Abstract
Independent component analysis (ICA) is a popular method for the analysis of functional magnetic resonance imaging (fMRI) signals that is capable of revealing connected brain systems of functional significance. To be computationally tractable, estimating the independent components (ICs) inevitably requires one or more dimension reduction steps. Whereas most algorithms perform such reductions in the time domain, the input data are much more extensive in the spatial domain, and there is broad consensus that the brain obeys rules of localization of function into regions that are smaller in number than the number of voxels in a brain image. These functional units apparently reorganize dynamically into networks under different task conditions. Here we develop a new approach to ICA, producing group results by bagging and clustering over hundreds of pooled single-subject ICA results that have been projected to a lower-dimensional subspace. Averages of anatomically based regions are used to compress the single subject-ICA results prior to clustering and resampling via bagging. The computational advantages of this approach make it possible to perform group-level analyses on datasets consisting of hundreds of scan sessions by combining the results of within-subject analysis, while retaining the theoretical advantage of mimicking what is known of the functional organization of the brain. The result is a compact set of spatial activity patterns that are common and stable across scan sessions and across individuals. Such representations may be used in the context of statistical pattern recognition supporting real-time state classification.
Keywords: fMRI, group ICA, bagging, clustering, bootstrap
I. INTRODUCTION
Functional magnetic resonance imaging (fMRI) is a 4D brain imaging method that captures snapshots of blood flow changes by detecting changes in the blood oxygenation level-dependent (BOLD) signal (Friston et al., 1995; Beckmann et al., 2006), an indirect measure of neuronal activations. Scans are costly to obtain, so analysis frequently involves small-sample studies where data compression and feature selection are required to separate signal from noise. The traditional approach to fMRI data analysis is explicitly model driven, using the general linear model (GLM; Friston et al., 1995) where the temporal fluctuations in MRI signal are compared statistically to time-course models of the stimuli or task conditions:
where Y is the fMRI data (x, y, z, and time), Xn is the task condition, n, timing convolved with the BOLD response function, β̂n is a column vector representing the estimated signal change for task condition, and ε is the residuals, or error. There are many limitations to fMRI when used in this manner: (1) specific hypotheses or explicit assumptions about the spatial, temporal characteristics of the fMRI data are required, and the results are therefore only confirmatory in nature, (2) the presence of noise (movement, changes in background noise, etc.) may either bias the parameter estimates or add to the residual error, (3) although the method might expose multiple areas of altered signal during a task, the functional roles of these regions as components of a system are difficult to infer from the brain “mapping” data and, (4) predictive accuracy of results detected in this way is largely untested—extrapolation to population results depends critically on hidden features such as the sphericity of the signal distribution (Friston et al., 1995; Beckmann et al., 2006; Poldrack, 2006).
Over the past several years, independent component analysis (ICA) has emerged as a popular model-free alternative for fMRI analysis that may expose unexpected properties of the data during task-driven activity or that may be useful in task-free situations where an a priori model based on an expected time course is not feasible (McKeown et al., 1998; Beckmann and Smith, 2005; Damoiseaux et al., 2006; Calhoun et al., 2009). Most recently, investigators have explored techniques that demonstrate the contributions of multiple brain regions to dynamic processes such as cognition. These studies typically operate within a single scan of a subject or perform simultaneous analysis over small groups of scan using high degrees of dimension reduction.
Our goal in this study is to develop a computationally efficient method of applying ICA to large datasets consisting of heterogeneous subject populations that avoids many of the limitations of existing ICA methods and which capitalizes on prior knowledge of the functional organization of the human brain. We begin by discussing the theories and limitations of ICA in brain imaging. Note that as ICA is sometimes utilized for machine learning, we will at times discuss methodological issues related to classification of either populations or mental states.
In the fMRI model, ICA is written as a decomposition of a data matrix XM × P into a product of statistically independent spatial maps Sn × P with a mixing matrix AM × N, given by X = AS where P is the spatial dimension of the data or number of voxels being observed, M is the length of time being sampled, and n is the number of components to be estimated (Hyvärinen et al., 2000). The spatial maps produced by the ICA estimation process are taken to be sources that mix linearly to produce neural activity. Because the algorithm is an optimization procedure, the results can vary because of dimension reduction levels used, selection of the optimizations procedure, and exogenous factors such as patient variability and composition (Ylipaavalniemi et al., 2008).
A. Sources of Variation in the ICA process
The ICA estimation process typically proceeds with a whitening and dimension reduction step through principal components analysis (P4CA), where the full data is projected onto its principal eigenvectors obtained through singular value decomposition X = U Σ V′. The data are then projected by Xreduced = U′X onto the N eigenvectors with the largest eigenvalues, and consequently are transformed from dimension M × P to dimension N × P, where M is the length of the time dimension and P is the number of voxels. The ICA algorithm is then run on the whitened data according to Xreduced = AS. The actual estimation can be performed by maximizing the non-Gaussianity of the projected data s = Wx, where xn × 1 is an N-dimensional (PCA reduced) observation, sn × 1 is a projection of the time series onto the space of n ≤ N underlying sources, and W ≈ A−1 is the pseudo-inverse of the mixing matrix defining the data Xreduced = AS. This compression limits the number of components to be estimated within the mixing matrix A, enhances computational feasibility, and reduces noise by removing components that explain little of its variance. However, this comes at the expense of increasing the bias in the estimation of the parameter maps by effectively lowering the sample size from which the high-dimensional random variables, the spatial maps, are estimated, an example of the “bias-variance tradeoff.”
