Abstract
Initial cell surface expression of the pre-B cell receptor induces proliferation. After 2 to 5 divisions, however, large pre-BII (Fraction C') cells exit cell cycle to become resting, small pre-BII cells (Fraction D). The mechanism by which pre-BII cells exit cell cycle, however, is currently unclear. The checkpoint at the Fraction C'-D transition is critical for immunoglobulin light chain gene recombination and to prevent malignant transformation into acute lymphoblastic leukemia. Here we demonstrate that inducible activation of pre-B cell receptor signaling induces cell-cycle exit through up-regulation of the transcriptional repressor BCL6. Inducible activation of BCL6 downstream of the pre-B cell receptor results in transcriptional repression of MYC and CCND2. Hence, pre-B cell receptor-mediated activation of BCL6 limits pre-B cell proliferation and induces cellular quiescence at the small pre-BII (Fraction D) stage.
Introduction
After successful completion of V(D)J recombination, pre-BI (Fraction C) cells express for the first time an immunoglobulin μ heavy chain as part of a pre-B cell receptor (pre-BCR) on the cell surface. Initiation of pre-BCR signaling induces proliferation and clonal expansion of large pre-BII cells (Fraction C'). After 2-5 cell divisions pre-BII cells become small (Fraction D) and exit cell cycle,1,2 the mechanism of which is currently unknown and the focus of this study. This checkpoint is critical for small resting pre-BII (Fraction D) cells to undergo immunoglobulin light chain gene rearrangement and, hence to differentiate into immature (Fraction E) B cells. In addition, failure to control proliferation at the large cycling pre-BII cell (Fraction C') stage predisposes to malignant transformation into acute lymphoblastic leukemia.3–7 We recently demonstrated that pre-BCR signaling and the Ikaros transcription factor cooperate to induce cell-cycle arrest in acute lymphoblastic leukemia cells.8 In addition, inducible differentiation of large cycling pre-BII (Fraction C') cells into small resting pre-BII (Fraction D) cells results in very prominent up-regulation of the BCL6 transcriptional repressor.9 At the Fraction D checkpoint, BCL6 serves as a survival factor for normal pre-B cells and protects from p53-mediated cell death, when pre-B cells sustain DNA double strand breaks during immunoglobulin light chain gene rearrangement.9 Likewise, BCL6 protects Ph+ ALL cells from p53-dependent cell death when treated with tyrosine kinase inhibitors (TKI), which represents a novel BCL6-dependent form of TKI-resistance.10 BCL6 was first identified as a proto-oncogene in germinal center-derived B-cell lymphoma11 and transcriptional repressor of p53 in germinal center B cells.12 In this study, we test the contribution of pre-BCR signaling and BCL6 to cell-cycle exit at the Fraction C' to Fraction D checkpoint.
Methods
Pre-B cell culture
Bone marrow from constitutive or inducible knockout and transgenic mice was harvested and bone marrow cells were cultured either in the presence of 10 ng/mL IL-7 on OP9 stroma layer or retrovirally transformed by BCR-ABL1.9 (For a list of genetic mouse models used in this study, see supplemental Table 4, available on the Blood Web site; see the Supplemental Materials link at the top of the online article.) All pre-B cells derived from bone marrow of mice were maintained in IMDM (Invitrogen) with GlutaMAX containing 20% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin, 50μM β-mercaptoethanol and 10 ng/mL recombinant mouse IL-7 (Peprotech) at 37°C in a humidified incubator with 5% CO2. All mouse experiments were subject to institutional approval by Childrens Hospital Los Angeles IACUC.
Retroviral transduction
Transfections of MSCV-based retroviral constructs encoding BCL6, MYC, BLNK, FoxO1CA, Cre, BCR-ABL1, μ-heavy chain and the respective empty vector controls were performed according to a detailed protocol provided in supplemental Table 3 and transduction efficiencies were verified as shown in supplemental Figure 1.
