goblet cells, have high baseline levels of ER stress, leading to activation of the unfolded protein response (UPR), which controls cellular programs that allow proper protein processing. The UPR is mainly cytoprotective, although it can signal apoptosis after sustained ER stress. Increased intestine epithelial ER stress and susceptibility to colitis have been observed in mice with overactivation of, or perturbations in, the UPR pathway, including Muc2 missense mutation, Agr2−/−, Ern2−/− (also known as Ire1b−/−), IEC-specific Xbp1−/− and Mbtps1-hypomorphic mice29,30. Similarly, studies in primary IECs from patients with IBD show activated ER stress responses, and hypomorphic variants of XBP1 have been associated with risk of IBD29. Overall, these results indicate that genetic variants that perturb mechanisms that protect against ER stress can affect intestinal homeostasis in IBD. In addition to its effects on cell viability, ER stress also activates autophagy and IL-23 release, suggesting that sustained ER stress may engage inflammatory circuits that are subsequently propagated by T cells31.
In addition to limiting bacterial translocation across the mucosal barrier, IECs promote intestinal homeostasis by regulating innate and adaptive immune responses. Illustrating this point, IECs produce intestinal alkaline phosphatase, which can mediate lipopolysaccharide detoxification. Resolvin-E1, which is generated in part through the action of epithelial cyclooxygenase-2, attenuates neutrophil transmigration and upregulates epithelial expression of intestinal alkaline phosphatase during the restitutive response, a process termed epithelial imprinting32. IECs can also modulate adaptive immune responses, driving the differentiation of anti-inflammatory T regulatory (Treg) cells by releasing the vitamin A metabolite retinoic acid and the cytokines thymic stromal lymphopoietin (TSLP) and transforming growth factor-β (TGF-β)33. Breakdown in such epithelial defence mechanisms could lead to pathological intestinal inflammation.
Checkpoints in the innate immune response
The physical barrier of the intestinal epithelium is complemented by a well-evolved mucosal innate immune system, which is populated by cells poised to defend against pathogenic incursions and curtail inflammatory responses to maintain a state of hyporesponsiveness to commensal bacteria. Dendritic cells, macrophages, innate lymphoid cells (ILCs) and neutrophils are crucial cellular components of the innate immune system during infection or inflammation. Supporting the notion that defective innate immune responses can lead to IBD, patients with innate immunodeficiencies such as chronic granulomatous disease and Hermansky–Pudlak syndrome, which is associated with defective responses to bacterial DNA motifs (CpG oligonucleotides) specifically in plasmacytoid dendritic cells, tend to develop IBD34. Similarly, patients with Crohn’s disease have defective innate immune responses, including attenuated macrophage activity in vitro, and impaired neutrophil recruitment and exogenous Escherichia coli clearance in vivo35.
Intestinal dendritic cells constitute a central interface for monitoring the environment and relaying signals to initiate appropriate adaptive immune responses33. Dendritic cell subsets are specialized and respond to endogenous and exogenous stimuli such as microbial motifs, fatty acids, oxidized lipids and vitamin D by selectively engaging pro-inflammatory, anti-inflammatory, epithelial restitutive or T-cell education programs, as well as inducing IgA production33,36. For example, Treg-cell differentiation can be promoted by tolerogenic dendritic cells induced by TSLP, TGF-β and retinoic acid, all of which are made by IECs and stromal cells; these dendritic cells express the integrin CD103 but not the chemokine receptor CX3CR1 (ref. 33). By contrast, dendritic cells expressing E-cadherin are a pro-inflammatory and page 298 subset that promotes TH17-cell differentiation (see ref. 37 for further details). Bacterial flagellins can override dendritic cell tolerogenic programs by stimulating TLR5 and inducing the release of pro-inflammatory mediators from hyporesponsive lamina propria CD11chigh dendritic cells, pointing to a broader role for flagellated bacteria in IBD38. This specific immunostimulatory role for TLR5 may be particularly relevant in IBD, as seroreactivity to the bacterial flagellin CBir1, observed in approximately 50% of patients with Crohn’s disease, correlates with a complicated clinical course.
Intestinal homeostasis is maintained in part by the actions of resident macrophages that have enhanced phagocytic and bactericidal activity and decreased production of pro-inflammatory cytokines. Specialized macrophage subsets are also involved; tumour-necrosis factor-α (TNF-α)-secreting and IL-1β-secreting Ly6Chigh monocytes are recruited in the initial phase of microbial challenge or tissue injury, whereas reparative IL-10-secreting, TGF-β-secreting and vascular-endothelial-growth-factor-secreting Ly6Clow monocytes are mobilized during the resolution phase of inflammation39. Neutrophils may also contribute to the resolution of inflammation, for example, by synthesizing anti-inflammatory mediators such as lipoxin A4. Studies showing impaired secretion of lipoxin A4 in mucosal tissues from patients with ulcerative colitis support the relevance of such mechanisms in IBD.
IL-22 is emerging as an important cytokine in epithelial homeostasis, showing protective activity in different models of colitis through its stimulatory effect on antimicrobial and reparative processes. Produced by several cell types, such as ILCs, lymphoid tissue induced (LTi) cells, TH17 cells and γδ T cells, most intestinal IL-22 at steady state is produced by ILCs expressing the transcription factor RORγt40,41. Studies in patients with Crohn’s disease have shown decreased frequencies of IL-22-secreting ILCs in the lamina propria42. Together, these findings suggest a central role for ILCs (and other IL-22-producing cells) in regulating intestinal homeostasis, which remains to be characterized in IBD.
NOD2 and IBD
NOD2 was the first gene to be associated with IBD, and thereafter several genes that interact epistatically with NOD2 signalling were also implicated. NOD2 recognizes the peptidoglycan product muramyl dipeptide (MDP), which modulates both innate and adaptive immune responses43 (Fig. 4). For example, MDP stimulation induces autophagy, which controls bacterial replication and antigen presentation, and acts on dendritic cells in conjunction with TLR ligands to promote TH17-cell differentiation44,45. NOD2 may also contribute to immune tolerance. These effects are impaired in cells from patients with the Crohn’s-disease-associated NOD2 mutation 3020insC. Furthermore, NOD2 can participate in distinct MDP-independent pathways such as regulation of the T-cell response and the type I IFN response to single-stranded RNA (ssRNA) stimulation, indicating that gut microbial ssRNAs may exist and have immunomodulatory properties46. The relative contributions of these cytosolic MDP-sensing pathways vary greatly between cell types (Fig. 4). Further studies are needed to uncover the effect of disease-associated NOD2 alleles in different cell-specific programs, and unravel the precise role(s) of NOD2 in IBD. Other families of innate immune receptors linked to intestinal inflammation and immunity include NOD-like receptors (NLRs) and RIG-I-like receptors (RLRs). These receptors recognize microbial motifs or damage-associated molecular patterns and can activate the inflammasome, thus appropriate regulation of these pathways is required for intestinal homeostasis. For example, mouse knockout studies of Nlrp3 or RIG-I (also known as Ddx58) show increased susceptibility to experimental colitis47. Conversely, sustained overactivation of NLRs can also have detrimental effects, as illustrated by activating mutations in NOD2 and NLRP3 giving rise to Blau syndrome and cryopyrinopathies, respectively.
