Abstract
Colorectal cancer is the second leading cause of death from cancer in the United States. Metastases in the liver, the most common metastatic site for colorectal cancer, are found in one-third of the patients who die of colorectal cancer. Currently, the genes and molecular mechanisms that are functionally critical in modulating colorectal cancer hepatic metastasis remain unclear. Here, we report our studies using functional selection in an orthotopic mouse model of colorectal cancer to identify a set of genes that play an important role in mediating colorectal cancer liver metastasis. These genes included APOBEC3G, CD133, LIPC, and S100P. Clinically, we found these genes to be highly expressed in a cohort of human hepatic metastasis and their primary colorectal tumors, suggesting that it might be possible to use these genes to predict the likelihood of hepatic metastasis. We have further revealed what we believe to be a novel mechanism in which APOBEC3G promotes colorectal cancer hepatic metastasis through inhibition of miR-29–mediated suppression of MMP2. Together, our data elucidate key factors and mechanisms involved in colorectal cancer liver metastasis, which could be potential targets for diagnosis and treatment.
Introduction
After lymph nodes, the liver is the most common site for colorectal cancer metastasis, and liver metastasis is a common cause of cancer-related mortality (1–4). Most colorectal cancer patients with hepatic metastasis are not candidates for surgical treatment, and their 5-year survival rate following diagnosis of hepatic metastasis is below 10% (2, 4). It is well established that 5-year survival rates exceed 90% in patients diagnosed with early stage colorectal cancer (5, 6). It is imperative that we uncover the underlying mechanisms and genetic alterations that predispose to the metastatic phenotype in colorectal cancer. Such an understanding has the potential to improve early detection and prevention in addition to helping with developing novel targeted therapies for late stage disease. Studies reveal that genomic instability in cancer cells leads to cellular heterogeneity, which may guide tumor cell aggression and specific organ colonization during the metastatic process (7, 8). Many studies have attempted to identify the metastasis-related genes in metastatic tumors from other types of cancer using global gene expression profiling analysis. For example, several genes have been identified to be involved in development of breast cancer metastasis to the lungs and brain, including Twist, Cox2, and MMP (9–11). Recent studies also revealed that MACC1, Notch and TGF-β/Smad signaling may be involved in the process of colon cancer liver metastasis (12–15). However, detailed molecular mechanisms that mediate colorectal cancer metastasis to liver have not been systemically characterized.
Results
Identification of a gene expression signature for colorectal cancer liver metastasis.
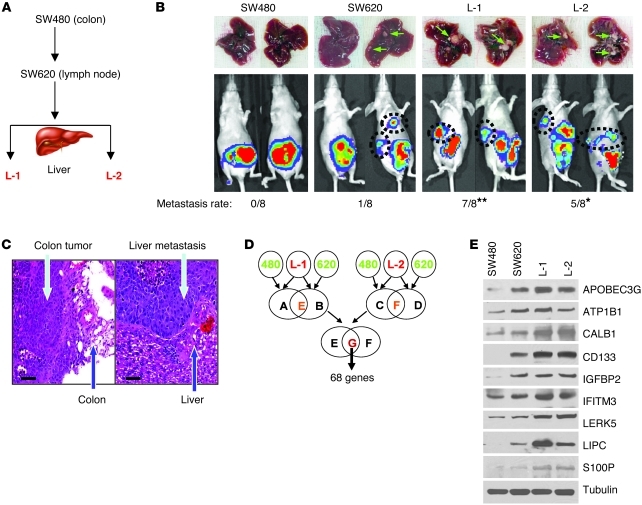
To study the genetic signatures that modulate human colorectal cancer hepatic metastasis, we performed a transcriptomic microarray analysis of the cell lines isolated from an orthotopic colorectal cancer liver metastasis animal model. The identified genes that potentially mediate liver metastasis were further validated by functional analysis both in vitro and in vivo and in human tumor tissues. First, we established an experimental platform using 2 well-known colorectal cancer cell lines, SW480 and SW620. These 2 cell lines were derived from different stages of colon cancer in the same patient; SW480 was isolated from the primary tumor, and SW620 was isolated from a lymph node metastasis (16). We first examined the capability of these 2 cell lines to form hepatic metastasis using an orthotopic colorectal cancer mouse model. SW480 failed to metastasize to the liver (0/8), whereas 1 case in the SW620 group (1/8) had 2 hepatic metastases. From these 2 hepatic metastases, we further established 2 cell lines (L-1 and L-2; Figure 1, A and B). After repeating the orthotopic experiments with L-1 and L-2, both cell lines showed significantly increased capability of forming hepatic metastasis (7/8 and 5/8, respectively; Figure 1B) compared with the parental SW620 cells. Representative histologic H&E staining of the orthotopic colon tumors and their hepatic metastasis was validated and is shown in Figure 1C. Thus, by in vivo selection, we successfully established a stable orthotopic colon cancer hepatic metastasis mouse model, which could provide highly metastatic cells in comparison with their parental cells under isogenic background.
Figure 1. Identifying a gene expression signature for colorectal cancer liver metastasis.
(A) Flow chart of in vivo selection process of colorectal cancer cell metastasis to the liver. The nonmetastatic colorectal cancer cells (SW480) and those with only lymph node metastasis (SW620) were inoculated into the colon of nude mice. Liver tumors were detected by the IVIS Imaging System (Caliper Life Sciences), and tumor cells (L-1 and L-2) were then isolated from the liver lesions and reinoculated to confirm their metastatic phenotype. (B) Representative images of the liver metastases (upper panels) and IVIS luciferase images in mice inoculated with colorectal cancer cells (lower panels). The hepatic metastasis rate of colorectal cancer cells is indicated at the bottom. *P < 0.05; **P < 0.01 compared with SW480 and SW620 cells. (C) Representative immunohistochemistry staining of colorectal primary tumor and hepatic metastases from the orthotopic transplanted nude mice. Scale bars: 50 μm. (D) Gene expression profiles reveal a set of 68 genes that are highly expressed in L-1 and L-2 cells as compared with SW480 and SW620 cells. (E) Immunoblotting analysis of gene expression levels in SW480, SW620, L-1, and L-2 cells.