In vitro pre-B cell differentiation assays
Differentiation of Rag2−/− tTA/μ-chain–transgenic pro-B cells was induced by removal of tetracycline from culture to induce μ-chain expression13 (verified in supplemental Figure 2). While this system induces pro-B to pre-B cell transition, the differentiation into κ-light chain expressing immature B cells was induced by removal of IL-7 from the cell culture medium.9
Quantitative RT-PCR
Total RNA from cells was extracted using RNeasy isolation kit from QIAGEN. cDNA was generated using a poly(dT) oligonucleotide and the SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with the SYBRGreenER mix (Invitrogen) and the ABI7900HT real-time PCR system (Applied Biosystems) according to standard PCR conditions. Primers for quantitative RT-PCR are listed in supplemental Table 1.
Western blotting and flow cytometry
Cells were lysed in CelLytic buffer (Sigma-Aldrich) supplemented with 1% protease inhibitor cocktail (Pierce), 1% Phosphatase inhibitor cocktail (Calbiochem) and 1mM PMSF. Protein samples were loaded on NuPAGE (Invitrogen) 4%-12% Bis-Tris gradient gels and transferred on PVDF membranes (Invitrogen). For the detection of mouse and human proteins by Western blot, primary antibodies were used together with the WesternBreeze immunodetection system (Invitrogen). The antibodies used for Western blotting and flow cytometry are listed in supplemental Table 2.
ChIP-on-chip assay
Human Ph+ ALL as well as B-cell lymphoma OCI-Ly1 and OCI-Ly7 cell lines were subjected to ChIP-on-chip analysis and qChIP single-locus validation according to the detailed protocol presented in supplemental Figure 3.
In vivo leukemia cell transplantation
BCR-ABL1 transformed murine pre-B acute lymphoblastic leukemia (B ALL) cells were transduced with BCL6-GFP or a GFP empty vector control. GFP+ cells were sorted 2 days after transduction and each 2 × 106 GFP+ cells were injected into sublethally irradiated (300 cGy) NOD/SCID mice. Three mice per group were injected via tail vein injection. Once the first mouse became sick, all mice were killed in both groups and analyzed.
Results and discussion
We have recently shown that signaling from the pre-BCR (eg, μ-heavy chain and BLNK adaptor molecule) cooperates with the Ikaros transcription factor to induce cell-cycle exit in acute lymphoblastic leukemia.8 Because inducible differentiation of large cycling pre-BII cells induces dramatic up-regulation of BCL6,9 we tested the contribution of pre-BCR signaling and BCL6 to cell-cycle exit at the Fraction C' to Fraction D checkpoint. This checkpoint involves 2 aspects, namely1 cell-cycle exit and2 down-regulation of the pre-B cell receptor.14
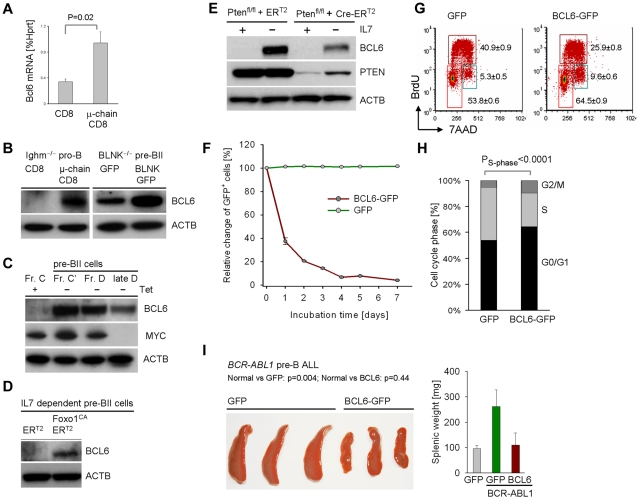
Interestingly, forced expression of pre-BCR components (μ-chain and BLNK) results in up-regulation of BCL6 both at the mRNA and protein level (Figure 1A-B). Likewise tetracycline dependent induction of pre-BCR signaling resulted in pre-BII cell differentiation (supplemental Figure 2) and strong up-regulation of BCL6 starting at the Fr C-C' checkpoint followed by down-regulation of Myc at the Fraction C'-D checkpoint (Figure 1C).
