Abstract
Type 1 diabetes (T1D) is an autoimmune disease that shows familial aggregation in humans and likely has genetic determinants. Disease linkage studies have revealed many susceptibility loci for T1D in mice and humans. The mouse T1D susceptibility locus insulin-dependent diabetes susceptibility 3 (Idd3), which has a homologous genetic interval in humans, encodes cytokine genes Il2 and Il21 and regulates diabetes and other autoimmune diseases; however, the cellular and molecular mechanisms of this regulation are still being elucidated. Here we show that T cells from NOD mice produce more Il21 and less Il2 and exhibit enhanced Th17 cell generation compared with T cells from NOD.Idd3 congenic mice, which carry the protective Idd3 allele from a diabetes-resistant mouse strain. Further, APCs from NOD and NOD.Idd3 mice played a central role in this differential Th17 cell development, and IL-21 signaling in APCs was pivotal to this process. Specifically, NOD-derived APCs showed increased production of pro-Th17 mediators and dysregulation of the retinoic acid (RA) signaling pathway compared with APCs from NOD.Idd3 and NOD.Il21r-deficient mice. These data suggest that the protective effect of the Idd3 locus is due, in part, to differential RA signaling in APCs and that IL-21 likely plays a role in this process. Thus, we believe APCs provide a new candidate for therapeutic intervention in autoimmune diseases.
Introduction
Type 1 diabetes (T1D) is an autoimmune disease that shows familial aggregation in humans and appears to be, at least partially, genetically determined. The NOD mouse serves as a good model for human diabetes, because the disease appears spontaneously and bears pathogenic features remarkably similar to those of human T1D. Genetic analyses of T1D in NOD mice and in humans have identified many susceptibility loci. One of these loci, insulin-dependent diabetes susceptibility 3 (Idd3), encodes several genes, of which only the cytokine genes Il2 and Il21 currently have known immunological importance. NOD mice carrying the Idd3 locus from diabetes-resistant C57BL/6 mice (NOD.Idd3 mice) are protected from autoimmune diabetes and EAE (1, 2). In this genetic interval, Il2 and Il21 are inherited as a haplotype. IL-2 and IL-21 are both T cell–derived cytokines that have antagonistic effects on the development of Foxp3+ Tregs and proinflammatory Th17 cells. IL-2 promotes Treg differentiation, whereas IL-21 suppresses it; in contrast, IL-21 supports the generation of Th17 cells, while IL-2 constrains Th17 differentiation (3–7). Thus, the Idd3 genetic interval may regulate the balance between pathogenic Th17 and protective Foxp3+ Tregs through the differential effects of IL-2 and/or IL-21.
Indeed, some studies suggest that the protective effect of the Idd3 genetic interval is due to increased production of Il2, which enhances Treg function, in NOD.Idd3 congenic mice (8). In this regard, we and others reported that the defective Treg function observed in NOD mice, relative to that of NOD.Idd3 mice, is determined by differences in NOD-derived versus NOD.Idd3-derived APCs (9, 10), rather than by intrinsic defects in the Tregs from NOD mice.
Although the role of IL-2 in the protective effect of Idd3 has received much attention, considerable evidence supports a role for IL-21 as well. It has been reported that IL-21 expression is elevated in NOD mice compared with that in diabetes-resistant strains of mice and that increased IL-21 expression is the key to susceptibility to autoimmune disease (11). In support of this, Il21 receptor–deficient (Il21r-deficient) NOD mice were reported to be protected against autoimmune diabetes (12–14).
The recently characterized Th17 cells have been shown to be highly pathogenic, and it has been argued that this subset is the primary mediator of tissue inflammation in many autoimmune diseases (15). The involvement of Th17 cells in diabetes is not well understood, but there is a growing body of research implicating Th17 cells in its pathogenesis (16–18). Using either neutralizing anti–IL-17 mAb or recombinant IL-25, Th17 cells have been demonstrated to have a pivotal role in T1D. The Th17-blocking therapies prevented diabetes development in NOD mice with a concomitant reduction in islet-specific T cell responses (19).
Here we show that T cells derived from NOD mice have a greater propensity to differentiate into Th17 cells compared with T cells from both NOD.Idd3 mice and NOD.Il21r KO mice. We further show that, in additional to intrinsic T cell differences, APCs have an important role in determining the balance of regulatory and pathogenic T cells. NOD- and NOD.Idd3-derived APCs differentially support Th17 cell polarization, and this difference in APC function is associated with retinoic acid (RA) signaling that may be regulated by IL-21. In the study of autoimmunity, APCs have received little attention relative to T cells. Our data suggest that APCs are key players and represent other avenues for therapeutic intervention in autoimmune diseases.
Results
NOD-derived cells have a greater potential to generate Th17 cells than NOD.Idd3-derived cells.
To compare the expression of Il21 and Il2 in naive T cells from NOD and NOD.Idd3 congenic mice, real-time PCR was performed. We found that T cells from NOD mice expressed more Il21 and fewer Il2 transcripts than NOD.Idd3-derived T cells upon in vitro activation using anti-CD3 plus anti-CD28 (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI46187DS1). Accordingly, when naive T cells were activated in the presence of self APCs, IL-21 and IL-17 production was elevated in NOD cultures compared with that in NOD.Idd3 cultures (Supplemental Figure 1B). Since recent data show that IL-2 and IL-21 have pivotal and opposing roles in the development of Th17 cells, we compared the generation of Th17 cells in vitro using naive T cells from NOD and NOD.Idd3 mice. In the absence of APCs, the polarization of Th17 cells, using a combination of IL-6 and TGF-β in NOD cultures, was on average twice as efficient as that in NOD.Idd3 cultures, as indicated by a higher frequency of IL-17+ T cells (Figure 1A). This defect is in line with the increased Il21 and decreased Il2 expression by T cells from NOD mice compared with that from NOD.Idd3 mice (Supplemental Figure 1). Furthermore, Th17 polarization was enhanced in both NOD and NOD.Idd3 cultures when neutralizing anti–IL-2 mAb was added. However, even at the highest dose of anti–IL-2 used, NOD.Idd3-derived T cells remained less efficient in polarizing to Th17 cells than NOD-derived T cells (Supplemental Figure 2). Thus, differential expression of IL-2 alone cannot account for the reduced potential of NOD.Idd3 T cells to differentiate into Th17 cells.