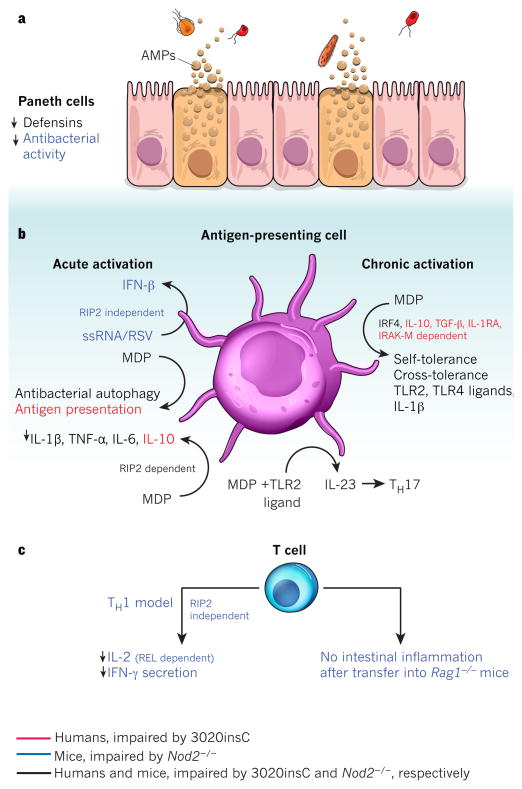
Figure 4. Cell-intrinsic functions of NOD2.
NOD2 is activated by the bacterial peptidoglycan muramyl dipeptide (MDP). Cell-specific NOD2 functions are shown, distinguishing between those functions impaired in cells from humans with the Crohn’s-disease-associated mutation 3020insC (red), from Nod2-deficient mice (blue), or from both (black). a, In Paneth cells, Nod2 deficiency leads to attenuated antibacterial activity in the intestinal crypts and decreased expression of α-defensin 4 (encoded by Defcr4, also known as Defa4) and α-defensin-related sequence 10 (DEFCR-RS10, also known as DEFA-RS10). b, MDP-stimulated release of pro-inflammatory NF-κB-dependent cytokines (such as IL-1β, TNF-α and IL-6), as well as secretion of IL-23 (which promotes TH17 differentiation) after co-stimulation with MDP and TLR2 ligands, is decreased in antigen-presenting cells from Nod2-deficient mice or 3020insC human donors. MDP stimulation also leads to NOD2-activated autophagy and antigen presentation. In mice, the activation of antigen-presenting cells by ssRNA or respiratory syncytial virus (RSV) stimulates secretion of type I interferon (IFN-β) in a NOD2-dependent, receptor-interacting protein-2 (RIP2)-independent fashion. In contrast to the pro-inflammatory effects, chronic NOD2 activation (right) by MDP induces both self-tolerance and cross-tolerance to IL-1β, and TLR2 and TLR4 ligands. This is dependent on IRF4 in mice and humans, and also on IL-10, TGF-β, IL-1RA and IL-1R-associated kinase M (IRAK-M) in humans. MDP-induced tolerance is lost in Nod2-deficient mice and in patients with the 3020insC variant. NOD2-dependent release of IL-10 after MDP stimulation has been demonstrated to be specific to humans and is impaired in 3020insC cells. MDP-stimulated release of several cytokines, including IL-10, IL-1β, TNF-α and IL-6, is dependent on RIP2. c, In mice, NOD2 mediates IFN-γ secretion and REL-dependent IL-2 production in T cells in response to Toxoplasma gondii infection. Also, Nod2 deficiency attenuates the ability of T cells to cause experimental colitis after transfer into Rag1-deficient hosts.
CARD9 and IBD
CARD9 is an IBD-implicated adaptor protein that integrates signals from many innate immune receptors that recognize viral, bacterial and fungal motifs. Depending on the stimulus, CARD9 interacts with distinct signalling complexes and activates different pathways to modulate cytokine environments appropriately48,49. In particular, recognition of fungal motifs in human dendritic cells leading to CARD9 and dectin-1 signalling results in the broad activation of members of the nuclear factor-κB (NF-κB) transcription factor family, whereas CARD9 and dectin-2 signalling selectively activates the IBD-implicated NF-κB factor REL, enhancing the production of TH17-polarizing cytokines such as IL-1β and the IL-23 p19 subunit50. Defective CARD9 function leads to the immune disorder mucocutaneous candidiasis, at least in part owing to failure to promote an adequate TH17 immune response. These data illustrate how innate immune signalling molecules, including NOD2 and CARD9, can act as central hubs to integrate diverse signals and selectively activate specific effector pathways; in the polymicrobial context of the gut, it seems reasonable that defects at such nodal points would constitute key predispositions to IBD.
Redox equilibrium in IBD
The reduction and oxidation (redox) state of the gut depends on an equilibrium between oxidants, such as free radicals, ROS or reactive nitrogen species, and antioxidant mechanisms, such as the glutathione peroxidase (GPX) and glutathione S-transferase enzymes. This redox state affects many signal-transduction pathways, such as NF-κB signalling and AMP activity51. Supporting the importance of antioxidant pathways in intestinal homeostasis, mice deficient in both Gpx1 and Gpx2 develop spontaneous colitis. IBD genetic studies have implicated loci containing GPX1 and GPX4, further highlighting the relevance of these mechanisms in disease (Fig. 2). Among the oxidants, ROS represent an important class of effector molecules generated by mitochondrial and non-mitochondrial sources. ROS are non-toxic at basal levels and are even required to maintain the intestinal stem-cell niche. In the context of innate immunity, ROS have important antimicrobial activity, and contribute to intracellular signalling, promoting the production of pro-inflammatory cytokines. Furthermore, ROS generated by epithelial cells after infection can transmit signals to adjacent cells in a paracrine manner, allowing the local coordination of chemokine production52. Genes within several IBD-associated loci may either regulate ROS production or protect against oxidative stress (Fig. 2). In particular, NOD2, CARD9 and IFN-γ-regulated leucine-rich repeat kinase 2 (LRRK2) all contribute to ROS production43,53,54. In addition to pro-inflammatory pathways, ROS are also involved in Treg-cell polarization and function55,56. Thus, understanding the role of disease variants will require a broader understanding of the cell- and tissue-specific effects of ROS.