The ICA algorithms are not closed-form equations; thus, they typically seek to estimate the components by optimizing a chosen objective function (Hyvärinen et al., 2000, Ylipaavalniemi et al., 2008). Because of this, they can be sensitive not just to noise in the data but also to variations in initial starting parameters that lead to local, rather than global, function optimization. There are multiple alternatives to define the objective function, such as minimizing mutual information or maximizing the negentropy or maximum likelihood. The individual criterion may have functional choices or parameter selection steps within them. In addition, choices must be made for whether the components should be estimated simultaneously or individually (deflation). Collectively, high levels of dimension reduction with PCA and the randomness within the optimization procedures can lead to variations in the estimated random variables, the spatial maps.
These sources of variation can be mitigated with the help of resampling methods. Permutation tests, cross-validation, and the bootstrap are established techniques to validate existing models or to estimate parameters of interest more precisely (Good, 1999; Yu, 2003). In the bootstrap, the data is resampled with replacement to estimate parameters or estimate the stability of parametric estimates (Efron, 1979; Efron and Tibshirani, 1994). In bagging, bootstrapped samples of the data are used to estimate the parameter of interest many times (Breiman, 1994; Ylipaavalniemi et al., 2008). The parameter estimates are then pooled and averaged to obtain a final estimate giving increased accuracy.
B. Single-Subject ICA
As methods developed for within-subject ICA analyses likely generalize to the group level, we review some of the relevant literature here. Resampling methods such as the bootstrap and clustering methods have been used to increase the stability of the ICA results, as the algorithm itself is a stochastic optimization procedure. Because of the computational costs of these techniques, they usually are applied to single-subject analysis, where the decomposition of the scans into spatial maps is performed within each scan, rather than across multiple scans or subjects. Single-subject ICA results have been clustered using self-organizing maps, where the similarity metric among components was taken to be a linear combination of both the spatial and temporal correlations of the time series (Esposito et al., 2005). This similarity matrix was then converted to a distance metric and the components were clustered using a supervised hierarchical clustering algorithm. By analyzing data within single subjects during an auditory task, consistent components were obtained by combining bootstrapping of data along with a hierarchical clustering algorithm (Ylipaavalniemi et al., 2008).
The technique known as ICASSO (Himberg et al., 2004) uses resampling methods to establish stable components within a single subject. It has been applied to group analysis as well for small sample sizes. Bootstrapping of the data is combined with perturbations in the initial starting condition of the ICA algorithm. The results from multiple iterations of ICA are then clustered using agglomerative clustering. Because of the heavy resampling, the results from ICASSO are not as sensitive to optimizing merely the local minima of fitting criterion, as are methods that perform the ICA estimation only once. Li et al. (2007) used the information theoretic criterion of the entropy rate to establish “independent” spatial regions. These regions were sub-sampled within and among individual scans to determine the number of scans present within a single subject. The subsampled data for each subject was clustered using ICASSO where it was observed that after between 20 and 30 components have been isolated, the stability of the clusters decreased sharply.
C. Group ICA
These single-subject ICA procedures have been extended to simultaneously analyze groups of scans, providing inference on common behavior across multiple runs or subject groups through group ICA (gICA). The gICA methods typically stack multiple fMRI scans across J subjects to obtain decomposition XMJ × P = AMJ × N SN × P. These gICA methods extract spatial maps SN × P common across subjects or runs. The scientific significance of such analyses at the group level is extremely high, and the results of such analyses have nominated a set of functionally plausible networks that can be interpreted, based on existing experimental results, in terms of cognitive process (Beckmann and Smith, 2004; Calhoun et al., 2009; Smith et al., 2009).
The best-studied approach to gICA is based on temporal concatenation of the time series of multiple subjects (Beckmann and Smith, 2005; Calhoun et al., 2009; Smith et al., 2009). In this case, the data first undergo a spatial normalization step that brings the individual brains into alignment, with an initial temporal dimension reduction by PCA, followed by ICA decomposition on the dimension-reduced and spatially transformed data. This analysis effectively recovers the components that survive a statistical thresholding procedure by virtue of their large magnitude, their frequency of occurrence (because the variance term is reduced) or, perhaps, their tendency to occur in isolation, which effectively reduces the noise term. Those components that occur in a minority of subjects are less likely to appear.
Damoiseaux et al. (2006) extended resampling techniques to group analysis by performing tensor probabilistic ICA along with bootstrapping of resting-state fMRI data, identifying 10 stable resting-state components that corresponded to identifiable phenomena. To allow operation on larger sample sizes, Calhoun et al. (2009) implemented a group-ICA procedure involving two steps of dimension reduction. The temporal dimensionality was reduced using PCA, then pooled over subjects, and then an additional PCA was applied prior to ICA estimation. Throughout this procedure the spatial dimensionality remained intact. Within-subject estimates of the spatial maps and time courses were recovered by multiplying by the inverse of the dimension-reducing matrices.
Conceptually, PCA works to determine the directions that account for the maximal variance within the data, by finding the eigenvectors of the covariance matrix that have the maximal eigen-values. When the objective is to discriminate between groups, PCA decomposition may be suboptimal because it would blur the differences between the groups by projecting the data upon the axis on which they are most similar. Methods such as Fisher’s linear discriminate find projections that instead maximize the differences between classes but are inefficient for data compression (Martinez et al., 2004).
Group ICA imposes a constraint that the spatial maps are common across subjects. However, the heterogeneity of neural activation across subject may be a defining characteristic of a disorder, so imposing common spatial components across different scans may blur the differences between patients and controls by requiring not only that the individual maps to be identical among different patients but also requiring that the spatial maps be universally present. The blind application of gICA methods may partition data into components that have no functional meaning and may split functionally connected components somewhat arbitrarily in the presence of noise. This has been shown to occur when ICA is performed at overestimated orders (Li et al., 2007). One population may move more during scanning than another, resulting in significant group differences but not results containing relevant neurobiological information. Running ICA on multiple fMRI scans may produce biased results if the initial starting conditions favor a particular patient group or session. Hence, gICA results may have greater variation than single-subject results, because the spatial components are optimized to best fit the pooled data. The total group variation is minimized at the expense of the best representation of the individual subjects’ spatial maps. We believe on a theoretical basis that the methods we develop here may suffer less from these limitations than those discussed above, although we do not present data to that effect in this article.