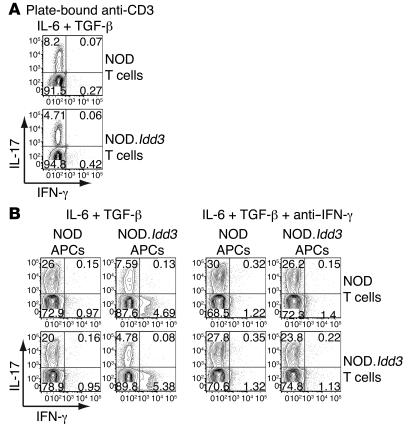
Figure 1. Cells from NOD mice generate Th17 cells more efficiently than cells from NOD.Idd3 mice.
Naive T cells were polarized with TGF-β and IL-6. After 3 days, cells were restimulated with PMA and ionomycin in the presence of monensin for 4 hours before cell surface and then intracellular staining. Values in each quadrant represent the percentage of 7-AAD–CD4+ cells. Cells were activated with (A) plate-bound anti-CD3 and soluble anti-CD28 or (B) with soluble anti-CD3 in the presence of APCs, with or without anti–IFN-γ mAb. Data shown are representative of 4 to 9 experiments.
Since we and others previously showed that APCs from NOD and congenic NOD.Idd3 mice have important effects on the suppressive activity of Tregs (9, 10) and that Tregs and Th17 cells have a reciprocal relationship (20), we examined the relative contribution of NOD- versus NOD.Idd3-derived APCs on Th17 cell differentiation. By swapping APCs, we found that NOD-derived APCs were more efficient at supporting the differentiation of Th17 cells, irrespective of the source of T cells (Figure 1B), while NOD.Idd3-derived APCs could not support polarization of Th17 cells as efficiently as NOD-derived APCs. Moreover, APCs from NOD.Idd3 mice enhanced production of IFN-γ, a prototypical cytokine of Th1 cells, even in the presence of Th17-polarizing cytokines IL-6 and TGF-β (Figure 1B). This increased production of IFN-γ in the presence of NOD.Idd3-derived APCs was coincident with reduced IL-17 production. This raises the possibility that IFN-γ induced by and/or produced by NOD.Idd3 APCs was responsible for inhibiting Th17 polarization. Indeed, the addition of neutralizing anti–IFN-γ mAb improved Th17 differentiation in all conditions and almost completely abolished the differences in Th17 differentiation observed between APCs derived from NOD and NOD.Idd3 mice (Figure 1B). This is consistent with previous reports that IFN-γ inhibits Th17 cell generation (16, 21, 22). Together, these data suggest that there is an APC-driven — as well as a T cell–intrinsic — difference in Th17 cell development between NOD and NOD.Idd3 mice. APCs from NOD.Idd3 congenic mice not only impaired Th17 differentiation but also promoted IFN-γ production by T cells, even under Th17 polarization conditions, thereby further inhibiting Th17 differentiation.
Th17 cells accumulate in NOD mice and can induce diabetes upon adoptive transfer.
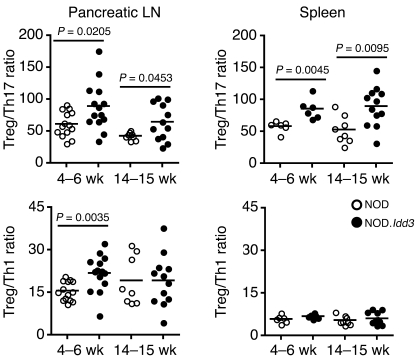
Given that NOD.Idd3 mice are protected from T1D and that both IL-2 and IL-21 can modulate the development of induced Tregs and Th17 cells, we assessed the balance of regulatory and pathogenic T cells in these mice. We and others have previously shown that the frequency of Tregs is not significantly different between NOD and diabetes-resistant NOD.Idd3 mice (8, 9, 23, 24). However, in prediabetic mice, we found that the ratio of Tregs/Th17 cells was higher in NOD.Idd3 mice than in NOD mice in both the splenic and pancreatic draining LN compartments (Figure 2). We also looked at the development of Th1 cells by staining for IFN-γ. Treg/Th1 ratios in the pancreatic LNs of 4- to 6-week-old mice were also higher in NOD.Idd3 congenic mice than those in NOD mice, but there was no difference in older mice or in the splenic compartment (Figure 2). These data indicate that the balance of Tregs/T effector cells is shifted toward effector cells, particularly Th17 cells, in NOD mice.
Figure 2. The Treg/T effector balance is skewed toward T effectors in NOD mice.
LN and spleen cells from Foxp3.gfp KI NOD (prediabetic) and NOD.Idd3 mice were stimulated with PMA and ionomycin for intracellular cytokine staining. Foxp3/GFP+ expression on CD4+ T cells was assessed in the same samples prior to restimulation. Each dot represents the ratio of Treg/cytokine+ 7-AAD–CD3+CD4+ T cells per individual mouse, and the means are indicated by the bars.
Th17 cells have become recognized as key orchestrators of many autoimmune disease processes (15), and indeed, NOD mice have higher frequencies of IL-17+ CD4 T cells in the pancreatic LNs as they progress toward diabetes (data not shown and ref. 17). Using T cells from mice with a TCR that is reactive to an islet antigen (BDC2.5 Tg mice), we thus directly compared the pathogenic capacity of Th1 and Th17 cells to induce T1D upon adoptive transfer. Upon transfer into either lymphopenic NOD/SCID mice or intact NOD mice, islet-specific Th1 and Th17 cells, generated in vitro, were equally efficient at inducing diabetes (Supplemental Figure 3A and ref. 18). Previous studies suggest that the diabetogenic effects of Th17 cells are due to and rely on the conversion of Th17 cells into IFN-γ–producing Th1 cells in vivo (17, 18). To address this issue, we generated islet-specific Th17 cells from IFN-γ–deficient mice and demonstrated that they were just as efficient as IFN-γ+/+ Th17 cells at inducing T1D upon adoptive transfer into NOD mice (Supplemental Figure 3B). Thus, Th17 cells can induce T1D independently of IFN-γ.