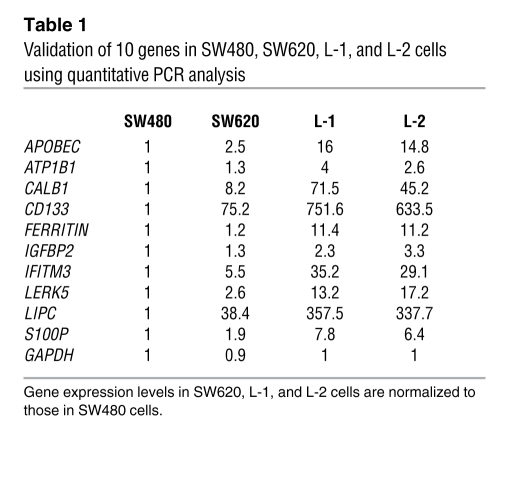
To identify genetic signatures that may be involved in colorectal cancer hepatic metastasis, we used an Affymetrix Human Genome U133A Array, which represents 14,500 well-characterized human genes to compare the gene expression profiles between the low metastatic (SW480 and SW620) and high metastatic (L-1 and L-2) cell lines. We identified 68 genes whose expression levels were induced in L-1 and L-2 cells when compared with SW480 and SW620 cells (Figure 1D, upregulated and downregulated genes; fold-change, UniGene ID, and P values are listed in Supplemental Tables 1 and 2; supplemental material available online with this article; doi: 10.1172/JCI45008DS1). Among these 68 genes, most were related to physiologic regulation (33%) and cellular metabolism (21%), and a few (2%) were associated with proliferation (Supplemental Figure 1A). To further narrow down the genes to be examined, we selected those genes from the microarray data with at least a 4-fold increase in their expression levels in the L-1 and L-2 cells when compared with both SW620 and SW480 cells; a total of 10 genes were identified (Supplemental Figure 1B). We then validated the expression of these 10 genes by RT-PCR (Supplemental Figure 2), semi-quantitative PCR (Table 1), and immunoblotting (Figure 1E). After validation of the mRNA and protein levels of the 10 genes, we excluded the ATP1B1 and IGFBP2 genes because their expression levels were not significantly increased in L-1 and L-2 cells compared with SW480 and SW620, and thus we focused on the functional examination of the remaining 8 genes.
Table 1 .
Validation of 10 genes in SW480, SW620, L-1, and L-2 cells using quantitative PCR analysis
Four genes are essential in mediating colorectal cancer liver metastasis.
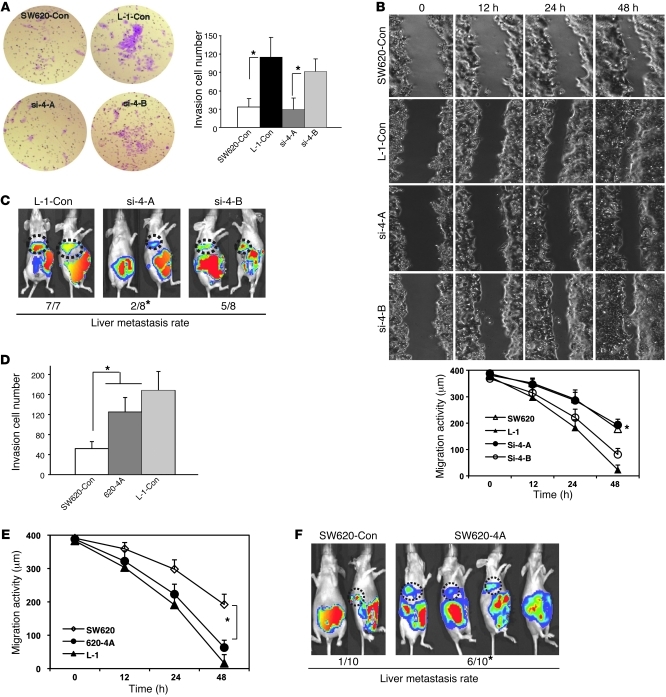
To further identify the genes that are functionally essential in mediating colorectal cancer hepatic metastasis, we generated retrovirus-encoded shRNAs (Supplemental Figure 3) and cDNA expression constructs to stably knock down or overexpress these target genes. We then examined their biological effects on metastasis. We randomly divided the 8 genes into 2 groups: group A (si-4-A), including APOBEC3G, CD133, LIPC, and S100P; and group B (si-4-B), including CALB1, IFITM3, FERRITIN, and LERK5. Four retroviral shRNA expression constructs against group A or group B genes were cointroduced into highly metastatic L-1 cells where their protein or mRNA levels were examined to validate the efficiency of the shRNAs (Supplemental Figure 4). Compared with SW620-control cells, knocking down group A genes suppressed the invasion and migration abilities of the L-1–control cells in vitro (Figure 2, A and B) and significantly decreased the frequency of hepatic metastasis in vivo (Figure 2C). In contrast, knocking down group B genes had no statistically significant effect on invasion, migration, and metastasis activity of L-1 cells (P > 0.05). These data suggest that group A contains 1 or multiple genes that can be crucial mediators for colorectal cancer hepatic metastasis.
Figure 2. Four genes are essential in mediating colorectal cancer liver metastasis.
(A) Images show the invasion activity of the hepatic metastasis L-1 cell line with knocked down group A and group B genes using retroviral shRNAs (si-4A, si-4B, upper panel). Cells migrating to the lower side of the Transwell filter were counted per HPF in 3 fields (lower panel). Each assay was repeated at least twice. Original magnification, ×200. *P < 0.05. (B) Images show the migration activity of the hepatic metastases L-1 cell line with knocked down group A genes using wound healing assay (upper panel). The distance of cell migration was calculated at 3 locations (lower panel). Con, control. (C) Representative images of the IVIS luciferase signal in the mice inoculated with colorectal cancer cells preinfected with 2 groups of retroviral shRNAs. The hepatic metastasis rate is indicated at the bottom. *P < 0.05 compared with L-1–control cells. (D) Invasion activity of the hepatic metastasis L-1 cell line overexpressing the group A genes. (E) Migration activity of the hepatic metastases L-1 cell line overexpressing the group A genes. (F) Representative images of the luciferase signal in mice inoculated with colorectal cancer cells expressing the group A genes. The hepatic metastasis rate is indicated at the bottom. *P < 0.05 compared with SW620-control cells. Bars show the mean value of the representative results from 3 experiments, each conducted in duplicate (± SD).