D. Bagged Clustering
We present here a method to generate group-wide independent components (ICs) using bagged clustering over single-subject ICA results from hundreds of scans. To do this, we first perform single-subject ICA and then align the resulting ICs into a common atlas space. The ICs correspond to the vectors si ∈ S, the set of spatial maps estimated during the single-subject ICA estimation procedure. We then project the aligned, individual ICs into a lower dimension by averaging over anatomically (and presumably functionally) constrained regions of interest (ROIs) defined by the Harvard-Oxford cortical and subcortical structural atlases. The spatially reduced ICs are pooled and clustered using bagging with k-means clustering. The bagged clustering is performed by first resampling 500 times from the pooled ICs with replacement using a predetermined number of clusters. The centroids from each iteration are then pooled and clustered again according to the bagging model, and the final centroids are taken to be representative of components universally present across the original data. The centroids are analogous to the intersection of all spatial components across all the data, as opposed to the union.
Here, these methods are applied to a large dataset, scanned using fMRI over two sessions during a cue-induced craving task to create our dictionary of group-wide ICs using data from both pre- and post-treatment sessions. Results are also compared to those obtained using the GLM in analyses reported using these data by Culbertson et al. (accepted for publication). We hypothesized that clusters would include key networks involved in craving and vision. Cue-induced cigarette craving is associated with enhanced brain activation in cortical and subcortical regions associated with attention (e.g., prefrontal cortex, especially medial and orbito-frontal cortices), emotion (e.g., amygdala, insula), reward (ventral tegmental area, ventral striatum), and motivation (striatum) (Brody, 2006; McClernon et al., 2009; Sharma et al., 2009; Culbertson et al., accepted for publication). The visual system is well known to be located in the posterior parts of the brain, with primary visual areas located in the posterior-most part of the occipital lobe (e.g., calcarine sulcus) and secondary visual areas in lateral occipital and parietal cortices. We also hypothesized that noise-related ICs would not contribute significantly to our clusters based in part on our averaging data across neurologically relevant neuroanatomical regions. Examples of noise-related ICs would include those associated with motion artifact, which in some cases can differ between conditions and subjects populations.
E. Anatomical Summarization
We argue that reducing the spatial domain is based rationally on knowledge of brain organization. It has long been recognized that the neocortex is made up of architectonically distinct regions responsible for the different brain functions. The original atlas of Brodmann identified 52 such regions (Brodmann, 1909/1994) each of which is thought to be specialized for certain functions. In this work, we use as spatial dimensions a series of accepted functional regions as instantiated in the Harvard–Oxford brain atlas. This atlas identifies 55 bilateral (55 × 2 hemispheres 5 110 regions) brain regions based on anatomical topology and includes both deep brain nuclei and neocortex. However, modern thinking is that while such regions may have separable individual functions, complex behavior requires the integrated actions of multiple primitive regions. Producing the complete range of human behavior depends on the dynamic groupings and regroupings of these brain regions in a coordinated manner to form networks. Each dynamic network may well be responsible for processing different aspects of tasks or behaviors. For example, visual thinking may engage both the classical early visual areas BA17, BA18, and BA19, as well as inferior temporal regions and prefrontal cortex. Appetitive behaviors might engage primarily deeper structures including the basal ganglia and medial forebrain bundle in addition to inferior frontal areas. More complex behaviors (e.g., modulating responsiveness to craving-related visual cues) might simultaneously engage the appetitive and visual systems. In our model, the regions themselves are anatomically static, whereas the networks are not.
The mathematical methods of ICA and clustering provide leverage to identify these dynamic networks—an interpretation consistent with the successful work of others in this field (Beckmann and Smith, 2004; Calhoun et al., 2009; Smith et al., 2009). With these ideas in mind, we propose that the intrinsic dimensionality of the brain can be studied in five dimensions, with the spatial dimension being relatively sparse.
II. METHODS
A. Subject Data
As part of a wider, placebo-controlled, study aimed at understanding the treatment effects of both group therapy and treatment with the medication bupropion HCl (only the relevant outlines of the subject population, treatment and experimental protocol are noted here, see Culbertson, 2010 for a more detailed description), 51 smokers were studied during fMRI scanning while performing a visually presented cue-induced cigarette craving induction task. Included in these analyses were 19 placebo-treated (mean age 40 years; SD 13; five female), 14 bupropion-treated (mean age 41 years; SD 10; three female), and 18 group therapy-treated participants (mean age 47 years; SD 10; six female), each scanned before and after 8 weeks of treatment.
B. Scanning
Briefly, participants were presented with three separate runs of the cue-induced craving task in each of two sessions. In each run, participants were presented with 45 s neutral or cigarette-related videotaped cues. The cigarette-related cue videos were presented twice during each run with participants being instructed either to allow themselves to crave or to resist craving.
C. Preprocessing
We preprocessed the fMRI scans using standard methods within the FMRIB FSL (FMRI Statistical Library; http://www.fmrib.ox.ac.uk/fsl; Jenkinson et al., 2002). Motion correction was performed using MCFLIRT, and we used model-free FMRI analysis using probabilistic ICA (MELODIC) to detect ICs within each scanning session for each subject, resulting in 21,256 ICs across all subjects. We used FLIRT to align these found ICs into Montreal Neurological Institute (MNI) space.
D. Data Compression
We segmented and averaged the magnitude of each IC over 110 anatomical ROIs specified by the probabilistic Harvard–Oxford cortical and subcortical structural atlases provided with FSL (Desikan et al., 2006). The regions were originally in MNI* space and transformed into individual scan native space before averaging, thereby compacting the 81,920 voxel dimensions into a compact 110 ROI dimension vector. Because the signs (positive or negative) of the ICs are assigned arbitrarily, we then align all ICs into a common geometric (not anatomical) “hemisphere” by choosing a single IC vector as a reference and by flipping the signs of all other ICs so that they each are of minimal distance to the reference IC. This is conceptually similar to taking the absolute value of a number, so that the total magnitude is the summary of the variable and not the direction (either positive or negative).