IL-21 signaling both in T cells and APCs contributes to the defect in Th17 cell generation and diabetes.
Since the Idd3 locus regulates expression of both Il2 and Il21, and perturbation of IL21R signaling completely protected NOD mice from autoimmune diabetes (12–14), we investigated the effects of IL-21 signaling on Th17 cell development using NOD.Il21r KO mice. In line with previous reports using Il21- and Il21r-deficient mice on diabetes-resistant backgrounds (5–7), Il21r-deficient T cells from NOD mice exhibited a defect in generating Th17 cells (Figure 3A), similar to that of NOD.Idd3-derived T cells (Figure 1A). Given our observations that IL-21 production is reduced in NOD.Idd3 mice and that NOD.Idd3-derived APCs do not promote Th17 differentiation, we next investigated the ability of NOD.Il21r KO APCs to support Th17 differentiation. Like NOD.Idd3-derived APCs, NOD.Il21r KO APCs did not support Th17 differentiation as well as NOD-derived APCs (Figure 3B). Furthermore, APCs from Il21r-deficient NOD.Idd3 mice (NOD.Idd3.Il21r KO mice) were even less efficient than NOD.Idd3-derived APCs at supporting Th17 generation (Figure 3B). These data show, for the first time to our knowledge, that IL-21 signaling in APCs conditions them for optimal Th17 generation. Moreover, these data support the hypothesis that decreased IL-21 signaling in APCs may partly underlie the disease resistance in NOD.Idd3 mice.
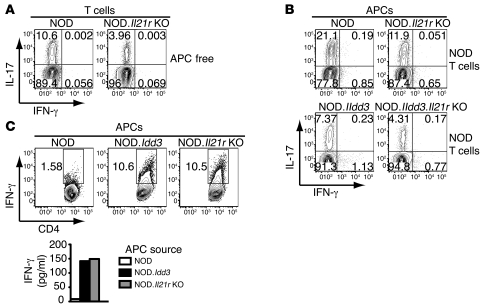
Figure 3. The defect in Th17 cell generation in NOD.Il21r KO mice is both T cell intrinsic and APC driven.
(A) Naive T cells from NOD or NOD.Il21r KO mice were activated with plate-bound anti-CD3, and (B and C) naive T cells from NOD mice were activated with soluble anti-CD3 in the presence APCs, (A and B) in the presence or (C) absence of IL-6 and TGF-β. Intracellular staining was performed after 3 days. Values in each quadrant or gate represent the percentage of 7-AAD–CD4+ cells. (C) Cell-conditioned media were assessed for IFN-γ production after 48 hours. Data are representative of 2 to 3 independent experiments.
It is of interest that APCs from mice with either reduced IL-21 signaling (NOD.Idd3 mice) or complete loss of IL-21 signaling (NOD.Il21r KO mice) were not able to support Th17 generation as well as APCs derived from NOD mice. Moreover, when NOD-derived T cells were activated under neutral conditions, they produced more IFN-γ when cocultured with APCs from NOD.Idd3 or NOD.Il21r KO mice than with APCs from NOD mice (Figure 3C). Together with the observation that NOD.Idd3-derived APCs could promote IFN-γ production by T cells, even in the presence of Th17-polarizing cytokines, and that neutralization of IFN-γ resulted in enhanced generation of Th17 cells (Figure 1B), these data indicate that APCs from NOD.Idd3 and NOD.Il21r KO mice both suppress IL-17 production by T cells, partly by promoting IFN-γ production.
NOD- and NOD.Idd3-derived CD11b+ cells differentially regulate Th17 polarization.
Previously, we showed that the differential suppressive activity of Tregs from NOD and NOD.Idd3 mice was regulated by a subset of APCs expressing CD11b and lacking the pan DC marker, CD11c (9). To identify the APC subsets associated with the differential Th17 cell polarization observed between NOD and NOD.Idd3 mice (Figure 1B), cocultures using APCs depleted of different subsets were set up. Upon depletion of CD11c–CD11b+ cells, the capacity of APCs from NOD and NOD.Idd3 mice to support Th17 polarization was equivalent (Figure 4A). Furthermore, the generation of Th17 cells was markedly more efficient when CD11b+ cells were depleted, indicating that this APC subset from NOD and NOD.Idd3 mice may also differentially constrain Th17 differentiation. In contrast, when CD11c+ cells were depleted from the APC pool, Th17 polarization in the presence of NOD.Idd3-derived APCs remained suboptimal compared with that in the presence of NOD-derived APCs, and the absence of CD11c+ APCs did not affect the Th17 polarization efficiency in either culture (Figure 4A). Thus, these data suggest that, similar to the regulation of Treg function (9), CD11b+ cells, but not CD11c+ cells, contribute to the APC-driven defect in Th17 cell development in NOD.Idd3 mice.
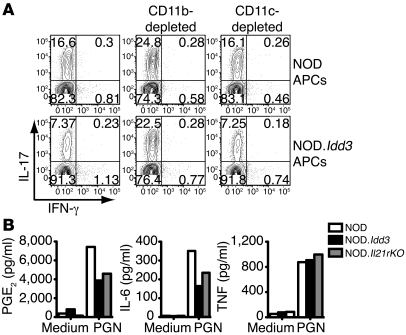
Figure 4. CD11b+ APCs regulate Th17 cell differentiation.
(A) NOD-derived naive T cells were polarized with IL-6 and TGF-β in the presence of APCs, with or without depletion of either CD11c–CD11b+ or CD11c+ cells, and, after 3 days, intracellular staining was performed to assess IL-17 and IFN-γ production. Values in each quadrant represent the percentage of 7-AAD–CD4+ cells. Data are representative of 3 independent experiments. (B) CD11b+ cells were stimulated with PGN for 48 hours, and supernatants were assayed for mediator production. Data represent the mean values from 2 to 3 experiments and are representative of 3 to 4 independent experiments.