Figure 1.
Pre-BCR signaling and Ikaros up-regulate BCL6 in normal and leukemic pre-B cells. Ighm−/− pro-B cells and Blnk−/− pre-BII cells were transformed with BCR-ABL1 and pre-BCR signaling was reconstituted by retroviral expression of BLNK-GFP, CD8/μ-chain or GFP and CD8 empty vectors. BCL6 mRNA levels were measured by quantitative RT-PCR in (A; n = 2) and protein levels were determined by Western blot in (B; n = 2) using β-actin as loading control. IL-7–dependent pre-B cells from Rag2−/− tTA/μ-chain-transgenic mice2 were cultured in the presence of tetracycline, removal of which induced activation of transgenic μ-chain expression under endogenous transcriptional control elements. Inducible differentiation was verified by flow cytometry (supplemental Figure 2) and BCL6 and MYC protein levels were measured by Western blot (C; n = 2). Normal IL-7–dependent pre-B cells were transduced with a constitutively active (CA) mutant of FoxO1 or an empty vector (transduction efficiency shown in supplemental Figure 1). After inducible activation with 4-hydroxy-tamoxifen (4-OHT), cells were subjected to Western blot analysis for BCL6 and β-actin as loading control (D; n = 2). Likewise, normal IL-7–dependent pre-B cells from Ptenfl/fl mice (supplemental Table 4) were transduced with 4-OHT–inducible Cre-ERT2 or an ERT2 empty vector control and subjected to antibiotic selection. Cre-mediated deletion of Pten and BCL6-up-regulation on IL-7 withdrawal was studied by Western blot using β-actin as loading control (E; n = 2). Pre-B ALL cells from Bcl6+/+ bone marrow were transduced with retroviral expression vectors for BCL6-GFP or a GFP empty vector control. The relative change of the percentage of GFP+ cells over time was measured by flow cytometry (F; n = 2). The cell-cycle profile of transduced cells was measured by BrdU incorporation and flow cytometry (G; n = 2) with statistical analysis (H). Pre-B ALL cells transduced with BCL6-GFP or GFP alone were sorted for GFP and transplanted into sublethally irradiated NOD/SCID mice via tail vein injection (3 mice per group). Spleens of recipient mice and their weight (right) are shown (I).
Pre-B cell receptor-mediated activation of Pten/FoxO1 induces BCL6 expression
Because recent work demonstrated that pre-BCR signaling via BLNK results in Pten-dependent activation of FoxO1,15 we tested the involvement of Pten/FoxO1 in pre-B cell receptor-induced up-regulation of BCL6 (see schematic in supplemental Figure 6). In a gain-of-function experiment, we could demonstrate that expression of a constitutively active FoxO1 mutant (FoxO1CA) was sufficient to induce BCL6 expression in normal pre-B cells even in the presence of IL-7 (Figure 1D). Conversely, a loss-of-function experiment showed that inducible deletion of Pten abrogates the ability of normal pre-B cells to up-regulate BCL6 in response to IL7 withdrawal (Figure 1E). Together we conclude that Pten-mediated activation of FoxO factors is required and active FoxO1 is sufficient for BCL6 expression downstream of the pre-BCR (supplemental Figure 6).