To further characterize whether overexpression of APOBEC3G, CD133, LIPC, and S100P are sufficient to induce hepatic metastasis, cDNA expression constructs expressing these 4 genes were introduced into low-metastatic SW620 cells. We found that overexpressing of these 4 genes increases the invasion and migration abilities of the SW620-control cells in vitro (Figure 2, D and E) and also significantly enhances the frequency of hepatic metastasis in vivo (Figure 2F). Together, these results suggest that APOBEC3G, CD133, LIPC, and/or S100P plays a role in promoting colorectal cancer hepatic metastasis.
Expression of the hepatic metastasis gene signature is enhanced in colorectal cancer with metastatic liver lesions.
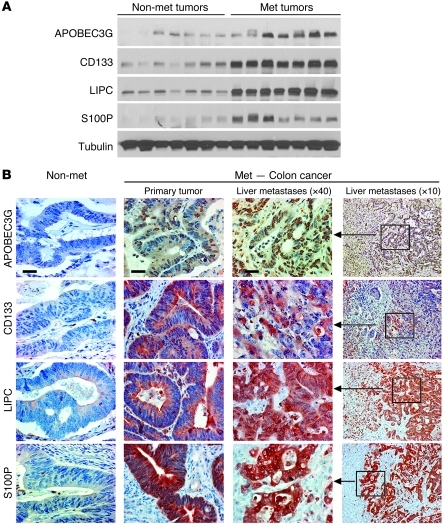
To determine the clinical correlation of our identified gene signatures with colorectal cancer hepatic metastasis, we examined the protein levels of APOBEC3G, CD133, LIPC, and S100P in 7 freshly isolated human colorectal cancer hepatic metastatic tumors and 7 nonmetastatic primary colorectal carcinomas. We showed that expression levels of these 4 genes are significantly increased in the metastatic tumors compared with the nonmetastatic primary tumors (Figure 3A).
Figure 3. Expression levels of the hepatic metastasis gene signature are enhanced in colorectal cancer with metastatic liver lesion.
(A) Protein expression levels of 4 genes were detected by immunoblotting in nonmetastatic human colorectal cancer samples and human colorectal cancer samples with hepatic metastases. Met, metastatic. (B) Representative images of protein expression levels of 4 genes in nonmetastatic human colorectal cancer samples and human colorectal cancer with hepatic metastases using immunohistochemistry staining. Scale bar: 50 μm.
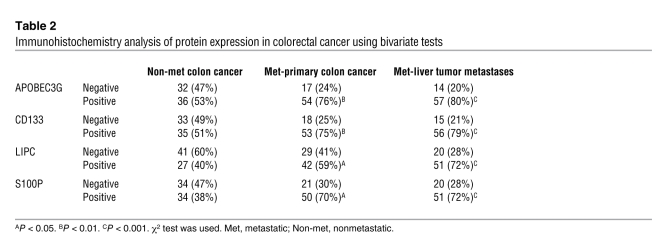
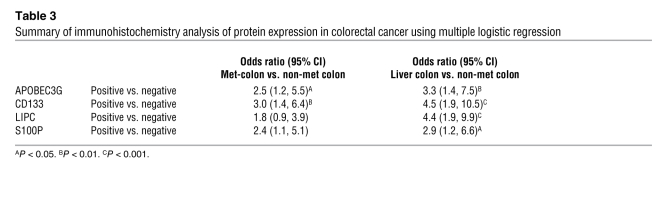
Furthermore, we performed immunohistochemical staining to investigate the protein expression levels of these 4 genes in 71 paired samples from human primary colorectal carcinomas along with their hepatic metastasis and 68 samples from the nonmetastatic primary colorectal carcinomas. The expression levels of all 4 genes were highly increased in the hepatic metastasis and their corresponding primary colorectal carcinomas compared with the nonmetastatic colorectal carcinomas; they were also significantly associated with metastatic colon cancer with liver metastasis (Figure 3B and Table 2). The results from multiple logistic regression models when all 4 genes entered the model (Table 3) simultaneously show that metastatic colon cancer patients were much more likely to have high expression of APOBEC3G (OR [Odds Ratio] = 2.5; 95% CI = 1.2–5.5), CD133 (OR = 3.0, 95% CI = 1.4–6.4), and S100P (OR = 2.4, 95% CI = 1.1–5.1) than the patients with no metastasis. Additionally, patients with liver metastasis were much more likely to have expression of all 4 genes (APOBEC3G: OR = 3.3, 95% CI = 1.4–7.5; CD133: OR = 4.5, 95% CI = 1.9–10.5; LIPC: OR = 2.4, 95% CI = 1.9–9.9; and S100P: OR = 4.9, 95% CI = 1.2–6.6) than the patients with no metastasis. Taken together, our data suggest that APOBEC3G, CD133, LIPC, and S100P are functionally essential for mediating colorectal cancer hepatic metastasis and may serve as signatures in primary colorectal carcinomas to predict a higher likelihood of recurrent cancer metastasis.
Table 2 .
Immunohistochemistry analysis of protein expression in colorectal cancer using bivariate tests
Table 3 .
Summary of immunohistochemistry analysis of protein expression in colorectal cancer using multiple logistic regression
APOBEC3G enhances colon cancer in vitro cell migration and invasion activity.
