Abstract
Therapies inhibiting receptor tyrosine kinases (RTKs) are effective against some human cancers when they lead to simultaneous downregulation of PI3K/AKT and MEK/ERK signaling. However, mutant KRAS has the capacity to directly activate ERK and PI3K signaling, and this is thought to underlie the resistance of KRAS mutant cancers to RTK inhibitors. Here, we have elucidated the molecular regulation of both the PI3K/AKT and MEK/ERK signaling pathways in KRAS mutant colorectal cancer cells and identified combination therapies that lead to robust cancer cell apoptosis. KRAS knockdown using shRNA suppressed ERK signaling in all of the human KRAS mutant colorectal cancer cell lines examined. However, no decrease, and actually a modest increase, in AKT phosphorylation was often seen. By performing PI3K immunoprecipitations, we determined that RTKs, often IGF-IR, regulated PI3K signaling in the KRAS mutant cell lines. This conclusion was also supported by the observation that specific RTK inhibition led to marked suppression of PI3K signaling and biochemical assessment of patient specimens. Interestingly, combination of RTK and MEK inhibitors led to concomitant inhibition of PI3K and MEK signaling, marked growth suppression, and robust apoptosis of human KRAS mutant colorectal cancer cell lines in vitro and upon xenografting in mice. These findings provide a framework for utilizing RTK inhibitors in the treatment of KRAS mutant colorectal cancers.
Introduction
Metastatic colorectal cancer remains largely incurable, and newer, targeted therapies have had only a modest impact on this disease. Colorectal cancers can be classified according to their major somatic genetic abnormalities, and each subtype will likely benefit from tailored targeted therapy approaches. Examples of highly effective targeted therapies in other advanced solid malignancies based on genotype are emerging. Some successful examples of effective therapies targeting receptor tyrosine kinases include EGFR tyrosine kinase inhibitors (TKIs) for EGFR mutant lung cancers (1–3), ALK inhibitors for EML4-ALK translocated cancers (4–7), HER2 inhibitors for HER2-amplified cancers (8–10), and imatinib for KIT and PDGFR mutant gastrointestinal stromal tumors (11). In each of these paradigms, inhibition of the corresponding tyrosine kinase invariably leads to concomitant downregulation of the PI3K/AKT and MEK/ERK signaling pathways. In fact, direct simultaneous inhibition of these pathways has been shown to be sufficient to recapitulate the effects of the TKI in vitro and in vivo (9). Thus, the downregulation of these pathways may underlie much of the therapeutic value of the TKIs.
In colorectal cancer, monoclonal antibodies against EGFR, such as cetuximab, are widely used. Although this antibody has activity in KRAS WT colorectal cancers, it provides no benefit to patients whose cancers are KRAS mutant (12–18). Thus, there is a clear need to establish treatment strategies for KRAS mutant cancers, as they have proven largely resistant to all targeted therapies to date (19, 20). Since KRAS can directly activate the MEK/ERK and PI3K/AKT signaling pathways by direct binding to RAF proteins and the PI3K subunit p110, respectively (19, 21–24), it has been postulated that therapies targeting receptor tyrosine kinases will generally fail to downregulate these pathways in KRAS mutant cancers and will therefore be ineffectual. However, simultaneous downregulation of MEK/ERK and PI3K/AKT may have potential therapeutic value in KRAS mutant cancers. Recent work by several laboratories including ours has shown that direct concomitant downregulation of PI3K and MEK induces impressive regressions of KRAS mutant cancers in vivo (25–28).
Since it has been difficult to identify drugs that specifically disrupt KRAS function, it remains unknown whether there are any therapeutic strategies that can simultaneously suppress both pathways other than combining PI3K and MEK pathway inhibitors. As the PI3K and MEK pathways are integral components of many cellular processes, it also remains unknown whether patients will tolerate simultaneous, complete inhibition of both pathways. Indeed, it is quite possible that toxicities may compromise the efficacy of this approach. In addition, recent data suggest that activation of other tyrosine kinase cascades may potentially be associated with resistance to combined PI3K and MEK inhibition (29). Therefore, alternative therapeutic strategies, perhaps those targeting receptor tyrosine kinases (RTKs), that indirectly suppress one or both pathways may prove both effective and more tolerable. Herein, we have conducted studies to elucidate the molecular regulation of the PI3K/AKT and MEK/ERK pathways in KRAS mutant cancers. Our findings have led to insights into the specific regulation of these pathways in colorectal cancers and suggest genotype-directed therapeutic approaches that may effectively suppress these pathways in KRAS mutant colorectal cancers.
Results
Regulation of ERK and PI3K in KRAS mutant colorectal cancers.
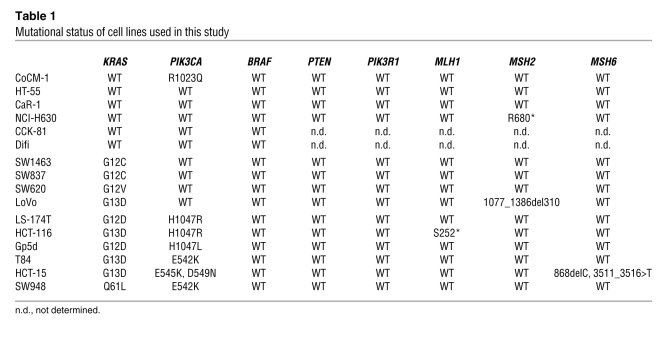
It has been hypothesized that since activated KRAS directly binds to RAF proteins and the PI3K subunit p110, the mutant form of KRAS leads to constitutive activation of both the MEK/ERK and PI3K/AKT pathways, accounting for the resistance of KRAS mutant tumors to TKIs (23, 30). Although many elegant studies in other model systems have demonstrated that RAS can bind PI3K and that PI3K is critical in KRAS-induced transformation (25, 31–33), the molecular mechanism of PI3K regulation in established KRAS mutant colorectal cancers has not been directly investigated. Thus, to determine whether KRAS constitutively activates both MEK and PI3K signaling in established colorectal cancers, we used RNA interference targeting KRAS using two independent KRAS shRNAs. As anticipated, ERK phosphorylation was suppressed by KRAS knockdown in the KRAS mutant cancer cell lines SW620 and SW837, but not the control KRAS WT cell line CoCM-1 (Figure 1A). However, to our surprise, knockdown of KRAS in mutant cancers did not suppress AKT phosphorylation. These experiments were performed in an additional 8 independent KRAS mutant colorectal cancer cell lines (total of 10), of which 6 had concurrent PIK3CA mutations. The specific mutations harbored in each cell line are shown in Table 1. As summarized in Figure 1B, ERK phosphorylation was suppressed in the KRAS mutant cancers, but AKT phosphorylation did not decrease, and in some cases appears to have increased modestly, upon KRAS knockdown (for raw data on Western blots, see Figure 1A and Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI57909DS1).