Autophagy and IBD
Genetic analyses have shown an unsuspected role for autophagy in innate immunity and IBD, implicating two component genes, ATG16L1 and IRGM, in IBD pathogenesis57–59. Autophagy is involved in intracellular homeostasis, contributing to the degradation and recycling of cytosolic contents and organelles, as well as to resistance against infection and the removal of intracellular microbes (Fig. 5). ATG16L1 is essential for all forms of autophagy, and the coding mutation T300A is associated with increased risk of Crohn’s disease. Despite ubiquitous expression of ATG16L1, defects associated with ATG16L1 polymorphisms have so far been described only within the gut, probably owing to the high microbial load in this tissue. Subsequent evidence for MDP stimulation of NOD2-activated autophagy illustrates a link between genetic risk loci, and highlights the importance of defining disease-associated pathways and the potential of new roles for known genes44,45. Epithelial cells and dendritic cells containing Crohn’s-disease-associated ATG16L1 and NOD2 variants show defects in antibacterial autophagy44,45,60. In dendritic cells, these defects are associated with an impaired ability to present exogenous antigens to CD4+ T cells44. These results illustrate a close relationship between NOD2, ATG16L1 and autophagy, affecting intracellular processing and communication with the adaptive immune system, suggesting that genetic polymorphisms may affect both pathways concomitantly.
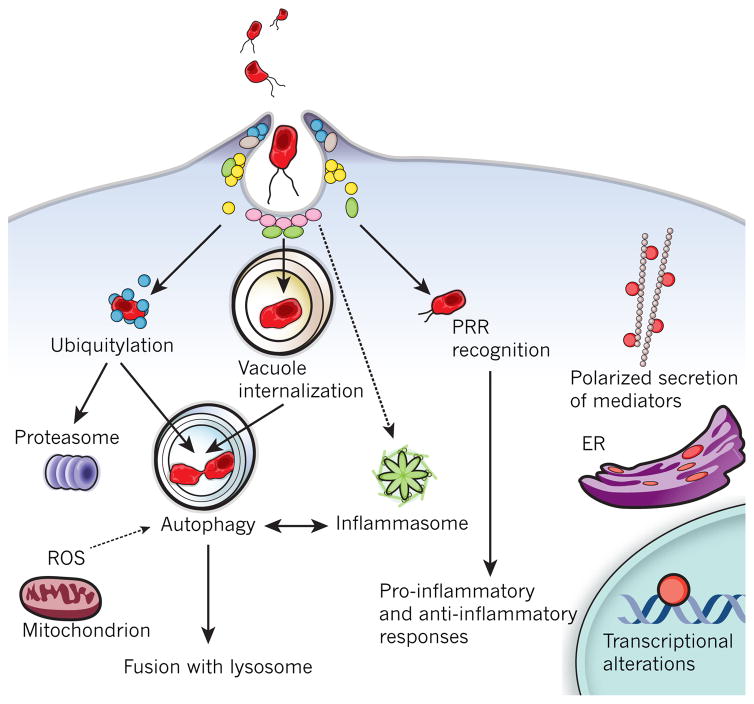
Figure 5. Intracellular defence programs in microbial recognition.
Host cells have evolved processes by which they restrict the availability of intracellular permissive niches to microbes. Microbial recognition by PRRs, such as NOD proteins and TLRs, activates key immediate host programs, leading to polarized secretion of pro-inflammatory mediators (directed to either the luminal or basolateral surface). Bacteria can either be maintained in subcellular compartments such as microbe-containing vacuoles, or escape into the cytoplasm, where they can be ubiquitylated and targeted for degradation. Both subsets can be targeted by the autophagy pathway, which is also regulated by other host defence mechanisms such as oxidative stress and inflammasome activation.
Abnormalities consistent with Crohn’s disease have been observed in mice with defects in autophagy, including hypomorphic Atg16l1 (Atg16l1HM) and IEC-specific Atg5-deficient mice27. Paneth cells either from Atg16l1HM mice or from patients with Crohn’s disease who have the ATG16L1 (T300A variant) allele show aberrant granule size, number and location, and reduced AMP secretion; notably, they also show gain of function, as evidenced by upregulated peroxisome proliferator-activated receptor signalling27. The landmark findings that gnotobiotic (germ-free) Atg16l1HM mice lost these Paneth cell anomalies and their sensitivity to dextrate sulphate sodium (DSS)-induced colitis, and that these abnormalities were restored by norovirus infection provide a definitive demonstration of how host–microbial interactions contribute to the pathophysiology of IBD61.
Effectors and regulators of adaptive immunity
Homeostasis in the gut involves a balance between anti-inflammatory and pro-inflammatory signals, such that inflammatory disease results from an inadequate Treg-cell response in the face of an overly exuberant response largely involving TH1 and TH17 cells in Crohn’s disease and TH2 cells in ulcerative colitis. Intestinal inflammation resulting from a failure to maintain this balance is exemplified by patients with immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome or with WAS (also known as WASP) deficiency, who have deficient Treg-cell function. Furthermore, TH-cell polarization in IBD is unlikely to be a simple divergence between a few disparate T-cell fates, but rather to include diverse sub-programs that can be selectively activated by antigen-presenting cells, cytokine milieu, microbial factors and metabolic programs. This notion is supported by the findings that T cells expressing both IFN-γ and IL-17 are detected during all stages of Crohn’s disease, and it evolves our understanding of pro-inflammatory and anti-inflammatory cell types and pathways.