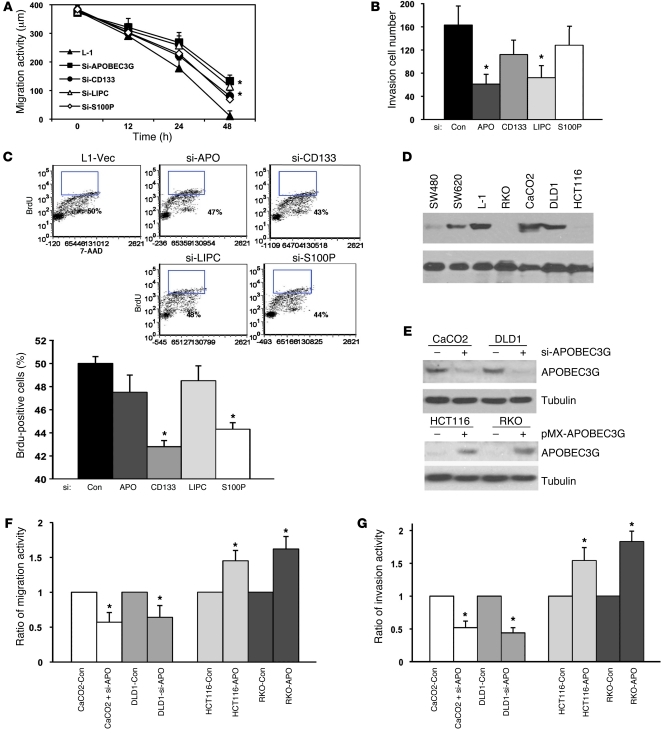
Next, we sought to determine which of these 4 genes (APOBEC3G, CD133, LIPC, and S100P) enhances colorectal cancer hepatic metastasis by affecting cancer cell migration and/or proliferation, 2 critical determinants of metastasis. Interestingly, knocking down either CD133 or S100P using shRNAs (si-CD133, si-S100P) substantially reduced proliferation of the metastatic cancer cells as measured by BrdU incorporation assay, with less significant suppression in cancer cell migration and invasion (Figure 4, A–C), suggesting that CD133 and S100P may mediate colorectal cancer hepatic metastasis mainly through enhancing the growth advantage of the metastatic cancer cells. Different from the effects of knocking down CD133 or S100P, knocking down either APOBEC3G or LIPC using shRNAs (si-APOBEC3G, si-LIPC) significantly inhibited in vitro migration and invasion of the L-1 cells with modest reduction in cell proliferation (Figure 4, A–C). However, knocking down either one of these genes was not sufficient to decrease the liver metastasis rate in the orthotopic animal model, if compared with knocking down all 4 genes (Supplemental Figure 5), indicating that the process of liver metastasis may require the cooperation/synergism of the 4 genes.
Figure 4. APOBEC3G enhances colon cancer cell invasion and migration.
(A) Cells were infected with retroviral shRNAs against specific targets as indicated (si-APOBEC3G, si-CD133, si-LIPC, si-S100P) and then subjected to cell migration assay. *P < 0.05 compared with L-1–control cells. (B) Cells were infected with retroviral shRNAs and then subjected to cell invasion assay. *P < 0.05 compared with L-1–control cells. APO, APOBEC3G. (C) Percentage of proliferative cells was determined by BrdU-FITC. *P < 0.05. (D) Immunoblotting of APOBEC3G in multiple colon cancer cell lines. (E) Immunoblotting of APOBEC3G expression (upper panel) in cells expressing retroviral APOBEC3G shRNA and pMX-APOBEC3G (lower panel). (F) Cells expressing APOBEC3G shRNA or pMX-APOBEC3G were subjected to cell migration assay. *P < 0.05 compared with control cells. (G) Cells expressing retroviral APOBEC3G shRNA or pMX-APOBEC3G were subjected to cell migration assay. *P < 0.05 compared with control cells. Bars show the mean value of the representative results from 3 experiments, each conducted in duplicate (± SD).
It is worthwhile to mention, among the 4 genes, only APOBEC3G, a central subunit of the RNA editing complex, has not been reported to play a role in tumor progression, whereas it most significantly produces cell migration and invasion (Figure 4, A and B). Therefore, we further investigated the underlying mechanism that links APOBEC3G overexpression to cell migration and cancer metastasis. To determine whether APOBEC3G expression levels in colon cancer cells are indeed involved in the regulation of cell migration activity, we examined APOBEC3G expression levels in 4 colon cancer cell lines. We found that APOBEC3G had relatively high expression levels in CaCO2 and DLD1 cells, while it had low expression levels in HCT116 and RKO cells (Figure 4D). Knocking down APOBEC3G in CaCO2 and DLD1 cells by shRNA (Figure 4E) significantly inhibited cell migration and invasion activity (Figure 4, F and G). In addition, ectopic expression of APOBEC3G in HCT116 and RKO significantly enhanced cell migration and invasion activity (Figure 4, F and G), suggesting that increased APOBEC3G expression promotes the mobility of colon cancer cells.
APOBEC3G enhances colon cancer cell migration and invasion through restoration of MMP2 from inhibition of miR-29.
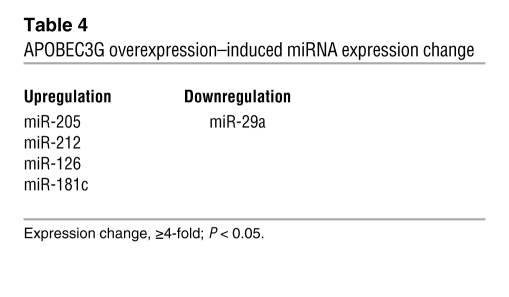
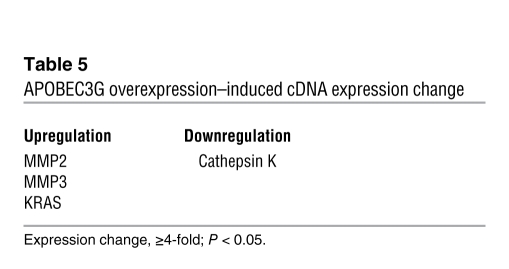
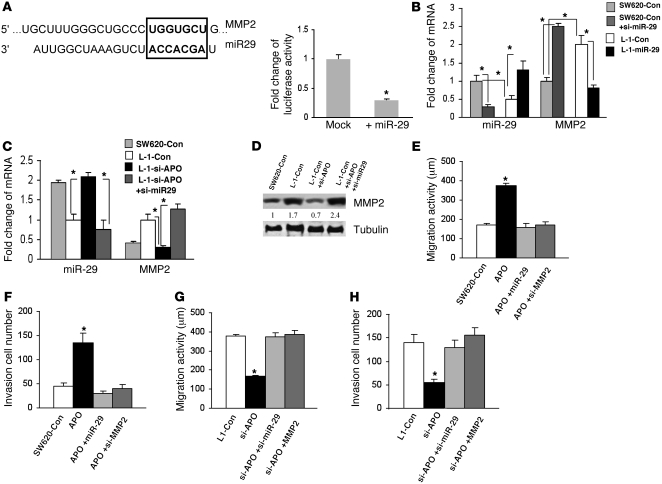
MicroRNAs (miRNAs) are 20–22–nt regulatory RNAs that participate in the regulation of various biological functions including tumor development and metastasis (17, 18). APOBEC3G has been implied to be involved in the regulation of miRNAs (19–23). Thus, we asked whether APOBEC3G promotes colorectal cancer liver metastasis through regulation of miRNAs; we screened a miRNA PCR array including approximately 90 miRNAs (SABiosciences). We found that miR-29 was downregulated and miR-126, 181c, 212, and 205 were upregulated in APOBEC3G-overexpressed cells (Table 4). To further determine the metastasis-related genes that could be controlled by APOBEC3G-regulated miRNAs, we used a human tumor metastasis PCR array (SABiosciences) that included 84 genes known to be involved in metastasis and found MMP2/3, KRAS, and Cathepsin K were regulated by APOBEC3G (Table 5). Then we used TargetScan software to search for the potential match between APOBEC3G-regulated miRNAs and their metastasis target genes. MMP2, a metastasis activator, was identified as one of the potential mRNA targets for miR-29 (Figure 5A). To further determine whether MMP2 is indeed a miR-29 target, we used a luciferase reporter linked with 3′ UTR of MMP2 (miR-29 binding site). We found that indeed ectopic expression of miR-29 significantly decreased MMP2 luciferase activity (Figure 5A). In addition, knocking down miR-29 using oligonucleotides in SW620 cells enhanced MMP2 expression, whereas overexpressing miR-29 in L-1 cells reduced MMP2 expression, suggesting MMP2 is suppressed by miR-29 (Figure 5B).