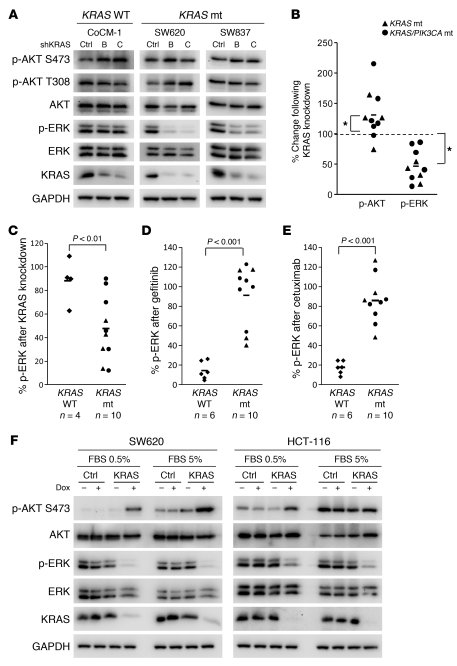
Figure 1. KRAS is required for MEK/ERK signaling in KRAS mutated colorectal cancers.
(A) KRAS WT (CoCM-1) and KRAS mutant (mt; SW620, SW837) cells were infected with 1 of 2 independent shRNAs targeting KRAS (B and C) or control shRNA (Ctrl). Three days after infection, protein lysates were prepared and probed with the indicated antibodies. (B) The levels of p-AKT and -ERK were quantified from each of the KRAS mutant cells following knockdown of KRAS (raw data in A and Supplemental Figure 1, A and B) and are presented as percentage of phosphorylated ERK or AKT relative to control shRNA–treated cells. Each point represents the results from a single cell line. Bars indicate mean values. *P < 0.05. (C) The levels of p-ERK were quantified from each of the KRAS mutant and WT cells following knockdown of KRAS (raw data in A and Supplemental Figure 1A) and are presented as percentage of phosphorylated ERK relative to control shRNA–treated cells. Bars indicate mean values. (D and E) The levels of phosphorylated ERK after treatment with gefitinib (D) and cetuximab (E) were quantified for each cell line examined (raw data shown in Supplemental Figures 2 and 3). Bars indicate mean values. (F) SW620 and HCT-116 KRAS mutant cells transduced with control or KRAS inducible shRNA were cultured in the presence or absence of doxycycline (Dox; 10 ng/ml) in either full serum (5% FBS) or low serum (0.5% FBS) for 72 hours. The cells were lysed, and Western blots were probed with the indicated antibodies.
Table 1 .
Mutational status of cell lines used in this study
We also evaluated the effect of KRAS knockdown in KRAS WT cancers (Figure 1A and Supplemental Figure 1B). In contrast to the KRAS mutant cancers, knockdown of KRAS did not suppress ERK signaling in these cells (Figure 1C). In the KRAS WT cancers, suppression of EGFR signaling by gefitinib or cetuximab suppressed ERK phosphorylation, but those treatments had no effect in KRAS mutant cancers (Figure 1, D and E, and Supplemental Figures 2 and 3). Of note, in a few KRAS mutant cancers, EGFR inhibition led to a modest suppression of ERK phosphorylation. In these cell lines, knockdown of KRAS still had a more profound impact on ERK phosphorylation than EGFR inhibition (Supplemental Figure 4). In total, these results confirm that the regulation of ERK signaling is different in KRAS WT and mutant cancers.
In the KRAS mutant cancers, we had expected that PI3K/AKT signaling would decrease with KRAS knockdown and thus were concerned that the studies evaluating the role of KRAS in regulating PI3K signaling could be affected by the presence of serum and its potential to activate PI3K. Thus, we also determined whether KRAS knockdown affected PI3K/AKT signaling in KRAS mutant cancers in the absence of serum. For these experiments, we employed doxycycline-inducible shRNA targeting KRAS. This enabled more acute KRAS knockdown in the presence or absence of serum. As shown in Figure 1F, KRAS knockdown led to a loss of ERK phosphorylation, but not AKT phosphorylation, in both low and normal serum conditions. For exclusion of off-target effects of the shRNA targeting KRAS, KRAS expression was rescued by mutant KRAS (G12V) cDNA refractory to silencing. As shown in Supplemental Figure 5, restoration of KRAS blocked the suppression of ERK phosphorylation induced by the KRAS shRNA, and it even abrogated the modest induction of AKT phosphorylation in the SW620 and HCT-116 cells with low serum. Thus, the impact of KRAS shRNA on ERK and AKT phosphorylation appears to be on-target and due to the loss of KRAS protein.
To further explore whether mutant KRAS activates PI3K, we used a transgenic model of lung cancer driven by the tetracycline-inducible mutant KRAS (34). In this model, lung tumors are induced by expression of a mutant KRAS, and removal of doxycycline leads to loss of expression of mutant KRAS and tumor regressions (25). We found that withdrawal of doxycycline led to suppression of ERK phosphorylation, but did not suppress PI3K signaling (Supplemental Figure 6, A and B), further substantiating the notion that mutant KRAS is dispensable for PI3K activation in KRAS mutant cancers.