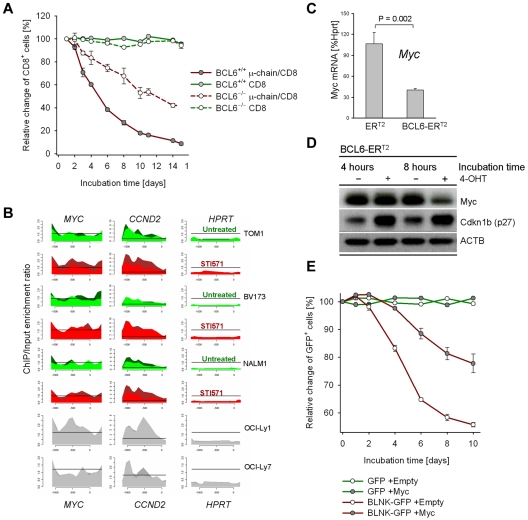
To mechanistically address the function of BCL6 up-regulation at this checkpoint, we overexpressed BCL6 in BCR-ABL1–transformed pre-B ALL cells with Fraction C' phenotype. BCL6 overexpression resulted in an increase in G0/G1 phase cells (Figure 1F-H). Interestingly, BCL6 overexpression also limited proliferation in vivo, when pre-B ALL cells were transplanted into sublethally irradiated NOD/SCID mice via tail vein injection (Figure 1I). We next tested whether BCL6 functions as an effector downstream of the pre-BCR to induce cell-cycle exit in proliferating pre-B cells. To this end, μ-chain expression was induced in BCR-ABL1–transformed Bcl6+/+ and Bcl6−/− pre-B ALL cells. While μ-chain induction causes rapid cell-cycle exit in Bcl6+/+ pre-B ALL cells, this effect was significantly attenuated in Bcl6−/− pre-B ALL cells (Figure 2A). We conclude that BCL6 mediates cell-cycle exit downstream of the pre-B cell receptor.
Figure 2.
Pre-B cell receptor–mediated up-regulation of BCL6 inhibits proliferation by transcriptional repression of Myc. Pre-B ALL cells from Bcl6+/+ and Bcl6−/− bone marrow were transduced with retroviral expression vectors for CD8/μ-chain or CD8 alone. The relative change of the percentage of CD8+ cells over time was measured by flow cytometry (A; n = 3). To identify BCL6-target genes, ChIP-on-chip analysis was performed for 3 human BCR-ABL1 pre-B ALL cell lines (BV173, Tom1, Nalm1) and 2 human diffuse large B-cell lymphoma cell lines (OCI-Ly1 and OCI-Ly7) as positive controls. High expression levels of BCL6 were induced by treatment of BCR-ABL1 pre-B ALL cell lines with 10μM STI571 for 24 hours (STI571). Recruitment to CCND2 and MYC (see supplemental Figure 3) promoters are shown and to the HPRT promoter as a negative control. The ChIP-on-chip data for Myc in Nalm1 cells is also published in Duy et al.9 (B) BCR-ABL1 pre-B ALL cells were transduced with 4-hydroxy-tamoxifen (4-OHT)–inducible vectors for BCL6-ERT2 and ERT2 empty vector control. Myc mRNA (C; n = 2) and protein (D; n = 2) levels on 4-OHT induction were measured by quantitative RT-PCR and Western blot, respectively. Induction of BCL6 also resulted in up-regulation of Cdkn1b (p27) protein levels (D). Overexpression of Myc was sufficient to partially rescue BLNK/BCL6-mediated inhibition of proliferation (E): Blnk−/− BCR-ABL1 pre-B ALL cells were transduced with Myc-Puro or a Puro-empty vector control and subjected to antibiotic selection. Subsequently, pre-BCR signaling was reconstituted by retroviral expression of BLNK-GFP or a GFP empty vector control and proliferation of the Myc-Puro versus Puro-transduced cells was measured as relative change of the percentage of BLNK-GFP+ or GFP+ cells (E; n = 2).