Thus, we compared CD11b+ APCs from NOD and NOD.Idd3 mice as well as NOD.Il21r KO mice for differences that could affect Th17 differentiation. As prostaglandin E2 (PGE2), a proinflammatory mediator, has been implicated in the development of Th17 cells (25–27), we examined the production of both IL-6 and PGE2 by CD11b+ APCs. Upon stimulation with a ligand for TLR2, peptidoglycan (PGN), CD11b+ cells from NOD mice produced more PGE2 and IL-6 than cells from both NOD.Idd3 and NOD.Il21r KO mice (Figure 4B). Reduced production of PGE2 and IL-6 in NOD.Idd3-derived and NOD.Il21r KO CD11b+ cells does not reflect overall dampened responsiveness to stimulation, as TNF production was comparable in all 3 strains (Figure 4B).
Given that PGE2 and IL-6 are known pro-Th17 mediators, decreased production of these mediators by NOD.Idd3-derived and NOD.Il21r KO APCs is in line with a defect in Th17 cell generation regulated by APCs in these mice. In support of our hypothesis that IL-21 influences mediator production by CD11b+ APCs in NOD and NOD.Idd3 mice and that the difference in mediator production is not caused by differential Il21r expression, we found that Il21r mRNA was similarly expressed on CD11b+ cells ex vivo from both strains (Supplemental Figure 4A). On the other hand, Il2ra expression was undetectable in CD11b+ cells ex vivo (Supplemental Figure 4A). Taken together, these data suggest that the differences in IL-21 signaling between NOD and NOD.Idd3 mice can influence the repertoire of proinflammatory mediators produced by APCs, which, in turn, affect Treg and Th17 cell generation and/or function. Thus, CD11b+ APCs may be differentially conditioned in diabetes-susceptible and diabetes-resistant mouse strains, and the observed differences in PGE2 and IL-6 production are merely products of greater underlying differences.
Gene profiling of CD11b+ cells reveals a link between altered RA signaling in APCs and Th17 differentiation.
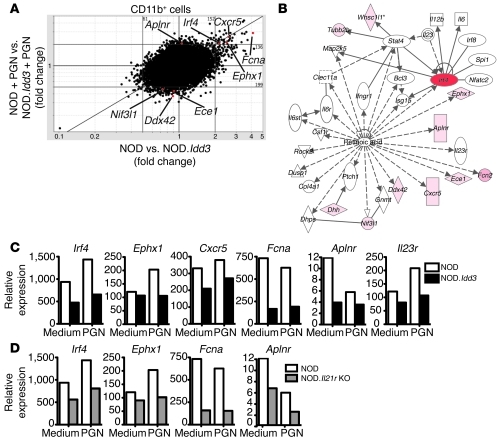
To further explore the effect mediated by the Idd3 locus on CD11b+ cells, we compared the gene profiles of NOD- and NOD.Idd3-derived CD11b+ cells using whole genome microarray analysis (GEO accession GSE24350). Comparisons between CD11b+ cells isolated from NOD and NOD.Idd3 mice yielded 213 and 335 transcripts with consistently more than 2-fold differential expression in duplicate comparisons of unstimulated and PGN-activated cells, respectively (Figure 5A).
Figure 5. Gene profiling of cells from NOD and NOD.Idd3 mice.
RNA was extracted from purified CD11b+ cells cultured for 4 hours with or without PGN. (A) Scatter plot shows gene profiling performed on duplicate CD11b+ cell samples from NOD and NOD.Idd3 mice cultured with or without PGN stimulation for 4 hours. Red dots indicate transcripts downstream of RA signaling. (B) The biological network showing differentially regulated genes in the RA pathway was produced with Ingenuity Pathways Analysis software. Genes expressed differentially more than 2 fold in CD11b+ cells from NOD and NOD.Idd3 mice are shaded pink, whereas more highly expressed genes are depicted by a darker pink and red. Indirect (dashed lines) and direct (solid lines) network interactions are shown. The asterisk denotes that this gene has multiple identifiers on the genechip. (C and D) Expression of RA-regulated genes was determined by real-time PCR and normalized to β-actin expression.
Ingenuity Pathways Analysis software was used to highlight transcriptional networks of genes differentially expressed between NOD- and NOD.Idd3-derived CD11b+ cells. Interestingly, some genes differentially expressed after TLR2 ligation in NOD- and NOD.Idd3-derived CD11b+ cells have been reported to be downstream of RA signaling (Figure 5B). All-trans RA, a vitamin A metabolite, has been reported to regulate the reciprocal relationship between Tregs and Th17 cells, whereby RA inhibits Th17 cell generation and enhances Treg induction (28). Previously, we showed that IFN regulatory factor 4 (IRF4), which is critical for Th17 cell generation (29, 30), is downregulated by RA (31). Microarray analysis showed that Irf4 expression in CD11b+ cells from NOD mice was higher than that in NOD.Idd3 mice, both in unstimulated and PGN-stimulated cells; this was confirmed by real-time PCR (Figure 5, A and C). In support of RA possibly modulating Irf4 expression in NOD and NOD.Idd3 mice, transcripts of RA receptors were detected in both CD11b+ and T cells from these mice (Supplemental Figure 4B). Moreover, Irf4 expression was also higher in acutely activated T cells from NOD mice than in those from NOD.Idd3 mice (Supplemental Figure 4C). Together with relatively higher Irf4 expression in NOD-derived CD11b+ cells, this is consistent with increased RA signaling in NOD.Idd3 mice. In addition, we showed previously that RA inhibits Il23r expression in T cells (31). Indeed, while this probe was not included on the Affymetrix Mouse Genome 430 2.0 microarray, real-time PCR showed that T cells as well as CD11b+ cells from NOD mice expressed elevated levels of Il23r transcript compared with those in cells from NOD.Idd3 mice (Supplemental Figure 4D and Figure 5C).