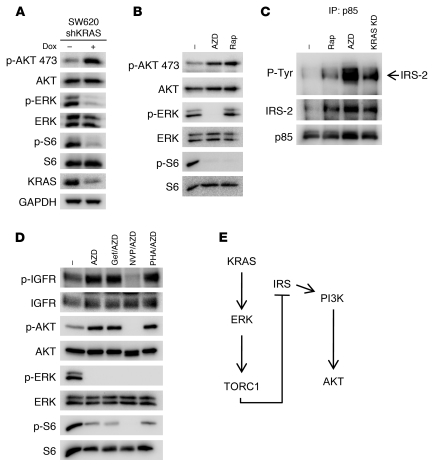
It was initially unexpected that knockdown of KRAS modestly increases AKT phosphorylation in some of the cell lines. Since KRAS knockdown suppressed both ERK and S6 phosphorylation (Figure 2A), we hypothesized that this increase in AKT phosphorylation in the SW620 cells may be due to the de-repression of the well-established feedback inhibition exerted by TORC1 on PI3K activation (35). Indeed, we observed that MEK/ERK regulates TORC1 in these cells and other KRAS mutant colorectal cell lines (Figure 2B and Supplemental Figure 7). Indeed, treatment with the MEK inhibitor led to an increased association between PI3K and IRS proteins (Figure 2C) and increased insulin-like growth factor I receptor (IGF-IR) phosphorylation (Figure 2D), consistent with the classical feedback regulation by TORC1. Concomitant treatment with an IGF-IR/insulin receptor (IR) inhibitor, NVP-AEW541, blocked the activation of AKT induced by MEK inhibition (Figure 2D). Accordingly, knockdown of KRAS (which suppressed ERK phosphorylation; Figure 2A) also induced an interaction between PI3K and IRS proteins (Figure 2C). These data suggest that this feedback (Figure 2E) might account for the modest increase in PI3K/AKT signaling following MEK inhibition. The results together indicate that KRAS appears to be a dominant regulator of MEK/ERK signaling, but not of PI3K/AKT signaling, in KRAS mutant colorectal cancers.
Figure 2. The increase in p-AKT induced by KRAS knockdown is consistent with loss of feedback inhibition from TORC1.
(A–C) SW620 cells infected with tetracycline-inducible KRAS shRNA were treated with 10 ng/ml doxycycline or vehicle for 72 hours. In parallel, parental SW620 cells were treated with DMSO, 50 nM rapamycin (Rap), or 1 μM MEK inhibitor AZD6244 (AZD) for 24 hours. (A and B) Protein lysates were probed with the indicated antibodies or (C) were subjected to immunoprecipitation with the anti-p85 antibody, followed by Western blot analyses with the indicated antibodies. KD, knockdown. (D) SW620 cells were treated with the indicated single-agent drug or drug combinations for 24 hours, and lysates were probed with the indicated antibodies. AZD, AZD6244 (1 μM); Gef/AZD, gefitinib (1 μM) and AZD6244 (1 μM); NVP/AZD, NVP-AEW541 (1 μM) and AZD6244 (1 μM); PHA/AZD, PHA-665752 (1 μM) and AZD6244 (1 μM). Note that the SW620 cells have undetectable EGFR, and no p-EGFR was detected. (E) Diagram of the feedback pathway from KRAS knockdown to PI3K activation.
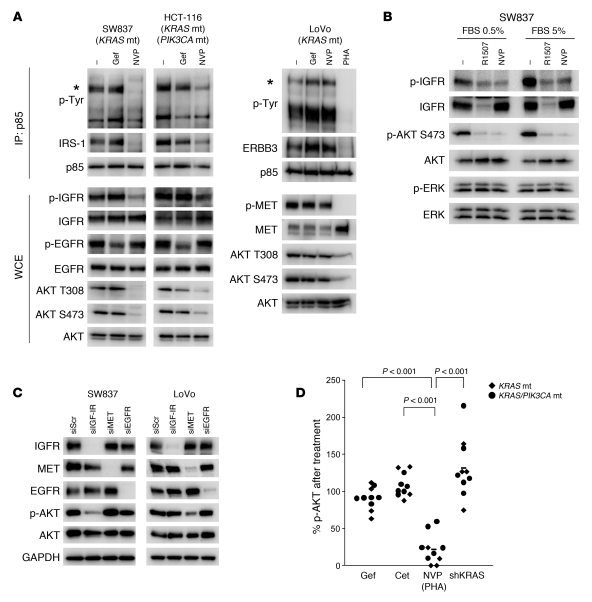
Since these results suggested that suppression of mutant KRAS is not sufficient to downregulate PI3K/AKT signaling, a better understanding of how PI3K/AKT signaling is activated in KRAS mutant colorectal cancers may suggest potential strategies to downregulate PI3K/AKT signaling in these cancers. Thus, we aimed to determine how PI3K is activated in established KRAS mutant cancers using a biochemical approach that takes advantage of the molecular mechanism of PI3K activation in many cancers. The regulatory subunit (p85) of class IA PI3K has two SH2 domains that engage phosphotyrosine proteins, such as RTKs or adaptors. This binding activates PI3K by recruiting it to the membrane, where its substrate resides, and by relieving a tonic intermolecular inhibition exerted by p85 onto p110 (36, 37). By immunoprecipitating p85, one can identify the phosphotyrosine proteins that are directly binding and activating PI3K. We have utilized this approach to identify how PI3K is activated in a number of cancer paradigms (38–40). In the KRAS mutant cancers, we observed that IRS proteins coprecipitated with PI3K in the SW837 (KRAS mutant) and HCT-116 (KRAS and PIK3CA double mutant) cells (Figure 3A). In these cells, these complexes were disrupted by a small molecule IGF-IR kinase inhibitor (NVP-AEW541), but not by an EGFR inhibitor (gefitinib), and accordingly, NVP-AEW541 led to loss of AKT phosphorylation. In LoVo cells, ERBB3 (~240 kDa) and growth factor receptor–bound protein 2 (GRB2)–associated binder 1 (Gab1) coimmunoprecipitated with p85. We previously observed these adaptors coprecipitating with PI3K in cancers that have MET-dependent activation of PI3K (39). Indeed, PHA-665752, a MET TKI, but not gefitinib or NVP-AEW541, disrupted both interactions and led to loss of AKT phosphorylation in these KRAS mutant cells (Figure 3A). These results are consistent with a previous report noting that LoVo cells have defective processing of the MET precursor resulting in its constitutive activation (41). Of the ten KRAS mutant cell lines examined, we found p85/IRS-1 or p85/IRS-2 complexes in 9 cell lines (data not shown). NVP-AEW541 has previously been shown to be highly specific for IGF-IR (42). Consistent with this notion, we observed that a humanized monoclonal anti–IGF-IR antibody, R1507 (43), downregulated IGF-IR and recapitulated the effects of NVP-AEW541 on AKT phosphorylation in low or normal serum (Figure 3B). Moreover, siRNA targeting IGF-IR, but not EGFR or MET, led to suppression of AKT phosphorylation in the SW837 cells (Figure 3C). MET siRNA led to suppression of AKT phosphorylation in the LoVo cells (Figure 3C). In contrast to the findings that neither KRAS knockdown nor EGFR inhibition suppressed AKT phosphorylation, inhibition of the RTK identified by PI3K immunoprecipitations effectively led to marked suppression of AKT phosphorylation in all of the KRAS mutant cancer cell lines (Figure 3D; individual Western blots are shown in Figure 3A, Figure 4A, Supplemental Figure 2, and Supplemental Figure 8A). Of note, although targeting RTKs effectively suppressed AKT phosphorylation in all of the KRAS mutant cancers, suppression of AKT phosphorylation was less impressive in 2 of the 6 KRAS/PIK3CA mutant cell lines, which might suggest that some cancers with concomitant PIK3CA mutations may be less susceptible to this approach. In total, these studies suggest that while ERK activation is primarily regulated by mutant KRAS, the PI3K/AKT pathway requires RTK-induced activation in KRAS mutant colorectal cancers, pointing to new potential therapeutic strategies.