Recent studies indicate that Treg cells and TH17 cells may arise from a common precursor, consistent with the observation that TGF-β helps to direct differentiation of both subsets62. The generation of two subsets with opposing activities from a common precursor is reminiscent of differential responses of precursor cells along morphogen gradients during development, suggesting that similar morphogen gradients may work in the gut in vivo. In this regard, TGF-β alone drives Treg-cell differentiation, with retinoic acid exerting a synergistic effect; the differentiation and function of mucosal Treg cells depends on the transcription factors BLIMP1 and IRF4 (ref. 63). Given the abundance of TGF-β in intestinal tissues, this may contribute to baseline homeostasis, for example, by promoting Treg-cell differentiation in naive lamina propria CD4+ T cells. However, in conjunction with other signals, including cytokines, metabolites and microbial signals, TH17-cell differentiation is promoted instead. Experiments demonstrating the crucial role of IL-23, IL-6 and IL-17 in the development of experimental colitis support a role for TH17 cells in disease propagation64. Illustrating some of the intercellular interactions that affect the TH17–Treg-cell axis, γδ T cells can drive the TH17 program and contribute to experimental colitis, and are in turn suppressed by Treg cells65,66. Furthermore, Treg cells can support the development of TH17 cells by maintaining decreased levels of IL-2 in the local milieu67.
Transcriptional programs helmed by the Treg- and TH17-cell-lineage-defining transcription factors FOXP3 and RORγt work together with a network of transcription factors, which can in turn respond to lineage-inducing cytokines and microbial factors. Transcription factors can mediate dichotomous functions depending on the cellular (and probably cytokine) context; for example, STAT3 drives TH17-cell differentiation, but is also required for anti-inflammatory IL-10 signalling through distinct pathways, inducing repressors such as strawberry notch homologue 2 (SBNO2). The aryl hydrocarbon receptor (AHR) is a nuclear receptor that is essential for IL-22 production and also enhances IL-17 production, albeit to a lesser extent than RORγt-driven pathways, illustrating how distinct transcription factors may drive separate functions in the same cell68. AHR responds to polycyclic hydrocarbons, suggesting that xenobiotic stimuli may modulate IL-22 and IL-17 production. Many of the genes required for Treg- and TH17-cell differentiation have been implicated in IBD (Fig. 3). CCR6, which lies in a locus associated with Crohn’s disease, encodes a chemokine receptor that is important for lymphocyte homing to the gut as well as for the development of intestinal lymphoid follicles — areas important for the production of T-cell-independent IgA, which affects the microbiota composition of the host33. Thus, the gut may use TGF-β pathways to poise T cells to carry out both pro- and anti-inflammatory programs depending on the local presence of cytokines and microbial products.
Illustrating the concept that hyper- or hypo-activation can affect the outcome of T-cell differentiation, defects in ITCH, a HECT-type E3 ubiquitin ligase involved in T-cell activation, lead to impaired Treg-cell polarization in mice and to autoimmunity in patients69. IBD-associated loci also contain other members of the ITCH pathway, including NDFIP1 and TNFAIP3. Defects in these proteins are associated with inappropriate T-cell activation, skewed TH-cell polarization and pathological intestinal inflammation, consistent with the hypothesis that NDFIP1 and TNFAIP3 are disease-contributory genes70.
The soluble mediators secreted by Treg cells are also released by other, FOXP3− regulatory T-cell subsets in the gut, such as T regulatory 1 (Tr1) cells. IL-10 release can be induced by IL-27 in several subsets, including both pro-inflammatory TH1 and anti-inflammatory Tr1 subsets. IL-27 is made by antigen-presenting cells, illustrating another homeostatic interaction between innate and adaptive immune cells71. Interestingly, GWAS implicate both IL10 and IL27 in IBD, suggesting that this may represent a central axis of immune regulation in the context of IBD.
In addition to the contribution of T-cell subsets, experimental evidence suggests that B-cell defects may contribute to the development of colitis in several ways, including impaired IgA production and antigen presentation, effects on early B-cell selection, and the perturbed production of pro- and anti-inflammatory mediators. Supporting the importance of IgA production in IBD, recent GWAS of selective IgA deficiency showed genes also implicated in IBD, namely ORMDL3, REL and PTPN22 (ref. 72).
Immunoglobulins can also have immune modulatory activity, which is highlighted by IBD genetic studies. Peripheral B-cell tolerance can be maintained by signalling through the lectin CD22, which binds immunoglobulin bearing α2,6-linked sialic acid73. This epitope is generated in part through the action of sialic acid acetylesterase (SIAE). Sequencing studies in small cohorts detected rare SIAE variants that disrupted enzyme function and secretion in patients with IBD and other autoimmune diseases73. In addition, sialylated IgG may signal through DC-SIGN (dendritic-cell-specific ICAM3-grabbing non-integrin) on myeloid cells, leading to increased expression of inhibitory FcγRIIβ (the gene encoding which lies in an ulcerative-colitis-implicated locus) on macrophages. These data demonstrate pathways by which immunoglobulins can exert anti-inflammatory activities and highlight components that genetic studies suggest may be perturbed in IBD.
The functional relevance of anti-inflammatory B regulatory (Breg) cells has been demonstrated in several mouse models of inflammatory diseases, including colitis; Breg cells from patients with SLE also show impaired function74. Breg cells differentiate after stimulation in the context of either anti-CD40 antibody or TLR ligands, and secrete anti-inflammatory cytokines such as TGF-β and IL-10. Defects in Breg-cell development or function might lead to failure to upregulate IL-10, leading to attenuated suppression of CD4+ T-cell production of IFN-γ and TNF-α, consistent with a broader role of Breg cells in autoimmunity and inflammatory disease.
Other cell types that help to regulate gut immunity include intraepithelial lymphocytes, which comprise many subsets such as CD8αα+ γδ T cells (which show both cytoprotective and cytolytic activities) and CD8αα+ αβ T cells (which are thought to be regulatory cells that require TGF-β for development)75. Enrichment analysis of expression profiles further suggests that a subset of IBD-implicated genes is expressed in natural killer T (NKT) cells, which can detect infection though microbial lipids presented by CD1d or through atypical endogenous lipids, such as isoglobotriaosylceramide, which accumulate after microbial–TLR signalling76. Furthermore, NKT cells from patients with ulcerative colitis produce IL-13 and show enhanced cytotoxicity, further indicating that a perturbed NKT-cell response to as yet unidentified bacterial ligands may be pro-colitogenic.
Genetic studies may offer insight into how this balance is disturbed, leading to pathological inflammation. Interestingly, perturbation by blocking cytotoxic T-lymphocyte antigen 4, an important inhibitory molecule expressed on activated T cells, commonly resulted in colitis in patients, again suggesting the poised state of activation of T cells in the gut77.
Mucosal ecology and immune responses in disease
The gut microflora is a community that has co-evolved with the host and confers beneficial effects, including helping to metabolize nutrients, modulate immune responses and defend against pathogens. However, dysregulation of normal co-evolved homeostatic relationships between gut bacteria and host immune responses can lead to intestinal inflammation. Indeed, accumulating evidence suggests that luminal flora is a requisite, perhaps even a central factor in the development of IBD.