BCL6 induces cell-cycle exit via transcriptional repression of Myc
Given that BCL6 functions as transcriptional repressor, we performed a genome-wide ChIP-on-chip analysis to identify BCL6 target genes in human Ph+ pre-B ALL cell lines (supplemental Figure 3). We focused our analysis on known promoters of cell-cycle progression and found that MYC, CCND2, MAX, CDK6, BRAF and BMI1 are all transcriptional targets of BCL6 (supplemental Figure 3; Figure 2B). These findings are consistent with down-regulation of MYC on pre-B cell receptor-mediated activation of BCL6 (Figure 1C). To test whether BCL6 not only directly binds the MYC promoter but also functions as transcriptional repressor of MYC in pre-B cells, we used a 4-hydroxy-tamoxifen (4-OHT)–dependent system for inducible expression of BCL616: 4-OHT–inducible activation of BCL6 resulted in down-regulation of MYC expression levels both at the mRNA and protein levels (Figure 2C-D)9 suggesting that BCL6 can directly repress MYC. However, we cannot rule out additional levels of regulation of MYC expression, considering that BCL6 is up-regulated at the Fr C-C' transition and MYC protein is down-regulated at the Fr C'-D checkpoint during pre-B cell differentiation (Figure 1C).
BCL6-mediated repression of Myc is functionally relevant as Cre-mediated reduction of Myc protein levels in BCR-ABL1–transformed Mycfl/fl pre-B ALL cells resulted in rapid cell-cycle exit (supplemental Figure 4). Conversely, overexpression of MYC was sufficient to partially rescue BLNK/BCL6-mediated inhibition of proliferation (Figure 2E). While reconstitution of pre-BCR signaling in BCR-ABL1–transformed Blnk−/− pre-B ALL cells rapidly induced cell-cycle exit (Figure 2E, supplemental Figure 5), concomitant overexpression of Myc significantly diminished the proliferation-inhibitory effect of BLNK (Figure 2E). Together, these findings establish that pre-BCR signaling induces cell-cycle exit via BCL6-mediated transcriptional repression of MYC.
We had recently shown that BCL6 represents a critical survival factor for TKI-treated Ph+ ALL cells10 and at the small resting pre-BII cell stage9 of normal B-cell development (Fraction D), when pre-B cells undergo Vκ-Jκ immunoglobulin light chain gene recombination. Here we identify BCL6 as a critical mediator of cell-cycle exit at the pre-BII checkpoint (transition from Fraction C' to D). This checkpoint is essential for Vκ-Jκ immunoglobulin light chain gene recombination. Induction of pre-B cell quiescence at this checkpoint serves as a major safeguard against malignant transformation toward acute lymphoblastic leukemia.7,17 Pre-BCR mediated activation of BCL6 establishes this checkpoint by transcriptional repression of Myc, Ccnd2, Cdk6, Bmi1 and other mediators of cell-cycle progression (supplemental Figure 3), suggesting a role for BCL6 in mediating pre-B cell quiescence.
Supplementary Material
Acknowledgments
The authors thank Riccardo Dalla-Favera (Columbia University, New York, NY) for sharing Bcl6−/− mice and wild-type controls with us. They thank Arthur L Shaffer and Louis M Staudt (NCI, Bethesda, MD) for sharing their inducible BCL6 constructs and Hans-Martin Jäck (Erlangen, Germany) and Hong Wu (UCLA, Los Angeles, CA) for bone marrow cells from Tet-μ transgenic and Ptenfl/fl mice, respectively. They also thank Ignacio Moreno de Alborán (CINES, Madrid, Spain) for providing bone marrow from Mycfl/fl mice.