Epoxide hydrolase 1 (EPHX1) is an enzyme that protects against oxidative stress; CXCR5 is a chemokine receptor; ficolin A (FCNA) is a lectin; and apelin receptor (APLNR) is a receptor for angiotensin. Genes encoding these proteins are reported to be regulated by RA (32–35) and were more highly expressed in NOD-derived than NOD.Idd3-derived CD11b+ cells, as determined by microarray and confirmed by real-time PCR (Figure 5, A and C). Interestingly, the expression of Irf4, Ephx1, Fcna, and Aplnr in NOD.Il21r KO CD11b+ cells showed a similar pattern to that observed in NOD.Idd3-derived CD11b+ cells (Figure 5D), suggesting a possible connection between the RA and IL-21 signaling pathways.
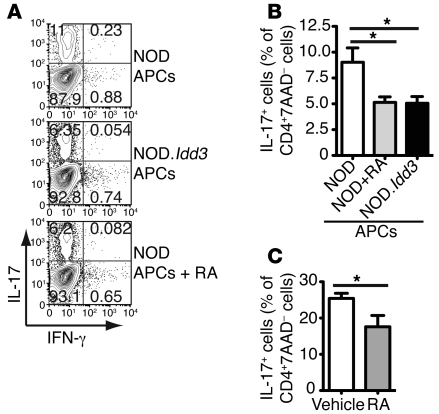
If differential RA signaling in NOD.Idd3- and NOD-derived APCs regulates Th17 differentiation, then increasing RA signaling in NOD-derived APCs could dampen Th17 differentiation to levels similar to those in cultures with NOD.Idd3-derived APCs. We therefore pretreated APCs from NOD mice with RA prior to coculture with naive T cells. When NOD-derived APCs were pretreated with RA, their ability to support Th17 polarization was dampened to the level observed in cultures with untreated NOD.Idd3-derived APCs (Figure 6, A and B). Furthermore, pretreatment of APCs in vivo by injecting RA into mice reduced their capacity to support generation of Th17 cells (Figure 6C). Lymphopenic mice (NOD/SCID) were used to avoid confounding effects resulting from stimulation of T cells with RA. Collectively, these data support the hypothesis that RA signaling is dysregulated in NOD mice relative to that of NOD.Idd3 mice and that this altered signaling acts on APCs to modulate Th17 differentiation in these 2 strains.
Figure 6. RA signaling in APCs dampens Th17 differentiation in NOD T cells.
Naive T cells from NOD mice were stimulated with soluble anti-CD3 plus TGF-β and IL-6 in the presence of APCs that were pretreated with or without RA. (A) After 3 days, Th17 polarization was assessed by intracellular staining. Values in each quadrant represent the percentage of 7-AAD–CD4+ cells. Scatter plots are representative of 3 independent experiments and summarized in B; the mean of 3 independent experiments are shown (mean ± SEM). *P < 0.05. (C) APCs were pretreated with RA in vivo in NOD/SCID mice. Values are representative of 2 independent experiments (mean ± SEM; n = 7). *P < 0.05.
Discussion
Our data demonstrate that naive T cells from NOD mice exhibit a greater propensity to differentiate into Th17 cells than NOD.Idd3-derived T cells and that the balance of Tregs/Th17 cells is tipped toward pathogenic Th17 cells in vivo in NOD mice carrying the diabetes-susceptible Idd3 allele. Moreover, we show that NOD- and NOD.Idd3-derived APCs, specifically, CD11c–CD11b+ cells, differentially support Th17 differentiation, and this difference is likely to be regulated by IL-21. Lastly, our data suggest that RA signaling may be dysregulated in NOD mice, relative to that in NOD.Idd3 mice, and that this leads to altered function of APCs.
We showed previously that NOD-derived CD11b+ APCs inhibit Treg function (9). We now present data that show that this same APC population also promotes Th17 cell differentiation in NOD mice relative to that of NOD.Idd3 congenic mice. Indeed, we show for the first time to our knowledge that there is a defect in Th17 cell generation in NOD.Idd3 mice that is not only T cell intrinsic but also mediated by APCs. Interestingly, these defects are mirrored in APCs from NOD.Il21r KO mice, suggesting that IL-21 signaling, in part, modulates APC function. IL-21 may act on APCs to induce production of mediators that, in turn, feedback onto T cells to further promote Th17 polarization. Indeed, PGE2 and IL-6, mediators that facilitate Th17 generation, are more highly produced by APCs from NOD mice relative to that produced by APCs from either NOD.Idd3 or NOD.Il21r KO mice upon activation. It is plausible that IL-21 signaling not only acts directly on T cells to induce Th17 cell development and inhibit Tregs but also acts indirectly by conditioning APCs to produce mediators that further amplify proinflammatory Th17 cell differentiation and suppress Treg function. For instance, IL-6 can inhibit Treg induction (20, 36) and induce further IL-21 production (7), completing a positive feedback loop to maintain high IL-21 expression in NOD mice and boosting the inherent proinflammatory bias. In addition, under our experimental conditions, the Th17 polarization supported by NOD-derived and NOD.Idd3-derived APCs could be equalized after neutralization of IFN-γ, suggesting that IFN-γ is another possible mediator produced by APCs and/or induced in T cells by APCs to inhibit Th17 cell generation. This represents a mechanism by which APCs can skew T cell differentiation away from Th17 and ultimately protect mice from autoimmune disease in diabetes-resistant strains.
Gene profiling of CD11b+ APCs from NOD and NOD.Idd3 mice shows that expression of many genes differs, indicating that the Idd3 locus either directly or indirectly affects the development and effector functions of CD11b+ cells. IRF4, a transcription factor essential for Th17 cell development (29), is downregulated in CD11b+ cells from NOD.Idd3 mice as well as NOD.Il21r KO mice, relative to that in cells from NOD mice. Given our recent data showing that RA inhibits Th17 cell development by inhibiting IL-6 signaling and downregulating Irf4 (31), it is conceivable that NOD.Idd3 mice have elevated RA signaling. In support of differential RA signaling in NOD and NOD.Idd3 mice, many genes reported to be regulated by RA signaling were differentially expressed by CD11b+ APCs from NOD and NOD.Idd3 mice. Since RA has been reported to tip the Treg/T effector cell balance toward regulatory cells (28, 31, 37), it is in line with NOD mice having a lower Treg/T effector ratio than NOD.Idd3 mice. This is also consistent with studies showing that RA treatment (38, 39) and vitamin A–rich diets (40) can inhibit the development of T1D. Moreover, RA and PGE2, an enhancer of Th17 cell development, are reported to be mutually antagonistic, whereby PGE2 inhibits synthesis of RA and vice versa (41, 42).