Efforts to correlate changes in enteric microbial communities with disease are complicated by the great interindividual variation, such that even monozygotic twins may share only 40% of faecal phylotypes78. However, clustering the abundance of genes in certain categories in a species-independent fashion shows high interindividual similarity, suggesting that the microbiome can be perceived as a conserved functional entity79. Differences in the abundance of both bacterial species and functional gene categories (such as bacterial motility, sugar and iron metabolism) can differentiate patients with IBD from healthy individuals, demonstrating IBD-related changes in gut microbial ecology80 (C. Huttenhower, personal communication).
Even within an individual, intestinal microbial communities are dynamic and influenced by host factors, dietary effects and the microbes themselves. Many of the specific examples have emerged from studies in mice. Illustrating how microbial communities can be affected by host responses, infection-induced inflammation results in an oxidative metabolic shift in Salmonella enterica serovar Typhimurium (S. Typhimurium), which the bacterium uses in conjunction with host-derived ROS to create a growth advantage over fermenting bacteria81. Microbe–host relationships are tightly interrelated, such that host factors can induce functional changes in the microflora that, in return, affect host biology. TLRs recognize microbial motifs and have a crucial role in determining mucosal susceptibility to injury and repair responses. Impaired TLR signalling due to Myd88 deficiency in non-obese diabetic (NOD) mice induces changes in microbiota community structure that protect against diabetes82. Conversely, impaired innate immune function in T-bet−/−Rag1−/− (also known as Tbx21−/−Rag1−/−) mice and Tlr5−/− mice leads to the generation of a pathogenic microbiota that causes colitis and metabolic syndrome, respectively, even in genetically normal hosts83,84. Similarly, the gut microbiota induces a dynamic IgA response, with qualitative and/or quantitative defects in IgA production resulting in impaired control of the microbial communities85,86. Deficiencies in activation-induced cytidine deaminase — an enzyme essential for somatic hypermutation and class-switching recombination during B-cell maturation — result in IgA deficiency, specific expansion of the anaerobic flora and segmented filamentous bacteria (SFB), and overstimulation of the mucosal immune system, with hyperplasia of mucosal lymphoid structures such as Peyer’s patches and lymphoid follicles. Similar observations in mice with selectively impaired somatic hypermutation point to the importance of affinity maturation in generating diversity in the IgA repertoire to control the intestinal microbial burden86.
Mice fostered on milk lacking sialyl(α2,3)lactose develop a distinct microbiota that confers transmissible resistance to DSS colitis, providing an example of dietary effects on the gut microbiota87. Dietary glycans can also be incorporated onto host cell membranes and can act as receptors for bacterial toxins. These findings demonstrate that host factors, both transient and genetic, can act together with dietary factors to modulate microbiota community structure and/or function, sometimes indelibly, in IBD-relevant ways.
The intestinal mucosa can monitor microbial ligands using pattern recognition receptors (PRRs), and microbial metabolites using G-protein-coupled receptors (GPCRs) and solute carriers. Short-chain fatty acids (SCFAs), generated by some microflora constituents and decreased in ulcerative colitis, can signal through the receptor GPR43 in neutrophils, with notable proresolving effects on inflammation88. Other examples of GPCRs with immune modulatory activity include GPR120, GPR65 (also known as TDAG8) and GPR35, which can be activated by ω-3 fatty acids, extracellular protons and kynurenic acid, respectively, with anti-inflammatory effects89. Of interest, kynurenic acid and other anti-inflammatory kynurenines are generated by the catabolism of tryptophan. Host levels of tryptophan are affected by the microbiota, suggesting how microbes can modulate the host immune response by metabolic effects. Other microbial metabolites can be transported into host cells. For example, the solute carrier SLC22A5 transports a quorum-sensing molecule from Bacillus subtilis, conferring resistance to oxidative stress in vitro90. The proton-coupled histidine/peptide cotransporter SLC15A4 is required for TLR7 and TLR9 signalling in plasmacytoid dendritic cells34. Furthermore, studies in Slc15a4−/− dendritic cells suggest that SLC15A4 contributes to TLR9 signalling by regulating endosomal histidine levels, and to NOD1 signalling by cytosolic delivery of NOD1 ligands such as the tripeptide motif L-Ala-γ-D-Glu-meso-diaminopimelic acid (TriDAP). Slc15a4 deficiency ameliorates susceptibility to DSS colitis in mice91. These findings and the presence of genes such as SLC22A5, GPR35 and GPR65 in IBD-risk loci suggest that PRRs, GPCRs and solute carriers help to maintain microbe–host relationships and intestinal homeostasis by transducing signals from microbial ligands and metabolites, which can in turn have immune modulatory effects on the host.
Microbial signals also shape innate and adaptive immune responses. Germ-free animals have underdeveloped Peyer’s patches, as well as fewer IgA-producing plasma cells and lamina propria CD4+ cells, illustrating the role of the microbiota in generating a mature mucosal adaptive immune response. The constituents of the microbiota can have important protective roles; for example, the impaired epithelial injury response in Myd88-deficient mice highlights the role of microbial stimulation in epithelial restitution92. Similarly, in mice, commensal bacteria activate expression of the transcription factor NFIL3, inhibit Il12b expression and protect against colitis, and CD14+ lamina propria cells from patients with IBD express less NFIL3 than healthy controls93. The microbiota also acts on the epithelium together with adaptive immune signals, inducing epithelial secretion of IL-25, which represses ILC secretion of IL-22, and thus IL-22-induced AMPs. This equilibrium in the healthy mucosa is abrogated by epithelial insults, leading to increased IL-22 and activation of antimicrobial programs40.
Microbial populations and ligands can have pro-inflammatory or anti-inflammatory effects. In mice, SFB promote TH17 differentiation and IgA production, whereas Clostridium clusters IV and XIVa and parasite-secreted proteins such as Heligmosomoides polygyrus excretory-secretory antigen promote Treg-cell differentiation94–97. Interestingly, patients with IBD show reduced representation of Clostridium clusters IV and XIVa, indicating one way in which anti-inflammatory Treg-cell effects might be diminished, leading to a predisposition to inflammation98. The common constituent of normal human microflora Bacteroides fragilis produces polysaccharide A, which suppresses IL-17 production and promotes the activity of IL-10-producing CD4+ T cells in mice99. The effects of microbial ligands and metabolites on adaptive immune function are exemplified by bacterial DNA signalling through TLR9 to limit Treg-cell differentiation and promote intestinal immune responses to oral infection, and by bacterial ATP promoting TH17 differentiation. Thus, the microflora shapes development and function of the mucosal immune system in a tightly correlated manner. Immune stimulatory effects of the microbiota are important to promote an effective response against potential pathogens, although dysregulated interactions, which might arise from perturbations in host, microbial or environmental factors, could lead to a loss of tolerance and promote intestinal inflammation.