E. Dictionary Creation
To create common ICs across individuals, we used bagged k-means clustering. A cluster can be represented by its centroid: the arithmetic average of all its members. In all, 21,256 compressed ICs were resampled with replacement, and this resampled data was then clustered into five groups. The number of components was determined for each subject uniquely using the Laplace Approximation to the Model Order (Minka et al., 2000). The centroids were then set aside. This was performed 500 times, and the 2500 centroids from the 500 runs were then clustered into five groups. The final centroids were then taken as the “dictionary” of ICs as an example of the most general, stable components representative of all the ICs.
F. General Linear Model Analysis
To compare results obtained using ICA-based methods with results using a GLM model-based approach, data were analyzed using the same methods as described by Culbertson et al. (accepted for publication). We used data from the first session (pretreatment). The contrast of interest here was visual cue-induced craving relative to rest (no cue). Variability in individual craving levels, obtained using self-report (on an analog scale of 1–5) were modeled and regressed out.
G. Determining the Number of Independent Components’ Dictionary Elements
Given the set of unlabeled data in feature space, we used two clustering methods to determine the intrinsic number of IC dictionary elements common across all subjects. First, we performed an iterative estimation using the expectation maximization (EM) algorithm for maximum likelihood clustering using a Gaussian mixture model with initial values from agglomerative hierarchical clustering (Fraley and Raftery, 1998). Additional clusters are iteratively considered until addition of new clusters no longer improves the model, as assessed by the Bayesian information criterion. Using the EM algorithm we found that four clusters were optimal for our IC data. Second, we applied the density-based spatial clustering of applications with noise (DBSCAN) clustering algorithm, which finds clusters based on how density-reachable and density-connected data elements are in feature space (Ester et al., 1996). DBSCAN found six clusters to be optimal. We therefore selected five as the number of common IC dictionary elements across subjects. This is consistent with Douglas et al. (accepted for publication), who found that 5.62 ICs were optimal for discriminating between belief and disbelief cognitive states across six different classification algorithms.
III. RESULTS
Using the clustering routine, every IC within a subject is categorized as belonging to one cluster, where the centroid of that cluster is taken to be the prototype of the activity over all ROIs. The discovered group ICs involved brain regions we a priori expected to be components of functional networks associated with the craving (e.g., ventral striatum, latral and medial orbito-frontal gyri, cingulate gyrus, insula, amygdala) and vision (e.g., calcarine sulcus, occipital pole, lateral occipital gyrus, precuneus, cuneus, lingual gyrus) based on prior literature (Fig. 1). Below, we list the relative magnitudes of each component within each a priori region listed above. Low and high relative magnitudes refer to those below and above the mean, respectively. If either hemisphere demonstrates high relative magnitudes, they are designated as high.
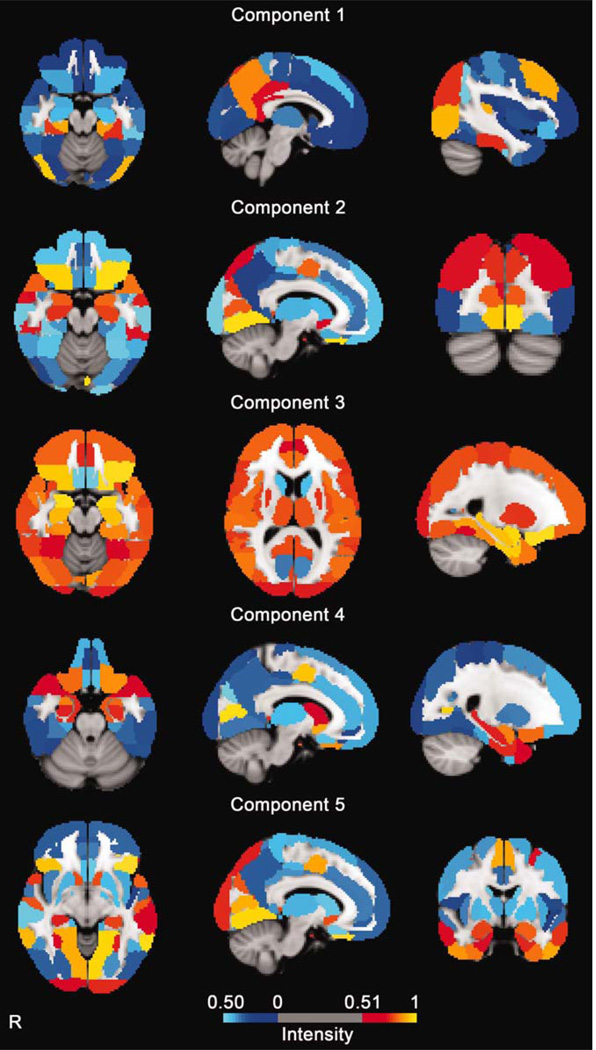
Figure 1.
Clusters discovered using ICA showing common components found in smokers who performed visual cue-induced craving, crave resist, and neutral control tasks while undergoing fMRI scanning. The maps represent in color the relative weighting of each of 110 brain regions expressed in the components. The amplitude values are expressed in normalized units. Areas of the brain containing principally myelinated axonal fibers (as opposed to neuronal cell bodies) are not shown in color and were not entered into the analyses. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
A. Craving Networks
Ventral striatum—[Component 1 (C1): magnitude low; C2: magnitude high; C3: magnitude low; C4: magnitude high; and C5: magnitude high].
Lateral orbito-frontal gyrus—(C1: magnitude low; C2: magnitude high; C3: magnitude high; C4: magnitude high; and C5: magnitude high).