Table 4 .
APOBEC3G overexpression–induced miRNA expression change
Table 5 .
APOBEC3G overexpression–induced cDNA expression change
Figure 5. APOBEC3G enhances colon cancer cell migration and invasion through restoration of MMP2 from the inhibition of miR-29.
(A) Sequence alignment for miRNA-29 and its mRNA target MMP2 using TargetScan software. Square shows miR-29 consensus target sequence (left). Luciferase activity of pMIR-Report construct containing 3′ UTR of MMP2 miR-29 binding site was measured in 293 cells overexpressing miR-29 (right). *P < 0.05. (B) miR-29 and MMP2 mRNA levels in SW620 cells expressing miR-29 antagomir (si-miR-29) and L-1 cells expressing miR-29 precursor using quantitative PCR. (C) miR-29 and MMP2 mRNA levels in SW620 cells, L-1 cells expressing retroviral APOBEC-shRNA (si-APO) and si-miR-29 using quantitative PCR. (D) Immunoblotting was performed in L-1 cells knocking down APOBEC3G or miR-29 to examine mRNA and protein expression of the indicated genes. Wound healing assay was performed in (E) SW620 cells and (G) L-1 cells expressing the indicated constructs or oligonucleotides to measure cell migrating distance for 48 hours. The number of invasive cells was determined in (F) SW620 cells and (H) L-1 cells expressing the indicated constructs or oligonucleotides. Bars show the mean value of the representative results from 3 experiments, each conducted in duplicate (± SD).
Furthermore, we inquired whether APOBEC3G expression could inhibit miR-29 activity and thereby derepress MMP2 expression. We found that, compared with SW620 cells, miR-29 is downregulated, whereas MMP2 is upregulated in L-1 cells. Knocking down miR-29 in SW620 cells led to increased MMP2 levels, whereas overexpression of miR-29 in L-1 cells led to decreased MMP2 levels (Figure 5B). Additionally, knocking down Apobec3G in L-1 cells resulted in increased miR-29 levels and decreased MMP2 mRNA and protein expression, which could be reversed by suppression of miR-29 (Figure 5, C and D). These data suggest that APOBEC3G may promote MMP2 expression through downregulating miR-29. Next, we asked whether APOBEC3G-mediated miR-29 downregulation and MMP2 upregulation contributed to cell migration activity and cell invasion. We found that overexpressing APOBEC3G enhanced SW620 cell migration and invasion, which was reversed by increasing miR-29 or silencing MMP2 (Figure 5, E and F, and Supplemental Figure 6, A and B). Knocking down APOBEC3G reduced L-1 cell migration and invasion, which was reversed by repressing miR-29 or increasing MMP2 (Figure 5, G and H, and Supplemental Figure 6, C and D). Moreover, ectopic expression of miR-29 in L-1 cells also inhibited cell migration and invasion activity, which was restored by overexpressing MMP2 (Supplemental Figure 6, E and F). However, knocking down either APOBEC3G or APOBEC3G and miR-29 does not affect cell apoptosis and viability (Supplemental Figure 7, A and B). Together, these results suggest that APOBEC3G overexpression may promote colon cancer metastasis through derepressing MMP2 from the inhibition of miR-29.
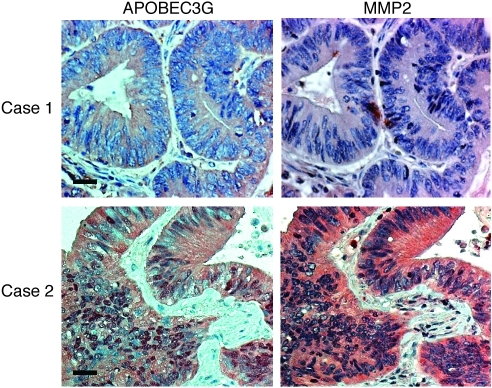
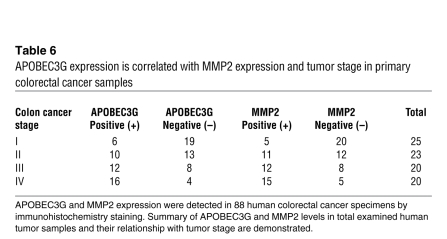
We then examined APOBEC3G and MMP2 expression in human colorectal cancer samples (Figure 6 and Table 6; total 88 cases, from stage I to IV) and found that MMP2 was correlated positively with APOBEC3G expression (P < 0.01). In addition, both high MMP2 expression and high APOBEC3G expression were associated with high tumor stage (P < 0.05), supporting the idea that APOBEC3G and MMP2 may be involved in colorectal cancer progression.
Figure 6. APOBEC3G expression is correlated with MMP2 expression and tumor stage in primary colorectal cancer samples.
Representative staining of APOBEC3G and MMP2 in colorectal cancer samples. Case 1 is a representative specimen with a low level or no expression of APOBEC3G and MMP2; case 2 is a representative specimen with a high level of APOBEC3G and MMP2. Scale bar: 50 μm.
Table 6 .