Most of the observations detailing the mechanisms of microbe–host interactions have been made in mice, and correlations in humans remain to be defined. Microbes associated with human IBD include Faecalibacterium prausnitzii, adherent-invasive E. coli, invasive Fusobacterium nucleatum and mucolytic bacteria such as Ruminococcus gnavus and Ruminococcus torques. Reduced levels of F. prausnitzii in resected ileal mucosa from patients with Crohn’s disease are associated with increased risk of endoscopic recurrence; F. prausnitzii stimulates IL-10 production in peripheral blood mononuclear cells, which may account at least in part for this protective effect100. Recent studies suggest that adherent-invasive E. coli exploits host defects in phagocytosis and autophagy arising from Crohn’s-disease-related polymorphisms to promote chronic inflammation in the susceptible host16. Patients with IBD have a compromised mucus layer and an epithelial surface that is densely coated with bacteria; the abundant presence of Ruminococcus strains in IBD mucosa raises the possibility that such microbes may contribute to the barrier defect observed in IBD, although whether their presence is causal or correlative remains unclear.
These findings show that the composition of the microbiota and its interaction with the host are emerging as underappreciated sources of gene–environment interactions and are crucial to understanding the context of IBD. For example, alterations in the microflora community structure, as might occur in the context of antibiotic therapy or infectious colitis, can promote the development of IBD or trigger disease flares in patients with IBD. Identifying the factors that shape microbial community structure and function within an individual and that influence its restoration after perturbations will be key to understanding IBD pathogenesis. Obtaining such knowledge will require identifying associations between microbiome and human genetic studies at the very least.
Future perspectives
GWAS and next-generation sequencing technologies have provided insight into genetic definitions of host susceptibility. GWAS have unequivocally identified numerous genomic regions containing IBD-risk factors, showing several features of the genetic architecture of Crohn’s disease and ulcerative colitis. First, IBD risk involves multigenic contributions, each with a relatively modest effect size. Second, genetic contributions to ulcerative colitis and Crohn’s disease overlap, suggesting shared mechanistic features. Third, within Crohn’s disease and ulcerative colitis, different clusters of risk loci are emerging, suggesting that these disease processes may comprise distinct pathological subsets beyond Crohn’s disease versus ulcerative colitis. Accordingly, there is a need to define clinically relevant parameters that might help to classify Crohn’s disease and ulcerative colitis further, including early-onset disease, stricturing disease, slow progressors, frequency of flares and response to therapeutics. Furthermore, given the importance of environmental factors in IBD risk, studies aimed at defining contributory environmental factors are greatly needed. Relevant approaches might include establishing prospective inception cohorts or following healthy, high-risk individuals, such as those with an affected first-degree relative.
An important adjunctive approach to GWAS is identifying rare variants, which frequently show larger effect sizes. The search for rare variants will help to prioritize the probable causal gene(s) within a locus (or loci) for experimental validation, identify disease-relevant pathways, and possibly identify domains important for protein function by leveraging natural mutations as a large forward genetic screen. Identifying and validating causal genes and assembling them into molecular pathways and cellular networks will require the use of patient samples and will considerably empower clinically relevant hypotheses. Given the diverse mechanisms that seem to participate in IBD, it will also become increasingly important to associate and stratify ‘-omic’ measurements of RNA, protein, small molecules, chemical DNA modifications and gut microbiota according to patient genotypes.
There is a clear need to generate quantitative and qualitative expression maps of allelic variants. This notion is reinforced by the many polymorphisms implicating gene deserts, which probably contain regulatory elements. Furthermore, alternative splicing is a major contributor to the diversity of our transcriptome and its relevance to IBD has already been demonstrated by findings in IL23R.
The gut has many tiers of defence against incursion by luminal microbes, including the epithelial barrier, and the innate and adaptive immune responses. These components are all tightly interrelated, and disease requires breakdown at several checkpoints. Generating models to systematically analyse the defects arising from genetic variants associated with IBD is crucial. However, these variants may show the disease-relevant defect under select conditions, such as high bacterial load found in the colon, and the accompanying cytokine milieu.
Viral infections are common, and key studies highlight their potential to exert important immune modulatory effects. Acute and/or chronic viral infections could interact with host-susceptibility factors in a manner that leaves either the cell or the cellular milieu poised to promote pathological intestinal inflammation after subsequent triggering events. Notably, these studies highlight the need to characterize all microbial constituents (viral, fungal, parasitic and bacterial) in the context of IBD. Other tools need to be developed to study the microbiota at the level of species, geographical location, genetic variations, transcriptional dynamics, as well as changes to proteins and metabolites. Indeed, bacterial metabolites are principal mediators of interactions between microbial species, as well as between microbe and host, as exemplified by SCFAs. Studies to identify bioactive metabolites and other small molecules may thus have diagnostic and therapeutic potential.
An important goal is to combine these various facets to understand how genetic traits are integrated and propagated through physiological networks in the context of interactions with other genes, cells, microbes and environmental stimuli to control intestinal homeostasis. Genetic studies are already used to predict sensitivity to IBD therapies such as 6-mercaptopurine and may also be useful in predicting responses to biological therapies. Combining the different aspects of IBD pathophysiology may allow us to develop a more holistic understanding of the disease, thus promoting advances in diagnostics and therapy.
Supplementary Material
Acknowledgments
R.J.X., A.G. and B.K. are supported by grants from the National Institutes of Health and the Crohn’s and Colitis Foundation of America. We apologize to those whose work is not cited owing to space constraints.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information: Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
The authors declare no competing financial interests.