Medial orbito-frontal gyrus—(C1: magnitude low; C2: magnitude low; C3: magnitude high; C4: magnitude low; and C5: magnitude low).
Cingulate gyrus—(C1: magnitude high; C2: magnitude low; C3: magnitude high; C4: magnitude low; and C5: magnitude low).
Insula—(C1: magnitude low; C2: magnitude low; C3: magnitude high; C4: magnitude low; and C5: magnitude low).
Amygdala—(C1: magnitude low; C2: magnitude high; C3: magnitude high; C4:magnitude high; and C5:magnitude high).
Striatum (caudate)–(C1: magnitude low; C2: magnitude low; C3: magnitude low; C4: magnitude high; and C5: magnitude low).
Striatum (putamen)—(C1: magnitude low; C2: magnitude low; C3: magnitude high; C4: magnitude; and C5: magnitude low).
B. Vision Networks
Calcarine sulcus (V1)—(C1: magnitude low; C2: magnitude high; C3: magnitude low; C4: magnitude high; and C5: magnitude high).
Occipital pole—(C1: magnitude low; C2: magnitude low; C3: magnitude high; C4: magnitude low; and C5: magnitude high).
Lateral occipital gyrus—(C1: magnitude high; C2: magnitude high; C3: magnitude high; C4: magnitude low; and C5: magnitude high).
Lingual gyrus—(C1: magnitude low; C2: magnitude high; C3: magnitude high; C4: magnitude low; and C5: magnitude high).
Precuneus—(C1: magnitude high; C2: magnitude low; C3: magnitude high; C4: magnitude low; and C5: magnitude low).
Cuneus—(C1: magnitude low; C2: magnitude high; C3: magnitude high; C4: magnitude low; and C5: magnitude high).
C. The General Linear Model
We hypothesized that we would also see similarities between results obtained contrasting video smoking-cue related brain activity with rest using the GLM and those found using our ICA clustering approach. We found the following (Fig. 2).
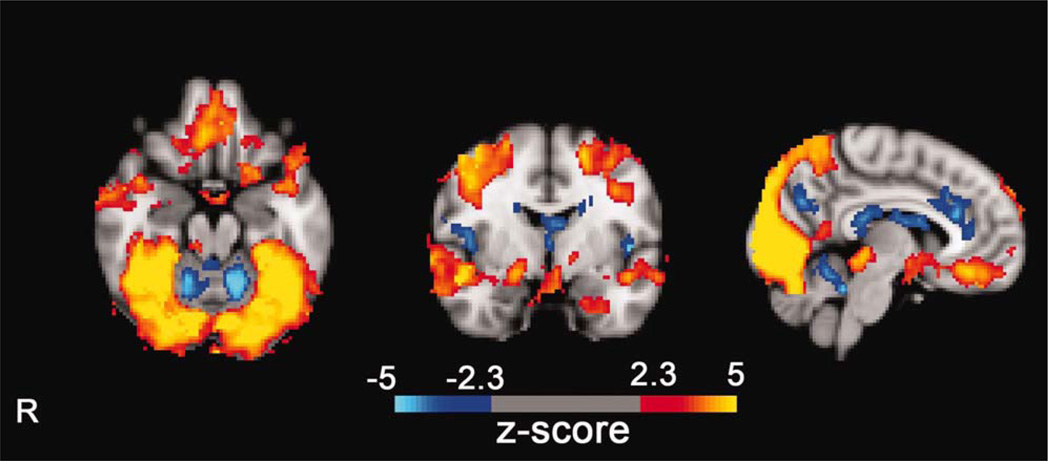
Figure 2.
Results using the standard univariate generalized linear modeling method to find areas of the brain active in smokers undergoing an fMRI scan. The results show the signal difference between a visual cue-induced craving task and a resting baseline. By convention, the color scale indicates z scores derived from t-statistics in the task contrast. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
C.1. Craving Networks
Results obtained contrasting the visual cue-induced craving condition with rest were similar to those obtained overall using ICA clustering, although involvement of ROIs was spread across five separate clusters rather than in a single statistical map, as was obtained using the GLM. One exception in the craving network was the putamen, in which we did not find significant involvement with visual cue-induced craving using the GLM, but did using ICA clustering. Another was the cuneus, part of the a priori vision network, which we also did not find significant involvement during visual cue induced craving using the GLM, but did using ICA clustering.
Ventral striatum—significant positive activity.
Lateral orbito-frontal gyrus—significant positive activity.
Medial orbito-frontal gyus—significant positive activity.
Cingulate gyrus—significant negative activity.
Insula—significant negative activity.
Amygdala—significant positive activity.
Striatum (caudate)—significant positive activity.
Striatum (putamen)—no significant activity.
C.2. Vision Networks
Calcarine sulcus (V1)—significant positive activity.
Occipital pole—significant positive activity.
Lateral occipital gyrus—significant positive activity.
Lingual gyrus—significant positive activity.
Precuneus—significant positive activity.
Cuneus—no significant activity.
IV. DISCUSSION
A. Advantages of Pooling Single-Subject ICA Results Via Clustering
Theoretically, ICA methods are constrained by their sensitivity to noise, the variance in the optimization results, and the computational limits on the amount of data they can handle (Ylipaavalniemi et al., 2008). These issues are exacerbated in gICA, where noise sources can vary across subjects, parameter choices can bias results in favor of a scan condition, and the large data sizes can make computation difficult without imposing dimension reduction first.
Standard gICA methods impose assumptions on the data that may be unrealistic; when analyzing large sets of scans that have different treatment groups, common spatial maps may not exist, and ill-chosen starting parameters may allow stochastic optimization in favor of a particular treatment group. While PCA is very useful for data reduction, and applied prior to ICA by many of the commonly used ICA algorithms (including methods implemented here), over-reducing data with PCA can be problematic. When data are reduced multiple times using PCA prior to gICA, the risk for bias in the estimates increases. In addition, temporal dimension reduction is inherently less efficient than summary statistics of spatial regions, as space occupies more dimensions than time.