Figure 3. Mechanism of PI3K activation in KRAS mutant cancers.
(A) KRAS mutant (SW837 and LoVo) or KRAS/PIK3CA mutant (HCT-116) colorectal cancers were treated with the indicated compounds for 6 hours (gef, gefitinib 1 μM; NVP, NVP-AEW541 1 μM; PHA, PHA-665752 1 μM), and the resulting protein lysates were immunoprecipitated with an anti-p85 antibody. The precipitated proteins were analyzed by Western blots with the indicated antibodies. Whole cell extracts were probed with the indicated antibodies. Asterisks indicate the IRS proteins for SW837 and HCT-116 cells, and ERBB3 for LoVo cells. (B) SW837 cells were grown in either normal serum (5% FBS) or low serum (0.5% FBS) and treated with vehicle, R1507 anti–IGF-IR antibody (25 μg/ml), or NVP-AEW541 (1 μM) for 6 hours. The cells were lysed and probed with the indicated antibodies. (C) The indicated cell lines were transfected with siRNA against IGF-IR, EGFR, or MET or control siRNA (siScr). Cells were lysed 72 hours after transfection. (D) Nine KRAS mutant cell lines were treated with 1 μM gefitinib, 10 μg/ml cetuximab, or 1 μM NVP-AEW541 for 6 hours. LoVo cells were treated with 1 μM PHA-665752 instead of the IGF-IR inhibitor. Cells were lysed, and phosphorylated and total AKT levels were determined (raw data for each cell line is shown in Figure 4A, Supplemental Figure 2, and Supplemental Figure 8A). The intensities of phosphorylated AKT (Ser473) and total AKT were quantified, and the amount of p-AKT relative to untreated cells is shown. p-AKT levels after KRAS knockdown are the same as in Figure 1B.
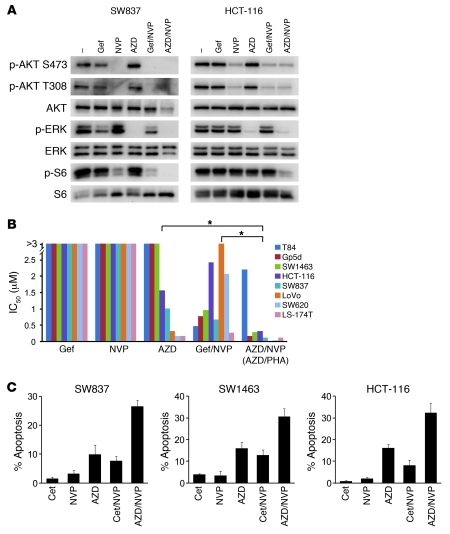
Figure 4. Combination strategies targeting PI3K/AKT and MEK/ERK effectively lead to cell death.
(A) Cells were treated with either DMSO (–) or the indicated drug or drug combinations for 6 hours. Drugs were used at the same concentrations as in Figure 3. AZD6244 (AZD) was used at 1 μM. Protein lysates were probed with the indicated antibodies. (B) IC50 values from 72-hour Syto60 assays were determined for each KRAS mutant cancer cell line using the indicated drugs and drug combinations (raw data are shown in Supplemental Figure 9, A and B). LoVo cells were treated with PHA-665752 and AZD6244 instead of NVP-AEW541 and AZD6244. The results for each cell line are represented as individual bars. *P < 0.01 between treatment groups. (C) KRAS mutant cells were treated with the indicated drugs and combinations for 72 hours. The percentage of cells undergoing apoptosis, as measured by annexin V positivity, is shown relative to untreated cells. The average ± SD of 3 independent experiments is shown.
Combination strategies that suppress PI3K/AKT and MEK/ERK signaling induce cell death.
One overall therapeutic strategy for colorectal cancers centers on simultaneously downregulating PI3K/AKT and MEK/ERK signaling. In KRAS mutant cancers, we had found that KRAS is an important activator of the ERK pathway and that RTKs are dominant regulators of the PI3K pathway. Therefore, we determined whether combined treatment with an RTK and MEK inhibitors would simultaneously suppress the PI3K/AKT and MEK/ERK signaling pathways. Indeed, treatment with a MEK inhibitor and IGF-IR inhibitor (or MET inhibitor in LoVo cells) led to a simultaneous marked loss of AKT and ERK phosphorylation (Figure 4A and Supplemental Figure 8A). Accordingly, the combination of MEK and IGF-IR inhibitors more potently decreased cell viability compared with single-agent EGFR, MEK, and IGF-IR inhibitors as well as combined EGFR and IGF-IR inhibition (P < 0.01) (Figure 4B and Supplemental Figure 9, A and B). Of note, T84 cells were unique in that they required two RTK inhibitors in combination with a MEK inhibitor to downregulate both PI3K and MEK/ERK signaling (Supplemental Figure 8B). As expected, the potency of the different therapeutic combinations correlated with their capacity to downregulate both PI3K/AKT and MEK/ERK signaling (Supplemental Figure 10).