This work is supported by grants from the National Institutes of Health/NCI through R01CA104348 (to A.M.), R01CA085573 (to B.H.Y.), R01CA137060, R01CA139032, R01CA157644 and R21CA152497 (to M.M.), grants from the Leukemia & Lymphoma Society (to M.M.), the William Laurence and Blanche Hughes Foundation and a Stand Up To Cancer–American Association for Cancer Research Innovative Research Grant IRG00909 (to M.M.). A.M. and M.M. are Scholars of The Leukemia & Lymphoma Society.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.N. and M. Muschen designed the research and interpreted the data; R.N. and P.R.-R. performed majority of the research including data collection. M. Mossner and C.D. helped in data collection; L.C. and H.G. performed the ChIP-on-chip experiments and analysis; H.J., H.Y. and A.M. contributed vital reagents and to critical reviewing of the manuscript; and M. Muschen wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Markus Müschen, Department of Laboratory Medicine, University of California San Francisco, 521 Parnassus Ave, San Francisco, CA 94143; e-mail: markus.muschen@ucsf.edu.
References
- 1.Decker DJ, Boyle NE, Koziol JA, Klinman NR. The expression of the Ig H chain repertoire in developing bone marrow B lineage cells. J Immunol. 1991;146(1):350–361. [PubMed] [Google Scholar]
- 2.Hess J, Werner A, Wirth T, Melchers F, Jäck HM, Winkler TH. Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc Natl Acad Sci U S A. 2001;98(4):1745–1750. doi: 10.1073/pnas.041492098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kersseboom R, Middendorp S, Dingjan GM, et al. Bruton's tyrosine kinase cooperates with the B cell linker protein SLP-65 as a tumor suppressor in Pre-B cells. J Exp Med. 2003;198(1):91–98. doi: 10.1084/jem.20030615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jumaa H, Bossaller L, Portugal K, et al. Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature. 2003;423(6938):452–456. doi: 10.1038/nature01608. [DOI] [PubMed] [Google Scholar]
- 5.Flemming A, Brummer T, Reth M, Jumaa H. The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol. 2003;4(1):38–43. doi: 10.1038/ni862. [DOI] [PubMed] [Google Scholar]
- 6.Nakayama J, Yamamoto M, Hayashi K, et al. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood. 2009;113(7):1483–1492. doi: 10.1182/blood-2008-07-166355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ta VB, de Bruijn MJ, ter Brugge PJ, et al. Malignant transformation of Slp65-deficient pre-B cells involves disruption of the Arf-Mdm2-p53 tumor suppressor pathway. Blood. 2010;115(7):1385–1393. doi: 10.1182/blood-2009-05-222166. [DOI] [PubMed] [Google Scholar]
- 8.Trageser D, Iacobucci I, Nahar R, et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206(8):1739–1753. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duy C, Yu JJ, Nahar R, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207(6):1209–1221. doi: 10.1084/jem.20091299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duy C, Hurtz C, Shojaee S, et al. BCL6 enables survival of Ph+ acute lymphoblastic leukemia cells upon BCR-ABL1 kinase inhibition. Nature. 2011;473(7347):384–388. doi: 10.1038/nature09883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye BH, Lista F, Lo CF, et al. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 1993;262(5134):747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 12.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432(7017):635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 13.Schuh W, Meister S, Herrmann K, Bradl H, Jack HM. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol Immunol. 2008;45(2):362–375. doi: 10.1016/j.molimm.2007.06.154. [DOI] [PubMed] [Google Scholar]
- 14.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 15.Herzog S, Hug E, Meixlsperger S, et al. SLP-65 regulates immunoglobulin light chain gene recombination through the PI(3)K-PKB-Foxo pathway. Nat Immunol. 2008;9(6):623–631. doi: 10.1038/ni.1616. [DOI] [PubMed] [Google Scholar]
- 16.Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13(2):199–212. doi: 10.1016/s1074-7613(00)00020-0. [DOI] [PubMed] [Google Scholar]
- 17.Ma S, Pathak S, Mandal M, Trinh L, Clark MR, Lu R. Ikaros and Aiolos inhibit pre-B-cell proliferation by directly suppressing c-Myc expression. Mol Cell Biol. 2010;30(17):4149–4158. doi: 10.1128/MCB.00224-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.