When the ICs are projected into ROI space, the computational complexity of the k-means algorithm decreases exponentially. The complexity of k-means, O(nmk+1 log n), is a function of the number of observations n, the number of clusters k, and the dimensionality of the data, m. Because we seek to cluster such a large number of observations (21,256), reducing the dimensionality of the observations from 81,920 voxels to 110 ROIs drastically reduces the complexity by a factor of 21,25681,810. Performing spatial data compression is also more advantageous because it allows more drastic compression than temporal reduction. As there are three spatial dimensions, their size is order of magnitudes larger than the number of time points they describe. Because of this, the spatial dimension is more practical to reduce as more redundancies would exist, and the k-means algorithm is able to estimate more precisely the lower-dimension centroids.
Performing clustering on ICs whose dimensionality has been reduced using ROI averages offers an interpretable means of compressing unwieldy spatial dimensions in fMRI data. Single-subject ICA has fewer assumptions of homogeneity and requires less dimension reduction prior to ICA estimation. More practically, the generalizations of homogeneous spatial responses to stimuli may not be suitable for studies where the architectures of the reactions may differ because of a treatment or a disease. Clustering these single-subject results allow the components individually to nominate clusters, instead of stipulating that a common set of components exist across a potentially variant group of scans.
B. Evaluating the Number of Independent Components’ Dictionary Elements
Determining the intrinsic structure, or number of clusters, in a dataset is difficult when only unlabeled data are available. The methods section above (“Determining the Number of Independent Components’ Dictionary Elements”) describes our approach in estimating the intrinsic structure of the data. We had an a priori belief that there would be at least five general components existing, as previous work by Douglas et al. (accepted for publication) had shown that 5.62 ICs were optimal for discriminating between belief and disbelief cognitive states averaged across six different machine classification algorithms. Therefore, we set the initial number of clusters within each bootstrapped run to be k = 5, and determined convergence of the clustering during the process by evaluating whether the number of clusters chosen during the iterative process was the same as the number of clusters estimated using the clustering of the pooled data. Note that five to six clusters was a starting point based on our stimulus and population parameters, but more may be estimated for a variety of reasons. For example, if one had a patient population and expected more brain networks to be engaged during a task in the patient than the control populations, it might be better to start the estimation with six or seven clusters. However, this determination does not mean that there exist only five functional networks common; rather, our initial parameter choice was based on a hypothesis that at least five common components would exist. This is analogous in determining the intersection of the subjects’ components instead of the union. Therefore, these results are presented as an efficient and compact way of describing the data but do not capture all the variation existing within it.
C. Interpretation of Centroids
The centroids are interpreted as a “dictionary” of cognitive states from which activation patterns were described. Conceptually these are a limited subset of all possible components that are most likely to be found across all subjects. We are particularly interested in the generalized use of identified functional networks as features in statistical pattern analysis and have shown that these networks can form a relatively sparse set of dimensions that achieve high classification accuracy (Anderson et al., 2010; Douglas et al., accepted for publication). For our intended applications in real-time classification, a small dimensional space consisting of cognitive networks likely to appear during normal behaviors helps to reduce classifier training by allowing a sparse description of activity.
Most importantly, to the extent that these functional networks are identifiable, the classification process also provides insight into the architecture of cognition by automatically nominating those systems that contribute to a given cognitive state, without imposing a prior model (e.g., Smith et al., 2009). By identifying the networks’ strength prospectively using a dictionary, this approach differs from the established implementations of ICA methods. The anatomical summarization allows stronger data compression than the typical temporal dimension reduction methods seen in many ICA methods, allowing the processing of larger numbers of scans than previously allowable. The dictionary’s creation can also potentially provide classification power in different populations performing tasks that induce similar mental states.
We consider our results as representative of the dynamic reorganization of the brain into processing modules responsible for different aspects of cognitive or behavioral processes as represented in the brain. Crudely, we conceive of the brain state at any moment as the superposition of the activities of multiple dynamic modules. Any given brain region likely participates in multiple functional modules depending on temporally local task demands, presumably resource limited by the capacity of the individual component regions. The need of the organism to operate concurrently on multiple tasks imposes the requirement that the functional modules are continuously engaged to varying degrees. The fact that we have shown good success in using IC dictionary elements as dimensions to form state classifiers (Anderson et al., 2009; Douglas et al., accepted for publication) supports our contention that the ICs represent functionally relevant systems and are in general support of our cognitive model based on superposed dynamic systems.
D. Biological Relevance of fMRI Independent Components as a Sparse Basis Set
To determine whether our approach was neurobiologically reasonable for finding functionally relevant networks using a model free approach, we used two metrics. First, we use a literature-based approach, where we identified regions that are thought to be part of two critical functional components of the task presented to these participants: craving and visual systems. We identified which of these regions were present in which of five components that resulted from our atlas-based clustering approach in aligning ICs across a large group. We found that generally our components did have the anticipated regional participation we expected to see. Craving-associated regions, such as the ventral striatum, medial and lateral orbito-frontal gyri, cingulate gyrus, amygdala, and to a lesser extent, the striatum and insula were present in some, if not the majority of the components discovered. Regions that play a part in the visual system, including the calcarine sulcus, occipital pole, lateral occipital gyrus, lingual gyrus, cuneus, and precuneus similarly were found in many of the components discovered. In fact, no component resulting from this method was absent at least a few regions in one or both of these networks.