Since induction of apoptosis may also be important for inducing tumor regressions (9, 44), and the decrease in cell viability measured in the assays in Figure 4B may have been due to cytostatic and/or apoptotic effects, we wished to directly determine whether the combinations leading to simultaneous suppression of AKT and ERK phosphorylation led to more substantial apoptosis. Indeed, we found that the combination of a MEK inhibitor and an IGF-IR inhibitor induced a strong apoptotic response, as the single agents and the combination of IGF-IR inhibitor with cetuximab were far less effective (Figure 4C). Similarly, in the KRAS mutant LoVo cells (which demonstrated MET-dependent activation of PI3K; Figure 3, A and C), the combination of the MET inhibitor PHA-665752 and AZD6244 induced remarkable growth inhibition and enhanced apoptosis (Supplemental Figure 9B). Taken together, our results provide a rationale for combinatorial therapy with an RTK (most often IGF-IR) and MEK inhibitor for KRAS mutant colorectal cancers.
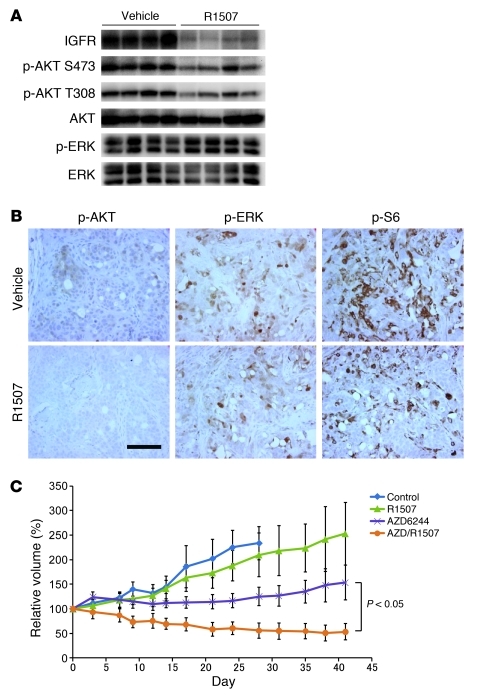
To determine whether appropriate RTK inhibition could suppress PI3K signaling in KRAS mutant cancers in vivo, the KRAS mutant cancer cell line SW837 was grown as xenograft tumors in mice. The tumor-bearing mice were treated with R1507 or vehicle for 11 days. As shown in Figure 5, A and B, R1507 led to suppression of AKT phosphorylation by Western blot analysis and immunohistochemistry (IHC). Of note, the suppression of AKT phosphorylation was more dramatic by IHC than by Western blot, and this likely reflects the specific suppression of PI3K signaling in the tumor cells, and not the surrounding stroma. These data further support the finding that the PI3K/AKT pathway can be downregulated in KRAS mutant cancers by targeting of the appropriate RTK.
Figure 5. The combination of an anti–IGF-IR antibody and a MEK inhibitor induces tumor regressions in a KRAS mutant colorectal cancer in vivo.
(A and B) Inhibition of IGF-IR suppresses PI3K/AKT signaling in SW837 tumor xenograft. SW837 KRAS mutant xenografts were treated with vehicle or R1507 for 11 days. The xenografts were assessed by Western blot analyses (A) and immunohistochemical analyses (B) with the indicated antibodies. Scale bar: 100 μM. (C) Xenograft tumors derived from the KRAS mutant SW837 colorectal cancer cells were developed. Once they achieved an average size of 400 mm3, the tumors were treated with the indicated drug regimens, and relative tumor volumes were plotted over time from the start of treatment (day 0) (mean ± SEM).
We next assessed whether the effect of combined PI3K/AKT and MEK/ERK signaling inhibition on cell proliferation and apoptosis observed in vitro would lead to greater therapeutic efficacy in vivo. Mice bearing SW837 tumor xenografts were treated with anti–IGF-IR antibody and MEK inhibitor, alone or in combination. Whereas inhibition of the MEK pathway alone via AZD6244 treatment slowed tumor growth, tumor regression was observed only with combination treatment (Figure 5C and Supplemental Figure 11).
PI3K immunoprecipitations from primary colorectal cancers.
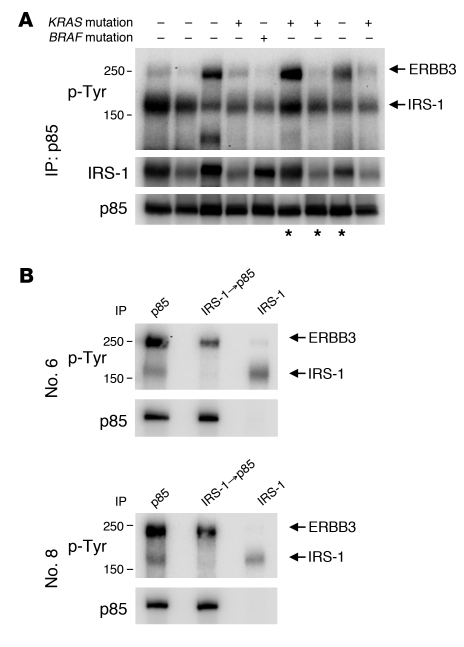
The above experiments were performed with cancer cell lines grown in vitro or as xenograft tumors. To determine whether there was evidence for the same regulation of PI3K in primary human cancers, we examined PI3K immunoprecipitations from resected human colorectal tumors. We detected IRS-1 associated with PI3K in all of 9 colorectal tumor samples, including 4 KRAS mutant and 1 BRAF mutant tumor (Figure 6A and Supplemental Figure 12A). We also identified interaction with ERBB3 in a subset of these cancers. We confirmed that IRS-1 was the major p-Tyr band running at approximately 160 kDa by immunoprecipitating IRS-1 with anti–IRS-1 antibodies prior to the p85 IP. As predicted, this resulted in loss of association with the major 160-kDa p-Tyr protein, but no change in association with ERBB3 (Figure 6B and Supplemental Figure 12B). We observed interactions between PI3K and IRS-1 in an additional 13 tumor samples, including 3 KRAS and three BRAF mutant cancers (Supplemental Figure 12C). These studies confirm that the IRS proteins and ERBB3 are the major activators of PI3K in human colorectal cancers, and also suggest that PI3K is activated by RTKs to a similar extent in KRAS WT and mutant cancers. Importantly, these results confirm that the IGF-IR/IR signaling pathways are actively engaging the PI3K signaling pathway in KRAS mutant human colorectal cancers and RTK inhibitors may potently suppress PI3K signaling in patients as it does in laboratory models.