References
- 1.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nature Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. This study identifies 39 new loci containing genes in previously identified Crohn’s-disease-relevant pathways, including IL10, IL27 and TYK2, and reports the important genetic overlap with loci implicated in other immune-related diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature Genet. 2011;43:246–252. doi: 10.1038/ng.764. This study identifies 29 new loci containing genes that reinforce the contribution of epithelial barrier function, intracellular defence and cytokine-dependent signalling in ulcerative colitis, and it also reports genetic overlap with Crohn’s disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spehlmann ME, et al. Epidemiology of inflammatory bowel disease in a German twin cohort: results of a nationwide study. Inflamm Bowel Dis. 2008;14:968–976. doi: 10.1002/ibd.20380. [DOI] [PubMed] [Google Scholar]
- 4.Yazdanyar S, Kamstrup PR, Tybjaerg-Hansen A, Nordestgaard BG. Penetrance of NOD2/CARD15 genetic variants in the general population. Can Med Assoc J. 2010;182:661–665. doi: 10.1503/cmaj.090684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monick MM, et al. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol. 2010;185:5425–5435. doi: 10.4049/jimmunol.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nature Genet. 2010;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janse M, et al. Three ulcerative colitis susceptibility loci are associated with primary sclerosing cholangitis and indicate a role for IL2, REL and CARD9. Hepatology. 2011;53:1977–1985. doi: 10.1002/hep.24307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang FR, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–2618. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 10.Waisberg M, et al. Genetic susceptibility to systemic lupus erythematosus protects against cerebral malaria in mice. Proc Natl Acad Sci USA. 2011;108:1122–1127. doi: 10.1073/pnas.1017996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glocker EO, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Meglio P, et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS ONE. 2011;6:e17160. doi: 10.1371/journal.pone.0017160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu RY, Gallagher G. A naturally occurring, soluble antagonist of human IL-23 inhibits the development and in vitro function of human Th17 cells. J Immunol. 2010;185:7302–7308. doi: 10.4049/jimmunol.1002410. [DOI] [PubMed] [Google Scholar]
- 15.Rosenstiel P, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Natl Acad Sci USA. 2006;103:3280–3285. doi: 10.1073/pnas.0505423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brest P, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease. Nature Genet. 2011;43:242–245. doi: 10.1038/ng.762. [DOI] [PubMed] [Google Scholar]
- 17.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muise AM, et al. Polymorphisms in E-cadherin (CDH1) result in a mis-localised cytoplasmic protein that is associated with Crohn’s disease. Gut. 2009;58:1121–1127. doi: 10.1136/gut.2008.175117. [DOI] [PubMed] [Google Scholar]
- 19.Sabath E, et al. Ga12 regulates protein interactions within the MDCK cell tight junction and inhibits tight-junction assembly. J Cell Sci. 2008;121:814–824. doi: 10.1242/jcs.014878. [DOI] [PubMed] [Google Scholar]
- 20.Scharl M, et al. Protection of epithelial barrier function by the Crohn’s disease associated gene protein tyrosine phosphatase N2. Gastroenterology. 2009;137:2030–2040.e5. doi: 10.1053/j.gastro.2009.07.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan SW, et al. Increased susceptibility to dextran sulfate sodium induced colitis in the T cell protein tyrosine phosphatase heterozygous mouse. PLoS ONE. 2010;5:e8868. doi: 10.1371/journal.pone.0008868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darsigny M, et al. Loss of hepatocyte-nuclear-factor-4α affects colonic ion transport and causes chronic inflammation resembling inflammatory bowel disease in mice. PLoS ONE. 2009;4:e7609. doi: 10.1371/journal.pone.0007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattin AL, et al. Hepatocyte nuclear factor 4α, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol. 2009;29:6294–6308. doi: 10.1128/MCB.00939-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 25.Pickert G, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu J, et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011;121:1657–1666. doi: 10.1172/JCI45538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biswas A, et al. Induction and rescue of Nod2-dependent Th1-driven granulomatous inflammation of the ileum. Proc Natl Acad Sci USA. 2010;107:14739–14744. doi: 10.1073/pnas.1003363107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaser A, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. This study illustrates the role of Paneth and goblet cells in intestinal homeostasis and how the control of ER stress is crucial for their maintenance, such that XBP1 deficiency leads to Paneth cell loss, reduced goblet cell numbers and increased susceptibility to experimental colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandl K, et al. Enhanced sensitivity to DSS colitis caused by a hypomorphic Mbtps1 mutation disrupting the ATF6-driven unfolded protein response. Proc Natl Acad Sci USA. 2009;106:3300–3305. doi: 10.1073/pnas.0813036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodall JC, et al. Endoplasmic reticulum stress-induced transcription factor, CHOP, is crucial for dendritic cell IL-23 expression. Proc Natl Acad Sci USA. 2010;107:17698–17703. doi: 10.1073/pnas.1011736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell EL, et al. Resolvin E1-induced intestinal alkaline phosphatase promotes resolution of inflammation through LPS detoxification. Proc Natl Acad Sci USA. 2010;107:14298–14303. doi: 10.1073/pnas.0914730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rescigno M. Intestinal dendritic cells. Adv Immunol. 2010;107:109–138. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- 34.Blasius AL, et al. Slc15a4, AP-3, and Hermansky–Pudlak syndrome proteins are required for Toll-like receptor signaling in plasmacytoid dendritic cells. Proc Natl Acad Sci USA. 2010;107:19973–19978. doi: 10.1073/pnas.1014051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith AM, et al. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn’s disease. J Exp Med. 2009;206:1883–1897. doi: 10.1084/jem.20091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amit I, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. This pioneering study uses computational analyses of time-course gene-expression profiles to characterize dendritic cell programs and predict signalling circuits engaged after stimulation by microbe-associated molecules, and it exerimentally validates key regulators. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siddiqui KRR, Laffont S, Powrie F. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 2010;32:557–567. doi: 10.1016/j.immuni.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uematsu S, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nature Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- 39.Swirski FK, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–616. doi: 10.1126/science.1175202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawa S, et al. RORγ+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nature Immunol. 2011;12:320–326. doi: 10.1038/ni.2002. [DOI] [PubMed] [Google Scholar]
- 41.Cella M, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. This study describes mucosal IL-22-secreting ILCs in humans and mice, capturing many key properties of these cells, including their ability to respond to IL-23 stimulation and promote epithelial restitutive effects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takayama T, et al. Imbalance of NKp44+NKp46− and NKp44−NKp46+ natural killer cells in the intestinal mucosa of patients with Crohn’s disease. Gastroenterology. 2010;139:882–892.e3. doi: 10.1053/j.gastro.2010.05.040. [DOI] [PubMed] [Google Scholar]