APOBEC3G expression is correlated with MMP2 expression and tumor stage in primary colorectal cancer samples
Discussion
Metastasis is a complex series of steps in which cancer cells leave the original tumor site and migrate to a distant organ. Certain cancers tend to spread to specific organ sites; however, the underlying mechanism is not completely understood. Different gene signatures may determine different organ targets with the involvement of metastatic microenvironment. For example, different sets of genes were identified to mediate breast cancer metastasis to either lung, bone, or brain (1, 9, 10, 24). A similar phenomenon was identified in colon cancer metastasis to lung and liver. Gene expression profiles that were either drawn from a colorectal cancer lung metastasis model using tail vein injection (25) or profiles generated from a cohort of colorectal cancer lung metastasis patient samples (26) exhibited distinct gene signatures of colon cancer metastasis. Though those findings do not overlap with our newly identified gene signature of colorectal liver metastasis, the metastasis process of colon cancer cells to lung and to liver may require different sets of gene activation in response to specific microenvironment.
Recent studies also revealed that certain genes and signaling pathways might play a role in colon cancer liver metastasis. Metastasis-associated in colon cancer-1 (Macc1) was identified as a key regulator of HGF-MET signaling and is able to enhance colon cancer cell migration in vitro and liver metastasis in mouse model (13). Another report indicates that the Notch pathway might be involved in colon cancer liver metastasis, as one metastasis suppressor gene AES (or GRG5) could repress colon cancer metastasis through Notch inhibition (14). In addition, TGF-β/Smad4 signaling was found to suppresses colon cancer metastasis in mice (12), while Smad7 could enhance colorectal cancer hepatic metastasis (15). Thus, the balance between Smad4/Smad7 and the TGF-β pathway in colorectal cancer may be critical for the metastatic process. It is speculated that the combination of some or all these signatures involved in colorectal cancer liver metastasis could better predict liver metastasis and patient survival. As shown in Figure 2 and Supplemental Figure 5, multiple genes are required to produce optimal effects in liver metastasis mediated by the group A genes. Since colon cancer is heterogeneous, it is conceivable that different sets of genes may cooperate with each other to induce liver metastasis.
In this current study, we separated 2 specific colorectal cancer cell lines with enriched metastatic activity to the liver by orthotopic injection and in vivo selection and further identified a set of genes, including APOBEC3G, CD133, LIPC, and S100P, which are essential for colorectal cancer liver metastasis. Among these genes, S100P has been shown to regulate calcium signal transduction, to mediate cytoskeletal interaction, and is correlated with poor prognosis in lung and breast cancer (27–29). Recently, studies have suggested that S100P may play a role in colon cancer growth and metastasis (30, 31), although the underlying molecular mechanism of S100P-mediated metastasis needs to be further investigated. CD133 is highly expressed in normal early progenitors (32) and has been used as a marker for a subpopulation of highly tumorigenic, undifferentiated cells in brain, colon, and other cancers (33, 34). Previous studies have indicated that high CD133 expression is related to a high incidence of metastasis in cholangiocarcinoma and melanoma, thereby identifying CD133 as a potential metastasis-related gene (35, 36). However, the role of CD133 in metastatic colon cancer and as a stem cell marker is still controversial, since evidence shows that CD133-negative colon cancer cells can still generate tumors and can be found in some liver metastasis of colon cancer (37, 38). In this study, we collected a relatively large cohort of human liver metastasis samples from colon cancer patients and found that CD133 expression was indeed correlated with incidence of liver metastasis (Figure 3B). Multivariate analysis also revealed that high expression of CD133 could be a molecular marker of liver metastasis in colon cancer patients (Table 3). However, questions regarding how CD133 is involved in metastasis and in which cancer stages, how CD133 expression is regulated, and what controls the transition of CD133+ to CD133– cells remain to be addressed.
Lipase C (LIPC) is a hydrolase enzyme and ligand/bridging factor for receptor-mediated lipoprotein uptake and is involved in lipoprotein and cytokine homeostasis (39, 40). A recent study also implicates a role for monoacylglycerol lipase in promoting tumor growth, migration, and invasion, as this lipase translates lipogenic phenotype to oncogenic signals in tumor cells (41). Consistent with that report, our current study demonstrates that LIPC, is involved in colon cancer liver metastasis. Together, these results suggest lipase could be a new family of metastasis-related genes.
Notably, to the best of our knowledge, this is the first time it has been shown that APOBEC3G, a gene involved in RNA editing, is able to promote tumor metastasis. Previous reports have shown that APOBEC3G can counteract the inhibition of protein synthesis mediated by various miRNAs through releasing target mRNA from bound miRNA in P bodies and thereby enhances expression level of the miRNA-targeted mRNA (19, 20). Our results further suggest that APOBEC3G may downregulate miR-29 expression and hamper miR-29 activity in repressing MMP2 (Figure 5). Though how APOBEC3G regulates miR-29 expression remains to be investigated, downregulation of miR-29 has been reported in aggressive leukemia and colon and breast cancer (42–44). This newly identified APOBEC3G/miR-29/MMP2 pathway may help open up a new avenue for understanding colorectal cancer liver metastasis and for the development of effective therapeutics for the treatment of advanced colon cancers.
Methods
Colorectal cancer hepatic metastasis animal model.
The parental SW480 and SW620 cell lines were obtained from ATCC. Cells were grown in DMEM/F12 medium supplemented with 10% FBS and stably transfected with luciferase. We used 6-week-old nude mice for the orthotopic tumor model. For liver metastasis formation, 5 × 105 viable cells were washed and harvested in PBS and then inoculated into the cecum of the nude mice after the laparotomy. The growth and metastasis of the tumors were monitored by weekly bioluminescence imaging using the IVIS imaging system (45) (Xenogen). Endpoint assays were conducted 12 weeks after the inoculation unless animal sacrifice was required because of significant morbidity.
Isolation of hepatic metastatic cells.
SW620 cells were inoculated in the cecum of the nude mice. Eight weeks later, 2 hepatic metastatic lesions (L-1 and L-2) were detected in 1 mouse by ex vivo bioluminescence imaging and then resected under sterile conditions. Half of the tissue was fixed with 4% paraformaldehyde for histologic analysis. The other half of the tissue was minced and placed in DMEM/F12-cultured medium with 0.125% collagenase for 3 hours and then centrifuged for 1 minute. Next, cells were resuspended in 0.25% trypsin for 15 minutes at 37°C and then grown in DMEM/F12-cultured medium with 10% FBS for further propagation.