Figure 6. PI3K immunoprecipitations from primary colorectal cancers.
(A) p85 was immunoprecipitated from 9 different human colorectal cancer specimens, followed by Western blotting for the indicated antibodies. The KRAS and BRAF mutational status of each cancer specimen is indicated. Asterisks indicate samples used for B and Supplemental Figure 12B. (B) Tumor lysates were immunoprecipitated with p85 (lane 1) or IRS-1 (lane 3). Supernatant from the IRS-1 IP was followed by p85 IP (lane 2). Note that immunoprecipitation of IRS-1 clears the lower, but not the upper, p-Tyr protein that coprecipitates with p85. Patients 6 and 8 correspond to the cancer specimens shown in lanes 6 and 8 of A, respectively.
Discussion
Therapies targeting receptor tyrosine kinases have emerged as effective treatments for a subset of advanced cancers. In such cancers, inhibition of the corresponding tyrosine kinase results in loss of PI3K/AKT and MEK/ERK signaling. Moreover, direct downregulation of PI3K/AKT and MEK/ERK is sufficient to recapitulate the induction of apoptosis induced by EGFR or HER2 TKIs in EGFR mutant and HER2-amplified cancers, respectively (9). This suggests that simultaneous loss of PI3K/AKT and MEK/ERK signaling may underlie many of the antitumor effects observed with tyrosine kinase inhibition. The presence of KRAS mutations is a well-validated biomarker for resistance to single-agent RTK inhibitors (12, 13, 30), and it is widely hypothesized that this resistance is the result of mutant KRAS directly activating downstream signaling (14, 45). Indeed, direct inhibition of MEK and PI3K has shown impressive activity in KRAS mutant cancers in vivo (25, 26). However, since the MEK/ERK and PI3K/AKT pathways are essential to many normal cellular processes and in many cell types, it is possible that toxicity will ultimately limit simultaneous full suppression of both of these pathways in patients. Thus, we aimed to determine how PI3K/AKT and MEK/ERK was regulated in KRAS mutated cancers. To our surprise, we found that specific RTK inhibition led to potent suppression of PI3K signaling in these cancers.
This finding points to an opportunity to effectively use RTK inhibitors in the treatment of KRAS mutant colorectal cancers. Combinations utilizing specific RTK inhibitors may potentially lead to reduced toxicity. However, since IGF-IR is also utilized for several cellular processes, clinical experience will be needed to verify this notion. Importantly, there may be greater therapeutic value from targeting RTKs, since their inhibition may suppress other prosurvival pathways in addition to PI3K/AKT.
In this study, we observed that knockdown of KRAS did not suppress PI3K signaling, but RTK inhibition did. This was a bit unanticipated, since many studies (including our own) have shown that PI3K signaling is important in KRAS-induced transformation and tumor maintenance (23, 25, 33, 46–48). It should be noted that many of these groundbreaking studies have assessed the role of PI3K signaling in KRAS mutant cancers but do not specifically determine the molecular mechanism of PI3K activation and, in particular, the specific role of KRAS in directly activating PI3K signaling in established human cancer models. However, in an elegant study by Downward and colleagues, p110 was mutated so it could not bind KRAS, and this blocked mutant KRAS–induced lung tumorigenesis (33). Those results suggested that direct binding of KRAS to p110 was critical for KRAS-induced tumorigenesis. In the current study, we also observed an important role for PI3K signaling in KRAS mutant cancers. We observed clear evidence that RTKs exert a more dominant role in the activation of PI3K signaling in established human KRAS mutant colorectal cancers. This is supported by a previous study that indicated that PI3K signaling in a KRAS mutant colorectal cancer cell line was highly responsive to exogenous insulin (49). Therefore, we hypothesize that KRAS may directly activate PI3K to levels sufficient to permit cellular transformation during initial tumorigenesis induced by mutant KRAS, but RTK-mediated activation may have an important role in maintaining and boosting PI3K signaling in established cancers. Most importantly from a translational medicine perspective, the finding that RTK inhibition abrogates PI3K signaling in established cancers provides a rationale to use RTK inhibitors to suppress PI3K signaling in established KRAS mutant colorectal cancers. We observed that this approach had therapeutic value in vitro and in vivo. Indeed, the observation that IRS-1 and ERBB3 are the major phosphotyrosine proteins bound to PI3K in human colorectal cancers increases confidence that the findings in cell lines will hold true in actual cancers.
Our study focused primarily on how to suppress PI3K and MEK signaling pathways as a concept to treat KRAS mutant cancers, but clearly KRAS has other important effectors that contribute to tumorigenesis. These studies do not evaluate the contribution of other KRAS effectors, such as the Ral pathway, that have been shown to be important for both tumorigenesis and tumor maintenance (50, 51). Indeed, suppression of PI3K and MEK may not be as highly effective for all KRAS mutant cancers. For example, we observed that one of the cell lines, SW620 KRAS mutant cancer, was effectively growth suppressed by combined IGF-IR/MEK inhibition, but did not undergo dramatic apoptosis (Supplemental Figure 13). In fact, direct PI3K and MEK inhibition did not induce cell death in these cells (Supplemental Figure 13), demonstrating that the success of targeting the PI3K/AKT and MEK/ERK pathways will not have uniform efficacy against all KRAS mutant colorectal cancers. Despite some of these potential limitations, this overall strategy is emerging as one of the most promising for this recalcitrant class of cancers. In fact, this approach may even have more therapeutic potential than directly targeting mutant KRAS in the subset of cancers that have lost sensitivity to KRAS loss (52).