Since IRF4 induces IL-6 (43), increased Irf4 in NOD mice is in line with elevated IL-6 protein production by CD11b+ cells and consequently Th17 cell generation in these mice. While Il6 mRNA was not found to be differentially expressed in stimulated NOD and NOD.Idd3-derived CD11b+ APCs in our microarray analyses, this may simply reflect the different kinetics of IL-6 protein and mRNA expression. Given that IL-6 induces IL-21 (7) and that IRF4 is required for IL-21–dependent generation and amplification of Th17 populations (30), this represents a convergence of pathways that lead to enhanced Th17 cell development. Furthermore, similar expression patterns of genes downstream of RA signaling in CD11b+ cells from both NOD.Idd3 and NOD.Il21r KO mice suggest that IL-21 may affect RA signaling. Reduced capacity of RA-treated APCs from NOD mice to promote the generation of Th17 cells further supports a link between RA signaling and APC-driven modulation of Th17 cell development.
Earlier studies have implicated defective APCs in the development of autoimmune diabetes in NOD mice (44–46), but the molecular mechanisms responsible for the difference were not identified. Moreover, recent studies on T1D patients show a defect in APCs, in that they produce more IL-6 and IL-1 (47). This is consistent with APCs having an important role in T1D development by altering the balance of pathogenic Th17 cells and protective Tregs. We previously reported that the reduced suppressive capacity of Tregs from NOD mice was mediated by APCs (9, 10), and here we provide evidence that APCs are important regulators of proinflammatory Th17 cell development and that IL-21 signaling is pivotal to the T cell–APC cross-talk that modulates Th17 differentiation, which, in turn, determines susceptibility to autoimmunity. Most of the differences between NOD and NOD.Idd3 mice are recapitulated in NOD.Il21r KO mice, suggesting an important role for IL-21, in addition to IL-2, in mediating the protective effects attributed to Idd3.
Clearly, RA-regulated pathways do not exist in isolation, and much of what we know about RA and its effects has been studied in other cell types. Further studies are required to elucidate how RA signaling operates in APCs and how this differs between NOD and NOD.Idd3 mice. Furthermore, understanding how IL-21 signaling converges with the RA pathway to modulate APC function would uncover important avenues for regulating T cell responses and autoimmune diseases.
Methods
Mice and reagents.
Female NOD and NOD.Idd3 (line 1098) mice were purchased from Taconic Farms as required. NOD.Il21r mice were backcrossed onto the NOD background using a speed congenic approach (13). NOD.Idd3.Il21r KO mice were generated by intercrossing NOD.Idd3 and NOD.Il21r KO mice. Foxp3.gfp knockin (KI) reporter mice were generated as previously described (20) and then backcrossed 12 times onto NOD and NOD.Idd3 backgrounds. All experiments were carried out in accordance with the guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School.
Beads for MACS were purchased from Miltenyi Biotec. All flow cytometry reagents were purchased from BioLegend, except CD3-APC-cy7 and 7-amino-actinomycin D (7-AAD), which were from BD Bioscience. Purified functional grade mAbs were obtained from BioXCell. Recombinant cytokines were purchased from R&D Systems.
T cell activation.
CD4+CD62Lhi44lo naive cells were purified from prediabetic NOD and age-matched NOD.Idd3 or NOD.Il21r KO mice by cell sorting using a FACSAria (BD Bioscience), after enrichment for CD4+ T cells by MACS. Naive T cells were activated with either 1 μg/ml plate-bound anti-CD3 (clone 145-2C11) and 2 μg/ml soluble anti-CD28 (clone PV-1) or with 1 μg/ml soluble anti-CD3 in the presence of self APCs irradiated with 33 Gy. Cytokine production in cell culture supernatants was determined either by ELISA or Cytometric Bead Array according to the manufacturer’s instructions (BD Biosciences) after 48 hours.
Real-time PCR.
Real-time PCR was performed on an ABI Systems 7500 Fast TaqMan machine using specific TaqMan probe sets purchased from ABI Systems and cDNA synthesized using the iScript Kit (Bio-Rad). Transcript expression was normalized against β-actin.
Th17 cell polarization.
Naive T cells were activated with anti-CD3 plus 3 ng/ml TGF-β and 30 ng/ml IL-6 with or without 20 μg/ml anti–IFN-γ (clone XMG1.2) in the presence of irradiated APCs. Unless otherwise stated, CD4-depleted splenocytes were used as APCs; for some experiments, CD11c+ or CD11c–CD11b+ cells were further depleted by cell sorting. APCs were pretreated with RA in vivo by injecting 0.5 mg RA in corn oil or corn oil alone i.p. into NOD/SCID mice on alternate days over 6 days; unfractionated splenocytes were used as APCs. For in vitro RA experiments, APCs (T cell–depleted splenocytes) were first pretreated with or without 10 nM RA (Sigma-Aldrich) for 20 hours. Cells were then washed, irradiated, and cultured with naive NOD-derived T cells under Th17-polarizing conditions. Polarized cells were analyzed by intracellular cytokine staining after 3 days.
Intracellular cytokine staining.
Cells were stimulated for 4 hours with 50 ng/ml PMA and 1 μg/ml ionomycin (both from Sigma-Aldrich) in the presence of 1 μl/ml monensin (Golgi Stop, BD Bioscience). Cell surface staining was followed by fixation and permeabilization with Cytofix/Cytoperm (BD Bioscience) and washed with Perm/Wash (BD Bioscience), as per the manufacturer’s directions for intracellular cytokine staining. Samples were acquired on a LSRII (BD Bioscience), and data analysis was performed with FlowJo software (TreeStar).