RNA isolation and microarray hybridization.
For each cell line, total RNA was extracted using the RNeasy Mini Kit (QIAGEN) and eluted with RNase-free water plus disposable tissue grinder (VWR). We used 5 mm of total RNA for first- and second-strand cDNA synthesis with an oligo-dT–containing T7 promoter. Synthesized double-stranded cDNA was then used as the template to generate biotin-labeled RNA using the One-Cycle Target Labeling and Control Reagents Kit (Affymetrix). Biotin-labeled RNA was broken into 35- to 200-base fragments in size using fragmentation buffer and then assessed by hybridization using the U133A GeneChip (Affymetrix). After hybridization, each array was stained in a GeneChip Fluidics Station 450 (Affymetrix) and then scanned for gene expression analysis using the GeneChip Operating System (Affymetrix). Statistical group comparison was performed to find genes with significant differences (P < 0.05) in mean expression levels between any 2 groups as shown in Figure 1D. The gene expression profiles of different cells were confirmed by RT-PCR, real-time PCR, and Western blotting using the following antibodies: rabbit anti-CD133 (Abcam), rabbit anti-Apobec3G (Proteintech Group), mouse S100P (BD Biosciences), mouse anti-IFITM3 (Abnova), mouse ATP1B1 (GeneTex), rabbit anti-Calb1 (Sigma-Aldrich), mouse anti-LipC (Santa Cruz Biotechnology Inc.), mouse anti-Lerk5 (Abnova), and rabbit anti-IGFBP2 (Abcam).
Real-time PCR.
Total RNAs were extracted from cells by using RNeasy Kit (QIAGEN). RNAs were reverse-transcribed by using Superscript II Kit (Invitrogen). Results were analyzed by the iCycler (Bio-Rad), and relative quantification of RNA levels was normalized to GAPDH as CT (difference of cycling threshold) = CT (target) – CT (control). Higher CT values indicate relatively lower expression RNA levels.
cDNA expression and shRNA expression constructs for retroviral infection.
cDNA of the specific genes was constructed in pMX-EGFP vectors. siRNAs against specific targets were annealed and inserted into pRsuper vectors (Orbigen). The indicated cell lines were infected with the supernatant of Phoenix Ampho cells (Orbigen) that had been transfected with the shRNAs with pRsuper vector or cDNAs with pMX vector for 48 hours. All procedures were performed as described in Orbigen’s user manual. The efficiency of the knockdown was confirmed by Western blotting with the specific antibodies. pMX-EGFP or pRsuper vectors were infected with the parental cells SW620 and/or L-1 as controls.
Luciferase assay and miR-29 oligonucleotides.
pMIR-report firefly luciferase plasmid (Ambion) was inserted with MMP2-derived miR-29 binding site into 3′ UTR. Reporters were transfected to 293 cells, and transfection efficiency was corrected by Renilla luciferase cotransfection. miR-29a precursor and miR-29a antagomir were purchased from Ambion and used according to the manufacturer’s instructions.
BrdU flow cytometry analysis.
Cells were transfected with different retroviral RNA interference (RNAi), as indicated in Figure 4, or stained with anti-CD133 antibody (Miltenyi Biotec), as indicated in Supplemental Figure 2. The cells were then fixed, permeabilized, and stained with BrdU and 7-AAD using the BrdU Flow Kit (BD Biosciences). Immunofluorescence was evaluated using FACS Canto II (BD Biosciences) and analyzed using the FCS3 Express Research Lite edition (De Novo Software).
Cell motility assay and invasion assay.
A cell motility assay using time-lapse microscopy was performed using the Axiovert 200 with a cell observer (Zeiss), AxioCam (Zeiss), with a cell observer (Zeiss) and AxioCam (Zeiss) at the incubator/heating stage. Cells infected with different retroviral RNAi against specific targeted genes were cultured on 12-well plates to about 80% confluence, and then the confluent cell monolayer was wounded using a vitreous pin that delivered a precise scratch. Cells were washed once with normal growth medium and then kept at a constant temperature of 37°C in 5% CO2 under microscopy. Images of the extent of wound healing were captured at intervals ranging from 30 minutes to 48 hours. Each assay was repeated at least twice. Cell invasion was examined as described before, with some modifications. Briefly, cells were infected with different retroviral shRNA against specific targeted genes, placed in the precoated upper chamber of the 24-well Transwell permeable plates (Corning), with a cell concentration of 1 × 105/well, and cultured with serum-free DMEM at 37°C in 5% CO2 for 48 or 72 hours. The lower chamber contained normal DMEM with 10% FBS. After incubation, the filters were fixed with 3% glutaraldehyde in PBS and stained with Giemsa (Fisher Scientific). The cells on the upper surface of the filter were removed by wiping with a cotton swab, and chemoinvasive activity was determined by counting the number of cells per 3 high-power fields (HPFs) (magnification, ×200) that had migrated to the lower side of the filter. Each sample was assayed in duplicate, and assays were repeated at least twice.
Immunohistochemical staining.
Surgically resected human colorectal cancer samples with paired hepatic metastasis samples were collected from Union Hospital, the Affiliated Hospital of Medical College, National Cheng Kung University Hospital, China Medical University and Hospital, and the University of Texas MD Anderson Cancer Center. Colorectal cancer tissue microarray slides (COC1502 and A203) were purchased from Pantomics and AccuMAX. For immunohistochemical staining of the human colorectal and hepatic tumor tissues, each sample was stained with specific antibodies and scored using the H-score method, which combines the immunoreactions intensity values and the percentage of tumor cell staining (46).
Accession number.
Microarray data are deposited in the GEO archive (GSE30687).
Statistics.
We used the χ2 test to analyze the relationship between hepatic metastasis and the expression levels of APOBEC3G, CD133, LIPC, and S100P in 146 primary colorectal cancer and hepatic metastasis samples. ANOVA was employed to compare the differences among multiple cell lines treated as indicated for metastasis analysis. Statistical analysis and graphs were performed using SPSS software, and the level of significance was set at P < 0.05.
Study approval.
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at MD Anderson Cancer Center. Resected human colorectal cancer samples with paired hepatic metastasis were obtained under the guidelines of the respective institutional review boards of Union Hospital, the Affiliated Hospital of Medical College, National Cheng Kung University Hospital, the China Medical University Hospital, and the University of Texas MD Anderson Cancer Center. Tumor samples were acquired from cancer patients with informed consent (IRB protocol LAB02-631, MD Anderson Cancer Center).