The second metric for the similarity of our IC components was their comparison to the results of the GLM, contrasting visual cue-induced craving with rest (no cue). For the most part, we found that these regions also were present in the GLM analysis. We did not see strong insular signal in our IC results, but similarly, we also did not see positive activation within the insula in the GLM contrast described above. So, while we expected the insula, based on the literature, to play a greater role in networks found using our methods, our results are consistent with those found using the GLM. The few differences we found between the ICA clustering and GLM methods in craving (putamen) and visual (cuneus) systems could be due to the general difference of how the two methods are implemented here. The ICA clustering method looks for components in all of the data, which contained three subtly different conditions, and therefore may be picking up on networks engaged during one of the other conditions (i.e., visual cue-induced crave resist and neutral cue conditions). We elected to use only one of these conditions (visual cue induced craving) because we believed it to be the dominant and most interesting brain state, and did not want to overly complicate the analysis of results with too many contrasts. We also believe the single contrast along with our general findings within a priori ROIs support the hypothesis that this ICA clustering method produces neurobiologically reasonable and relevant findings.
There are certain clear limitations both in our cognitive model and in the implementation suggested here. First, the atlas regions are arguably somewhat arbitrary, as they were not nominated in any way by the individual subject data. These atlas regions were defined primarily on structural, rather than functional, features, yet are interpreted here as functional elements. On the other hand, the atlas regions are based on the accumulated evidence of literally thousands of studies that already give putative functional assignments to most of the atlas ROIs. We believe therefore that the use of the atlas is a principled means by which to bring prior knowledge into the exploratory data analysis methods. It is clear, too, that some of the atlas regions are large enough to support multiple, separable, functional subregions. In the future, for our method to be optimally successful as a tool for neuroscience discovery, it will be necessary to further refine the atlas elements that define our spatial dimensions.
From a cognitive science perspective, the notion that brain states are made up of a superposition of individual dynamic modules seems plausible, but the idea that this superposition should be linear is much less so. However, the structural model for ICA represents all such superpositions as linear. As such, it is on its own unlikely to form the basis for a very satisfying description of the full range of interesting cognitive states. However, under the small perturbations that are present in the resting states, the presumption of first-order linearity may well be appropriate. Unfortunately, it is less likely to be so under the complex behavioral conditions such as cue-induced craving that were present in our experimental data. However, this problem of linearity is not intrinsic to our conceptual model. The latter might be extended to correct for higher order effects.
V. CONCLUSIONS
By performing ICA within each subject, and then pooling the results with clustering, less temporal dimension reduction is necessary, thus reducing the bias in the overall estimates. If ICA results are processed within each scan, there is no need to make assumptions about common functional networks operating across the group or identical patterns of physiological artifacts as motion. This method allows the ICA results to be reflective of the individual scan, and to adapt the spatial maps based on signal found only within that time period, for that unique subject. By performing bagged clustering, the pooled data are allowed to nominate the networks most generalizable across large numbers of scans. In this manner, the functional networks nominated through this process become more representative of the underlying cognitive processes; the signal (spatial map) is isolated uniquely for each subject, yet the common elements present across all scans are extracted without stipulating that every particular element be found within every scan. Cohesively, these methods differ from existing fMRI gICA methods by relaxing the assumptions of commonality while still allowing large-sample data analysis.
Projecting the individual ICs into a lower-dimensional space based on anatomically defined ROIs improves algorithmic efficiency and enables analysis of larger numbers of scan session results. By using the average IC magnitudes within-regions, our method may also be somewhat more robust against problems of movement and misregistration than voxel-based approaches. Averaging over ROIs avoids interpreting data from nonbiologically relevant components by suppressing noise and by using a priori functionally significant regions to reduce the data. The clusters contain centroids that are functionally plausible but may also be useful to identify artifacts such as motion or noise.
Resampling techniques lead to more stable results (Efron, 1979; Breiman, 1994; Ylipaavalniemi et al., 2008) and can help group-ICA methods that have multiple sources of variation within the data and the estimation procedures. Choices of starting parameters or the randomness of the ICA optimization procedure leads to variations in the final estimates. Performing heavy temporal dimension reduction can exacerbate this by estimating high-dimensional sources from low-dimension recordings. In addition, noise can exist in the scan conditions or even patient behavior within or between runs, deteriorating estimates even further with unpredictable physiological artifacts such as motion or respiration. Resampling helps create more stable estimates by averaging over this noise.
Although not the subject of this report, the more general aims of our current research are to develop practical means of real-time state classification using automated machine-learning methods based on meaningful dimensions of the input data. We propose using ICA on functional MRI input data to identify a set of basis functions to be used in subsequent analysis. We have shown in prior work that a small number of automatically detected ICs can be used as bases for clinical segmentation of neuropsychiatric populations (Anderson et al., 2010) or can be applied on a statement-by-statement basis to determine if a subject believes or disbelieves propositions (making an interesting form of lie detector (Douglas et al., accepted for publication). The literature on ICA in human imaging data suggests that the revealed components may express functionally meaningful networks. If so, it is possible to consider cognitive or clinical state classification based on ICs as suggesting that these states can be thought of as superpositions of the activities of functional networks that themselves may be identifiable based on prior knowledge of patterned regional activation. In other data acquisition methods, such as electrophysiology, there may be more parsimonious and meaningful basis functions that share the characteristics of reduced dimensionality driven by functionally meaningful components.
ACKNOWLEDGMENTS
We are very grateful for the training data for FIRST, particularly to David Kennedy at the CMA, and also to: Christian Haselgrove, Centre for Morphometric Analysis, Harvard; Bruce Fischl, Martinos Center for Biomedical Imaging, MGH; Janis Breeze and Jean Frazier, Child and Adolescent Neuropsychiatric Research Program, Cambridge Health Alliance; Larry Seidman and Jill Goldstein, Department of Psychiatry of Harvard Medical School; Barry Kosofsky, Weill Cornell Medical Center.