There are currently numerous approaches under investigation to identify strategies to treat cancers based on their genetics. In this study, we have devised strategies to effectively and simultaneously downregulate the PI3K/AKT and MEK/ERK pathways to kill cancer cells. Our findings point to clear therapeutic strategies. Our studies predict that a combination of MEK (or RAF) inhibitors and IGF-IR inhibitors will inhibit MEK/ERK and PI3K/AKT signaling in KRAS mutant cancers. Previous work by our laboratory and others has shown that IGF-IR inhibition may have a role in the treatment of several cancer types (40, 49, 53). Indeed in the current study, we observed that this approach may also demonstrate efficacy in KRAS WT cancers, because PI3K signaling was suppressed by IGF-IR inhibition in some of these cancers as well. However, in many instances, both IGF-IR and EGFR contributed to PI3K signaling in these cancers (Supplemental Figure 14). Indeed, the importance of EGFR signaling in KRAS WT cancers for the regulation of both ERK and PI3K signaling may limit that approach in those cancers, and in some cases we observed more marked effects from combined EGFR and IGF-IR inhibition than MEK and IGF-IR inhibition (e.g., Difi cells; Supplemental Figure 15).
The therapeutic approach currently being suggested for KRAS mutant cancers is obviously a simplification that does not account for the heterogeneity among KRAS mutant cancers. For example, as demonstrated by our studies on LoVo cells, this approach would not be effective for the rare KRAS mutant cancers with other genetic abnormalities, such as MET amplification and HER2 amplification. Should clinical trials assessing this combination be developed, it will also be important to determine whether cancers harboring activating mutations in both PIK3CA and KRAS are less sensitive to a combination of IGF-IR and MEK inhibitors, even though cell lines with these mutations were sensitive to this combination in laboratory short-term assays. Despite these limitations, we believe that this approach represents a promising initial genotype-directed strategy for the treatment of advanced KRAS mutant colorectal cancer.
Methods
Cell lines and reagents.
NCI-H630, CaR-1, CCK-81, SW948, HCT-15, and HCT-116 cells were cultured in RPMI 1640 (Cellgro; Mediatech Inc.) with 5% FBS. HT-55, CoCM-1, SW837, SW1463, LoVo, Gp5d, LS-174T, and T84 cells were cultured in F12/DMEM (Invitrogen) with 5% FBS. SW620 cells were cultured in DMEM (Cellgro; Mediatech Inc.) with 5% FBS. Difi cells were grown in F12/DMEM with 10% FBS. Cell lines were obtained from the Center for Molecular Therapeutics at the Massachusetts General Hospital Cancer Center. The mutation status was obtained from the Wellcome Trust Sanger Institute Cancer Genome Project Web site ( http://www.sanger.ac.uk/genetics/CGP/). The following drugs were used: NVP-AEW541 (Selleck Chemicals), AZD6244 (Otava), PHA-665752 (Tocris Biosciences), rapamycin, and gefitinib (American Custom Chemicals Corp. and LC Laboratories). Compounds were dissolved in DMSO to a final concentration of 10 mmol/l and stored at –20°C. Cetuximab (2 mg/ml; ImClone Systems Inc.) was purchased from the pharmacy at Massachusetts General Hospital and stored at 4°C. R1507 was provided by Roche and also stored at 4°C.
Survival assays.
Assessment of cell viability was performed as follows. Cells were seeded in 96-well plates so that the control cells would reach approximately 80% confluency at the end of the assay. On the following the day, the cells were treated with increasing concentrations of the indicated drugs for 72 hours. Treatments with each concentration was performed 6 times (n = 6). Cell viability was determined using the Syto60 assay as previously described (9). The data were graphically displayed using GraphPad Prism version 5.0 (GraphPad Software). IC50 was determined by a nonlinear regression model with a sigmoidal dose response in GraphPad.
Immunoprecipitation and immunoblotting.
Lysates were prepared as previously described (38, 39). Antibodies against p-Akt (Ser473 and Thr308), p-p42/44 MAP kinase (Thr202/Tyr204), p42/44 MAP kinase, p-S6 ribosomal protein (Ser235/236 and Ser240/244), S6 ribosomal protein, p-MET (Tyr1234/1235), MET (25H2), IRS-2, p–IGF-IRβ receptor (Tyr1135/1136), IGF-IRβ receptor, p-Tyr100, p-RSK (Ser380), and RSK were from Cell Signaling Technology. Antibodies against ERBB3, AKT, KRAS, and EGFR were purchased from Santa Cruz Biotechnology Inc. The phospho-specific EGFR (Tyr1068) antibody was from Abcam. The anti–IRS-1 and anti-p85 antibodies used for immunoprecipitations were from Millipore.
Western blot quantification.
Western blot images were captured using GeneSnap image acquisition software and analyzed using GeneTools manual band quantification (Syngene).
Lentiviral shRNA experiments.
shControl and shKRAS have been previously described (52). The tetracycline-inducible shRNA vector used in this study was provided by Novartis (54). The inducible shKRAS encodes the same sequence as shKRAS C. Lentiviral preparation and infections were performed as previously described (52). shRNA transduced cell lines were grown in each medium supplemented with 5% Tet-approved FBS (Clontech) in the presence of 2 μg/ml puromycin. In the tetracycline-inducible system, expression of shRNA was induced by growing cells in the presence of 10 ng/ml doxycycline (Sigma-Aldrich).
siRNA knockdown.
Cells were seeded into 6-well plates at a density of 1.5 × 105 cells/well. Twenty-four hours later, cells were transfected with ON-TARGETplus SMARTpool siRNA against EGFR, IGF-IR, or MET (Dharmacon) or Silencer negative control #1 siRNA (Ambion) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. Transfected cells were cultured at 37°C for 72 hours before analysis.