Supplementary Material
Acknowledgments
We would like to thank Stephanie A. Miller, Jeng C. Cheng, and Alyson Todd for editing the manuscript. This work was supported by the Kadoorie Charitable Foundation, the CMUH/MDACC Sister Institution Foundation (to M.-C. Hung), the DOH-TD-C-111-005 Cancer Research Center of Excellence, Taiwan (to L.-Y Li, L.-B. Jeng, and M.-C. Hung), and the DOD postdoctoral fellowship (W81XXWH-10-1-0598 to C.-W. Li). We would also like to thank the late Tiong Loi Ang for his contributions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(11):4526–4536. doi:10.1172/JCI45008.
References
- 1.Chiang AC, Massague J. Molecular basis of metastasis. N Engl J Med. 2008;359(26):2814–2823. doi: 10.1056/NEJMra0805239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curley SA, Izzo F, Abdalla E, Vauthey JN. Surgical treatment of colorectal cancer metastasis. Cancer Metastasis Rev. 2004;23(1–2):165–182. doi: 10.1023/A:1025875332255. [DOI] [PubMed] [Google Scholar]
- 3.Manfredi S, Bouvier AM, Lepage C, Hatem C, Dancourt V, Faivre J. Incidence and patterns of recurrence after resection for cure of colonic cancer in a well defined population. Br J Surg. 2006;93(9):1115–1122. doi: 10.1002/bjs.5349. [DOI] [PubMed] [Google Scholar]
- 4.Manfredi S, Lepage C, Hatem C, Coatmeur O, Faivre J, Bouvier AM. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang GJ, Rodriguez-Bigas MA, Skibber JM, Moyer VA. Lymph node evaluation and survival after curative resection of colon cancer: systematic review. . J Natl Cancer Inst. 2007;99(6):433–441. doi: 10.1093/jnci/djk092. [DOI] [PubMed] [Google Scholar]
- 6.Dashwood RH. Early detection and prevention of colorectal cancer (review). Oncol Rep. 1999;6(2):277–281. [PubMed] [Google Scholar]
- 7.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 8.Weinberg RA. Leaving home early: reexamination of the canonical models of tumor progression. Cancer Cell. 2008;14(4):283–284. doi: 10.1016/j.ccr.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 9.Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhang B, et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138(3):969–980. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein U, et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15(1):59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 14.Sonoshita M, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19(1):125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 15.Halder SK, Rachakonda G, Deane NG, Datta PK. Smad7 induces hepatic metastasis in colorectal cancer. Br J Cancer. 2008;99(6):957–965. doi: 10.1038/sj.bjc.6604562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leibovitz A, Stinson JC, McCombs WB 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36(12):4562–4569. [PubMed] [Google Scholar]
- 17.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25(46):6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 18.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12(12):580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282(46):33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H. The Inhibitory effect of apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and Its family members on the activity of cellular microRNAs. Prog Mol Subcell Biol. 2010;50:71–83. doi: 10.1007/978-3-642-03103-8_5. [DOI] [PubMed] [Google Scholar]
- 21.Gallois-Montbrun S, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81(5):2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhoads RE. miRNA regulation of the translational machinery. Preface. Prog Mol Subcell Biol. 2010;50:v–vi. [PubMed] [Google Scholar]
- 23.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2(5):e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–549. doi: 10.1016/S1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 25.Smith JJ, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo MA, et al. Gene expression profiling of liver metastases from colorectal cancer as potential basis for treatment choice. Br J Cancer. 2008;99(10):1729–1734. doi: 10.1038/sj.bjc.6604681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, et al. Induction of metastasis by S100P in a rat mammary model and its association with poor survival of breast cancer patients. Cancer Res. 2006;66(2):1199–1207. doi: 10.1158/0008-5472.CAN-05-2605. [DOI] [PubMed] [Google Scholar]
- 28.Saleem M, et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc Natl Acad Sci U S A. 2006;103(40):14825–14830. doi: 10.1073/pnas.0606747103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beer DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 30.Jiang L, et al. Targeting S100P inhibits colon cancer growth and metastasis by lentivirus-mediated RNA interference and proteomic analysis. Mol Med. 2011;17(7–8):709–716. doi: 10.2119/molmed.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuentes MK, et al. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Dis Colon Rectum. 2007;50(8):1230–1240. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 32.Peichev M, et al. Expression of VEGFR-2 and AC133 by circulating human CD34(+) cells identifies a population of functional endothelial precursors. Blood. 2000;95(3):952–958. [PubMed] [Google Scholar]
- 33.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 34.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 35.Shimada M, et al. CD133 expression is a potential prognostic indicator in intrahepatic cholangiocarcinoma. J Gastroenterol. 2010;45(8):896–902. doi: 10.1007/s00535-010-0235-3. [DOI] [PubMed] [Google Scholar]
- 36.Rappa G, Fodstad O, Lorico A. The stem cell-associated antigen CD133 (Prominin-1) is a molecular therapeutic target for metastatic melanoma. Stem Cells. 2008;26(12):3008–3017. doi: 10.1634/stemcells.2008-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shmelkov SV, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.LaBarge MA, Bissell MJ. Is CD133 a marker of metastatic colon cancer stem cells? J Clin Invest. 2008;118(6):2021–2024. doi: 10.1172/JCI36046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watt MJ, Steinberg GR. Regulation and function of triacylglycerol lipases in cellular metabolism. Biochem J. 2008;414(3):313–325. doi: 10.1042/BJ20080305. [DOI] [PubMed] [Google Scholar]
- 40.Wong H, Schotz MC. The lipase gene family. J Lipid Res. 2002;43(7):993–999. doi: 10.1194/jlr.R200007-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010;140(1):49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cummins JM, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103(10):3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calin GA, Croce CM. Chronic lymphocytic leukemia: interplay between noncoding RNAs and protein-coding genes. Blood. 2009;114(23):4761–4770. doi: 10.1182/blood-2009-07-192740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie X, et al. Targeted expression of BikDD eradicates pancreatic tumors in noninvasive imaging models. Cancer Cell. 2007;12(1):52–65. doi: 10.1016/j.ccr.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Ding Q, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19(2):159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.