Grant sponsors: This work is supported by the National Institute on Drug Abuse under DA026109 to M.S.C and by WCU (World Class University) program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (R31-2008-000-10008-0) to AY. The grant funding that supported the data collection was: the National Institute on Drug Abuse (A.L.B. [R01 DA20872]), a Veterans Affairs Type I Merit Review Award (A.L.B.), and an endowment from the Richard Metzner Chair in Clinical Neuropharmacology (A.L.B.). We thank Michael Durnhofer for maintaining the systems necessary for these data analyses.
Footnotes
MNI (Montreal Neurological Institute) space defines a standardized diffeomorphic mapping of individual brains onto an atlas space defined by averaging the brains of a test population. This procedure is conventional in the brain imaging literature.
REFERENCES
- Anderson A, Dinov ID, Sherin JE, Quintana J, Yuille AL, Cohen MS. Classification of spatially unaligned fMRI scans. NeuroImage. 2010;49:2509–2519. doi: 10.1016/j.neuroimage.2009.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imaging. 2004;23:137–152. doi: 10.1109/TMI.2003.822821. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Smith SM. Tensorial extensions of independent component analysis for multisubject fMRI analysis. NeuroImage. 2005;25:294–311. doi: 10.1016/j.neuroimage.2004.10.043. [DOI] [PubMed] [Google Scholar]
- Beckmann CF, Jenkinson M, Woolrich MW, Behrens TEJ, Flitney DE, Devlin JT, Smith SM. Applying FSL to the FIAC data: Model-based and model-free analysis of voice and sentence repetition priming. Hum Brain Mapp. 2006;27:380–391. doi: 10.1002/hbm.20246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. Bagging predictors (Technical Report 421) Berkeley: University of California; 1994. [Google Scholar]
- Brody AL. Functional brain imaging of tobacco use and dependence. J Psychiatric Res. 2006;40:404–418. doi: 10.1016/j.jpsychires.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodmann K. Brodmann’s Localisation in the Cerebral Cortex. London, UK: Smith-Gordon; 1909/1994. [Google Scholar]
- Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. NeuroImage. 2009;45:163–172. doi: 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson CS, Bramen J, Cohen MS, London ED, Olmstead RE, Gan JJ, Costello MR, Shulenberger S, Mandelkern MA, Brody AL. Effect of bupropion treatment on brain activation induced by cigarette-related cues in smokers. Arch Gen Psychiatry. 2011 Jan 3; doi: 10.1001/archgenpsychiatry.2010.193. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Consistent resting-state networks across healthy subjects. Proc Natl Acad Sci USA. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Douglas PK, Harris S, Yuille A, Cohen MS. Performance comparison of machine learning algorithms and number of independent components used in fMRI decoding of belief vs. disbelief. doi: 10.1016/j.neuroimage.2010.11.002. (accepted for publication in Neuroimage) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Proceedings of the 2nd international conference on knowledge discovery and data mining pages; Portland, Oregon. 1996. pp. 226–231. [Google Scholar]
- Esposito F, Scarabino T, Hyvärinen A, Himberg J, Formisano E, Comani S, Tedeschi G, Goebel GR, Seifritz E, Disalle F. Independent component analysis of fMRI group studies by self-organizing clustering. NeuroImage. 2005;25:193–205. doi: 10.1016/j.neuroimage.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- Efron B. Bootstrap methods: Another look at the jackknife. Ann Stat. 1979;7:1–26. [Google Scholar]
- Fraley C, Raftery AE. How many clusters? Which clustering method? Answers via model-based cluster analysis. Comput J. 1998;41:578–588. [Google Scholar]
- Friston KJ, Frith CD, Frackowiak RS, Turner R. Characterizing dynamic brain responses with fMRI: A multivariate approach. Neuroimage. 1995;2:166–172. doi: 10.1006/nimg.1995.1019. [DOI] [PubMed] [Google Scholar]
- Good PI. Resampling Methods: A Practical Guide to Data Analysis. Birkhauser, Basel, Switzerland: 1999. [Google Scholar]
- Himberg J, Hyvärinen A, Espositeo F. Validating the independent components of neuroimaging time series via clustering and visualization. NeuroImage. 2004;22:1214–1222. doi: 10.1016/j.neuroimage.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Hyvärinen A, Oja E. Independent component analysis: Algorithms and applications. Neural Networks. 2000;13:411–430. doi: 10.1016/s0893-6080(00)00026-5. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Bannister PR, Brady JM, Smith SM. Improved optimisation for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Li YO, Adali T, Calhoun VD. Estimating the number of independent components for functional magnetic resonance imaging data. Hum Brain Mapp. 2007;28:1251–1266. doi: 10.1002/hbm.20359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez K. PCA versus LDA. IEEE Trans Pattern Anal Mach Intell. 2004;23:228–233. [Google Scholar]
- McClernon FJ. Neuroimaging of nicotine dependence: Key findings and application to the study of smoking-mental illness comorbidity. J Dual Diagn. 2009;5:168–178. doi: 10.1080/15504260902869204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ. T.J. Sejnowski analysis of fMRI data by blind separation into independent spatial components. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minka TP. Automatic choice of dimensionality for PCA. Tech. Rep., NIPS. 2000 [Google Scholar]
- Poldrack RA. Can cognitive processes be inferred from neuroimaging data? Trends Cogn Sci (Regul Ed) 2006;10:59–63. doi: 10.1016/j.tics.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Sharma A, Brody AL. In vivo brain imaging of human exposure to nicotine and tobacco. Handb Exp Pharmacol. 2009;192:145–171. doi: 10.1007/978-3-540-69248-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylipaavalniemi J, Vigário R. Analyzing consistency of independent components: An fMRI illustration. NeuroImage. 2008;39:169–180. doi: 10.1016/j.neuroimage.2007.08.027. [DOI] [PubMed] [Google Scholar]
- Yu CH. Resampling methods: Concepts, applications, and justification. Practical Assess Res Eval. 2003;8 [Google Scholar]