- 43.Shaw MH, Kamada N, Warner N, Kim YG, Nunez G. The ever-expanding function of NOD2: autophagy, viral recognition, and T cell activation. Trends Immunol. 2011;32:73–79. doi: 10.1016/j.it.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nature Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 45.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nature Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 46.Sabbah A, et al. Activation of innate immune antiviral responses by Nod2. Nature Immunol. 2009;10:1073–1080. doi: 10.1038/ni.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaki MH, et al. The NLRP3 inflammasome protects against loss of epithelial integrity and mortality during experimental colitis. Immunity. 2010;32:379–391. doi: 10.1016/j.immuni.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsu YMS, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nature Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 49.Poeck H, et al. Recognition of RNA virus by RIG-I results in activation of CARD9 and inflammasome signaling for interleukin 1β production. Nature Immunol. 2010;11:63–69. doi: 10.1038/ni.1824. [DOI] [PubMed] [Google Scholar]
- 50.Gringhuis SI, et al. Selective C-Rel activation via Malt1 controls anti-fungal TH-17 immunity by dectin-1 and dectin-2. PLoS Pathogens. 2011;7:e1001259. doi: 10.1371/journal.ppat.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroeder BO, et al. Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature. 2011;469:419–423. doi: 10.1038/nature09674. [DOI] [PubMed] [Google Scholar]
- 52.Dolowschiak T, et al. Potentiation of epithelial innate host responses by intercellular communication. PLoS Pathogens. 2010;6:e1001194. doi: 10.1371/journal.ppat.1001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gardet A, et al. LRRK2 is involved in the IFN-γ response and host response to pathogens. J Immunol. 2010;185:5577–5585. doi: 10.4049/jimmunol.1000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu W, Hsu YMS, Bi L, Songyang Z, Lin X. CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nature Immunol. 2009;10:1208–1214. doi: 10.1038/ni.1788. [DOI] [PubMed] [Google Scholar]
- 55.Efimova O, Szankasi P, Kelley TW. Ncf1 (p47phox) is essential for direct regulatory T cell mediated suppression of CD4+ effector T cells. PLoS ONE. 2011;6:e16013. doi: 10.1371/journal.pone.0016013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kraaij MD, et al. Induction of regulatory T cells by macrophages is dependent on production of reactive oxygen species. Proc Natl Acad Sci USA. 2010;107:17686–17691. doi: 10.1073/pnas.1012016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rioux JD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nature Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hampe J, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nature Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 59.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nature Genet. 2008;40:1107–1112. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn’s disease associated ATG16L1 variant. PLoS ONE. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. This seminal paper reports that a Crohn’s disease susceptibility gene requires interaction with environmental (microbial) cues to manifest a disease phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou L, et al. TGF-β-induced Foxp3 inhibits TH17 cell differentiation by antagonizing RORγt function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nature Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 64.Ghoreschi K, et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Do JS, Visperas A, Dong C, Baldwin WM, III, Min B. Generation of colitogenic Th17 CD4 T cells is enhanced by IL-17+ γδ T cells. J Immunol. 2011;186:4546–4550. doi: 10.4049/jimmunol.1004021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park SG, et al. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity. 2010;33:791–803. doi: 10.1016/j.immuni.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, et al. Foxp3+ regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity. 2011;34:409–421. doi: 10.1016/j.immuni.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 68.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from TH-17, TH1 and TH2 cells. Nature Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 69.Lohr NJ, et al. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. Am J Hum Genet. 2010;86:447–453. doi: 10.1016/j.ajhg.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ramon HE, et al. The ubiquitin ligase adaptor Ndfip1 regulates T cell-mediated gastrointestinal inflammation and inflammatory bowel disease susceptibility. Mucosal Immunol. 2010;4:314–324. doi: 10.1038/mi.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cox JH, et al. IL-27 promotes T cell-dependent colitis through multiple mechanisms. J Exp Med. 2011;208:115–123. doi: 10.1084/jem.20100410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ferreira RC, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nature Genet. 2010;42:777–780. doi: 10.1038/ng.644. [DOI] [PubMed] [Google Scholar]
- 73.Surolia I, et al. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 2010;466:243–247. doi: 10.1038/nature09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blair PA, et al. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity. 2010;32:129–140. doi: 10.1016/j.immuni.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 75.Konkel JE, et al. Control of the development of CD8αα+ intestinal intraepithelial lymphocytes by TGF-β. Nature Immunol. 2011;12:312–319. doi: 10.1038/ni.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darmoise A, et al. Lysosomal α-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity. 2010;33:216–228. doi: 10.1016/j.immuni.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boasberg P, Hamid O, O’Day S. Ipilimumab: unleashing the power of the immune system through CTLA-4 blockade. Semin Oncol. 2010;37:440–449. doi: 10.1053/j.seminoncol.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 78.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci USA. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. This paper shows how S. Typhimurium can use ROS generated during inflammation to form an electron acceptor that confers a growth advantage, illustrating how gut environment can affect microbial communities by providing a selective advantage to some bacterial species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wen L, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vijay-Kumar M, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei M, et al. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nature Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- 87.Fuhrer A, et al. Milk sialyllactose influences colitis in mice through selective intestinal bacterial colonization. J Exp Med. 2010;207:2843–2854. doi: 10.1084/jem.20101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. This paper highlights how commensal populations can modulate the host immune response, identifying a bacterial metabolic product (SCFA) and its receptor (GPR43), and shows that this pathway can ameliorate experimental colitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mogi C, et al. Involvement of proton-sensing TDAG8 in extracellular acidification-induced inhibition of proinflammatory cytokine production in peritoneal macrophages. J Immunol. 2009;182:3243–3251. doi: 10.4049/jimmunol.0803466. [DOI] [PubMed] [Google Scholar]
- 90.Fujiya M, et al. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe. 2007;1:299–308. doi: 10.1016/j.chom.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 91.Sasawatari S, et al. The solute carrier family 15A4 regulates TLR9 and NOD1 functions in the innate immune system and promotes colitis in mice. Gastroenterology. 2011;140:1513–1525. doi: 10.1053/j.gastro.2011.01.041. [DOI] [PubMed] [Google Scholar]
- 92.Brandl K, et al. MyD88 signaling in nonhematopoietic cells protects mice against induced colitis by regulating specific EGF receptor ligands. Proc Natl Acad USA. 2010;107:19967–19972. doi: 10.1073/pnas.1014669107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kobayashi T, et al. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J Immunol. 2011;186:4649–4655. doi: 10.4049/jimmunol.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. This important study shows how particular components of the gut microbiota can modulate the adaptive immune response, with SFB promoting differentiation of pro-inflammatory TH17 cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 96.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2010;331:337–341. doi: 10.1126/science.1198469. This study demonstrates that Clostridium clusters IV and XIVa affect the adaptive immune system by inducing a TGF-β-rich milieu and promoting differentiation of anti-inflammatory Treg cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grainger JR, et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-β pathway. J Exp Med. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 100.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.