CD11b+ APC cultures.
CD11b+ cells were isolated from CD11c-depleted splenocytes by positive selection using MACS. CD11c–CD11b+ cells were stimulated with PGN, and supernatants were harvested after 48 hours and assayed for cytokine production by cytometric bead array and PGE2 production using a Singleplex Multibead Kit from Assay Designs, both in accordance with the manufacturer’s instructions. For transcript profiling, CD11b+ cells were isolated by positive selection using MACS, and then CD11c–CD11b+ cells were purified by flow cytometry. Purified cells were stimulated with or without PGN for 4 hours.
Microarray analysis.
RNA was isolated using the RNeasy Mini Kit (Qiagen), including a genomic DNA elimination column, according to the manufacturer’s instructions. cRNA probes were synthesized from RNA isolated from 2 independent experiments, and each was hybridized to Affymetrix Mouse Genome 430 2.0 microarrays at the Partners Center for Personalized Genetic Medicine. Microarray data have been deposited at the Gene Expression Omnibus database ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE24350; accession number GSE24350). Analysis of microarray data was performed using GenePattern software (Broad Institute). Transcripts with a coefficient of variance of greater than 0.55 for unstimulated cells and 0.7 for PGN-stimulated cells in the duplicate samples were excluded. Network analysis was performed using Ingenuity Pathways Analysis software (Ingenuity Systems Inc.).
Statistics.
Statistical analysis was performed using unpaired, 2-tailed Student t tests, 1-way ANOVA, or Dunnett’s multiple comparison test and compared with the NOD reference group (Figure 6B). P values of less than 0.05 were considered statistically significant.
Study approval.
The experimental methods were reviewed and approved by the Institutional Animal Care and Use Committee at Harvard Medical School.
Supplementary Material
Acknowledgments
This work was supported by NIH grants NS054096 (to A.C. Anderson) and AI044880 and NS038037 (to V.K. Kuchroo); National Multiple Sclerosis Society grant RG 4374-A-1 (to A.C. Anderson); and JDRF Center on Immunological Tolerance at Harvard grants 7-2005-1329 and 4-2007-1057 (to V.K. Kuchroo). V.K. Kuchroo is the recipient of a Javits Neuroscience Investigator Award from the NIH (NS30843). S.M. Liu is a NHMRC C.J. Martin Research Fellow. We thank R. Chandwaskar, D. Rainbow, and A. Solomon for technical assistance; D. Kozoriz for cell sorting; S. Davis for help with microarray analysis; and L. Wicker for helpful discussions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(11):4303–4310. doi:10.1172/JCI46187.
Sue M. Liu’s present address is: Immunology Research Program, Garvan Institute of Medical Research, Darlinghurst, NSW, Australia.
References
- 1.Wicker LS, Todd JA, Prins JB, Podolin PL, Renjilian RJ, Peterson LB. Resistance alleles at two non-major histocompatibility complex-linked insulin-dependent diabetes loci on chromosome 3, Idd3 and Idd10, protect nonobese diabetic mice from diabetes. J Exp Med. 1994;180(5):1705–1713. doi: 10.1084/jem.180.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Encinas JA, et al. QTL influencing autoimmune diabetes and encephalomyelitis map to a 0.15-cM region containing Il2. Nat Genet. 1999;21(2):158–160. doi: 10.1038/5941. [DOI] [PubMed] [Google Scholar]
- 3.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3(+) CD25(+) CD4(+) regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J Exp Med. 2005;201(5):723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26(3):371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 5.Korn T, et al. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448(7152):484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nurieva R, et al. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448(7152):480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, et al. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8(9):967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 8.Yamanouchi J, et al. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39(3):329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson AC, Chandwaskar R, Lee DH, Kuchroo VK. Cutting edge: the Idd3 genetic interval determines regulatory T cell function through CD11b+CD11c- APC. J Immunol. 2008;181(11):7449–7452. doi: 10.4049/jimmunol.181.11.7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alard P, Manirarora JN, Parnell SA, Hudkins JL, Clark SL, Kosiewicz MM. Deficiency in NOD antigen-presenting cell function may be responsible for suboptimal CD4+CD25+ T-cell-mediated regulation and type 1 diabetes development in NOD mice. Diabetes. 2006;55(7):2098–2105. doi: 10.2337/db05-0810. [DOI] [PubMed] [Google Scholar]
- 11.King C, Ilic A, Koelsch K, Sarvetnick N. Homeostatic expansion of T cells during immune insufficiency generates autoimmunity. Cell. 2004;117(2):265–277. doi: 10.1016/S0092-8674(04)00335-6. [DOI] [PubMed] [Google Scholar]
- 12.Spolski R, Kashyap M, Robinson C, Yu Z, Leonard WJ. IL-21 signaling is critical for the development of type I diabetes in the NOD mouse. Proc Natl Acad Sci U S A. 2008;105(37):14028–14033. doi: 10.1073/pnas.0804358105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datta S, Sarvetnick NE. IL-21 limits peripheral lymphocyte numbers through T cell homeostatic mechanisms. PLoS One. 2008;3(9):e3118. doi: 10.1371/journal.pone.0003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutherland AP, et al. IL-21 is required for the development of type 1 diabetes in NOD mice. Diabetes. 2009;58(5):1144–1155. doi: 10.2337/db08-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cua DJ, et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421(6924):744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 16.Jain R, et al. Innocuous IFNgamma induced by adjuvant-free antigen restores normoglycemia in NOD mice through inhibition of IL-17 production. J Exp Med. 2008;205(1):207–218. doi: 10.1084/jem.20071878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Orozco N, Chung Y, Chang SH, Wang YH, Dong C. Th17 cells promote pancreatic inflammation but only induce diabetes efficiently in lymphopenic hosts after conversion into Th1 cells. Eur J Immunol. 2009;39(1):216–224. doi: 10.1002/eji.200838475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bending D, et al. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J Clin Invest. 2009;119(3):565–572. doi: 10.1172/JCI37865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emamaullee JA, et al. Inhibition of Th17 cells regulates autoimmune diabetes in NOD mice. Diabetes. 2009;58(6):1302–1311. doi: 10.2337/db08-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bettelli E, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441(7090):235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 21.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 22.Park H, et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6(11):1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Alise AM, et al. The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc Natl Acad Sci U S A. 2008;105(50):19857–19862. doi: 10.1073/pnas.0810713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sgouroudis E, Albanese A, Piccirillo CA. Impact of protective IL-2 allelic variants on CD4+ Foxp3+ regulatory T cell function in situ and resistance to autoimmune diabetes in NOD mice. J Immunol. 2008;181(9):6283–6292. doi: 10.4049/jimmunol.181.9.6283. [DOI] [PubMed] [Google Scholar]
- 25.Boniface K, et al. Prostaglandin E2 regulates Th17 cell differentiation and function through cyclic AMP and EP2/EP4 receptor signaling. J Exp Med. 2009;206(3):535–548. doi: 10.1084/jem.20082293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chizzolini C, et al. Prostaglandin E2 synergistically with interleukin-23 favors human Th17 expansion. Blood. 2008;112(9):3696–3703. doi: 10.1182/blood-2008-05-155408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napolitani G, Acosta-Rodriguez EV, Lanzavecchia A, Sallusto F. Prostaglandin E2 enhances Th17 responses via modulation of IL-17 and IFN-gamma production by memory CD4(+) T cells. Eur J Immunol. 2009;39(5):1301–1312. doi: 10.1002/eji.200838969. [DOI] [PubMed] [Google Scholar]
- 28.Mucida D, et al. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317(5835):256–260. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 29.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8(9):958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 30.Huber M, et al. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc Natl Acad Sci U S A. 2008;105(52):20846–20851. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao S, et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181(4):2277–2284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Battle TE, Levine RA, Yen A. Retinoic acid-induced blr1 expression promotes ERK2 activation and cell differentiation in HL-60 cells. Exp Cell Res. 2000;254(2):287–298. doi: 10.1006/excr.1999.4766. [DOI] [PubMed] [Google Scholar]
- 33.Kim HJ, Lotan R. Identification of retinoid-modulated proteins in squamous carcinoma cells using high-throughput immunoblotting. Cancer Res. 2004;64(7):2439–2448. doi: 10.1158/0008-5472.CAN-03-2643. [DOI] [PubMed] [Google Scholar]
- 34.Tice DA, et al. Synergistic induction of tumor antigens by Wnt-1 signaling and retinoic acid revealed by gene expression profiling. J Biol Chem. 2002;277(16):14329–14335. doi: 10.1074/jbc.M200334200. [DOI] [PubMed] [Google Scholar]
- 35.Zhong JC, et al. Effects of all-trans retinoic acid on orphan receptor APJ signaling in spontaneously hypertensive rats. Cardiovasc Res. 2005;65(3):743–750. doi: 10.1016/j.cardiores.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 36.Korn T, et al. IL-6 controls Th17 immunity in vivo by inhibiting the conversion of conventional T cells into Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2008;105(47):18460–18465. doi: 10.1073/pnas.0809850105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill JA, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi cells. Immunity. 2008;29(5):758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stosic-Grujicic S, Cvjeticanin T, Stojanovic I. Retinoids differentially regulate the progression of autoimmune diabetes in three preclinical models in mice. Mol Immunol. 2009;47(1):79–86. doi: 10.1016/j.molimm.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Van YH, Lee WH, Ortiz S, Lee MH, Qin HJ, Liu CP. All-trans retinoic acid inhibits type 1 diabetes by T regulatory (Treg)-dependent suppression of interferon-gamma-producing T-cells without affecting Th17 cells. Diabetes. 2009;58(1):146–155. doi: 10.2337/db08-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zunino SJ, Storms DH, Stephensen CB. Diets rich in polyphenols and vitamin A inhibit the development of type I autoimmune diabetes in nonobese diabetic mice. J Nutr. 2007;137(5):1216–1221. doi: 10.1093/jn/137.5.1216. [DOI] [PubMed] [Google Scholar]
- 41.Stock A, Booth S, Cerundolo V. Prostaglandin E2 suppresses the differentiation of retinoic acid-producing dendritic cells in mice and humans. J Exp Med. 2011;208(4):761–773. doi: 10.1084/jem.20101967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim BH, Kang KS, Lee YS. Effect of retinoids on LPS-induced COX-2 expression and COX-2 associated PGE(2) release from mouse peritoneal macrophages and TNF-alpha release from rat peripheral blood mononuclear cells. Toxicol Lett. 2004;150(2):191–201. doi: 10.1016/j.toxlet.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Mudter J, et al. The transcription factor IFN regulatory factor-4 controls experimental colitis in mice via T cell-derived IL-6. J Clin Invest. 2008;118(7):2415–2426. doi: 10.1172/JCI33227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dahlen E, Hedlund G, Dawe K. Low CD86 expression in the nonobese diabetic mouse results in the impairment of both T cell activation and CTLA-4 up-regulation. J Immunol. 2000;164(5):2444–2456. doi: 10.4049/jimmunol.164.5.2444. [DOI] [PubMed] [Google Scholar]
- 45.Piganelli JD, Martin T, Haskins K. Splenic macrophages from the NOD mouse are defective in the ability to present antigen. Diabetes. 1998;47(8):1212–1218. doi: 10.2337/diabetes.47.8.1212. [DOI] [PubMed] [Google Scholar]
- 46.Serreze DV, Gaskins HR, Leiter EH. Defects in the differentiation and function of antigen presenting cells in NOD/Lt mice. J Immunol. 1993;150(6):2534–2543. [PubMed] [Google Scholar]
- 47.Bradshaw EM, et al. Monocytes from patients with type 1 diabetes spontaneously secrete proinflammatory cytokines inducing Th17 cells. J Immunol. 2009;183(7):4432–4439. doi: 10.4049/jimmunol.0900576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.