Apoptosis analysis.
Cells were seeded at approximately 30%–40% confluence in 6-cm plates. After overnight incubation, media was aspirated and replaced with media with or without indicated drugs. After 72 hours, media was collected. Cells were washed with PBS and trypsinized. PBS wash and trypsinized cells were added to the collected media in a single tube. Cells were pelleted, washed once with PBS, and resuspended in Annexin binding buffer (BD) at approximately 1 × 106 cells/ml. Cells were stained with propidium iodide (BD) and Annexin V Cy5 (BioVision) according to the manufacturers’ protocols and assayed on an LSR II flow cytometer (BD). All experiments were performed in triplicate. LoVo cells were stained with propidium iodide and were quantified for subG0/G1 population. For cell cycle analysis, cell were fixed in 70% ethanol, treated with propidium iodide (50 μg/ml) and RNase A (50 μg/ml), and subjected to FACS analysis. The subG0/G1 population was determined by Modfit LT version 3.0 (Verity Software House) (9).
Xenograft mouse studies.
For xenograft experiments, a suspension of 5 × 106 cells was injected subcutaneously into the left flank of 6- to 8-week-old male athymic nude mice. The mice were maintained in laminar air flow units under aseptic conditions, and the care and treatment of experimental animals were in accordance with institutional guidelines. Mice were randomized to 3 treatment groups (n = 5 per group) once the mean tumor volume reached approximately 400 mm3. AZD6244 was dissolved in 0.5% methylcellulose (Sigma-Aldrich) and 0.4% polysorbate (Tween 80; Sigma-Aldrich) and administered at 25 mg/kg b.i.d. by oral gavage. R1507 was diluted with PBS and administered at 18 mg/kg twice per week via i.p. injection. Tumors were measured twice weekly using calipers, and volume was calculated using the following formula: length × width2 × 0.52. Mice were monitored daily for body weight and general condition. According to institutional guidelines, mice were sacrificed when their tumor volume reached 2,000 mm3 or became excessively ulcerated. For pharmacodynamic analyses in SW837 xenografts, tumor-bearing mice (average volume, approximately 200 mm3) were administered R1507 (36 mg/kg) or saline vehicle by i.p. injection every 3 days for 11 days (4 total injections). Mice were sacrificed 24 hours after the final treatment. Tumor tissue was excised and snap-frozen in liquid nitrogen for immunoblotting or fixed in formalin for immunohistochemical studies.
For the experiments with the genetically engineered mouse model of mutant KRAS-induced lung cancers, we utilized Tet-op-mutant Kras mice as described previously (25). Tet-op-mutant KRAS mice were crossed with CCSP-rtTA mice. Double transgenic mice were administered a doxycycline diet (c11300-2000i, Research Diets) until tumors developed. The doxycycline was removed, and lungs were harvested 24 and 48 hours later.
IHC.
Immunohistochemical analyses assessing p-AKT (473), p-ERK, and p-S6 were performed as previously described (25).
Identification of KRAS, BRAF, and PIK3CA mutations in primary colorectal tumor.
Primary human colorectal cancers were obtained from the Brigham and Women’s Hospital’s tissue bank after receipt of approval from the institutional IRB. The presence of mutations in KRAS (exon 2), BRAF (exon 15), and PIK3CA (exons 9 and 20) were determined by direct sequencing. Genomic DNA was harvested using the QIAGEN DNA extraction kit. Nested primer sets used for amplification of exon 2 of KRAS were (F) 5′-GAATGGTCCTGCACCAGTAA-3′ and (R) 5′-GTGTGACATGTTCTAATATAGTCA-3′; (nested F) 5′-GTCCTGCACCAGTAATATGC-3′ and (nested R) 5′-ATGTTCTAATATAGTCACATTTTC-3′. Primers used for amplification of exon 15 of BRAF were (F) 5′-TCATAATGCTTGCTCTGATAGGA-3′ and (R) 5′-CTTTCTAGTAACTCAGCAGC-3′. Primers used for amplification of exon 9 of PIK3CA were (F) 5′-ATCATCTGTGAATCCAGA-3′ and (R) 5′-TTAGCACTTACCTGTGAC-3′. Primers used for amplification of exon 20 of PIK3CA were (F) 5′-TGACATTTGAGCAAAGACC-3′ and (R) 5′-GTGTGGAATCCAGAGTGA-3′.
Statistics.
The Mann-Whitney U test was used to compare KRAS WT and mutant cells with respect to the impact of cetuximab, gefitinib, or KRAS shRNA on signaling (Figure 1, C–E). Two-tailed Student’s t test was performed for Figure 1B, Figure 5C, and Supplemental Figure 10. Differences with P values less than 0.05 were considered statistically different. One-way ANOVA by ranks and Tukey’s multiple comparison tests were used for testing equality of population medians among treated groups (Figure 3D, Figure 4B, and Supplemental Figure 14C). In ANOVA analysis, a P value less than 0.01 was considered statistically significant. All statistical calculations were performed using NCSS software. Synergy was assessed by CalcuSyn software (Biosoft), using the Chou-Talalay method (55).
Study approval.
All animal procedures were approved by the Subcommittee on Research Animal Care at Massachusetts General Hospital.
Supplementary Material
Acknowledgments
This study is supported by grants from the NIH Gastrointestinal Cancer SPORE P50 CA127003 (to J.A. Engelman, J.A. Meyerhardt, and L.C. Cantley), K08 grant CA120060-01 (to J.A. Engelman), R01CA137008-01 (to J.A. Engelman), R01CA140594 (to J.A. Engelman and K.-K. Wong), 1U01CA141457-01 (to J.A. Engelman), R01CA122794 (to K.-K. Wong), R01CA137181 (to K.-K. Wong), 1RC2CA147940 (to K.-K. Wong), R01GM41890 (to L.C. Cantley), National Cancer Institute Lung SPORE P50CA090578 (to J.A. Engelman and K.-K. Wong), the American Association for Cancer Research (to J.A. Engelman), the V Foundation (to J.A. Engelman), American Cancer Society RSG-06-102-01-CCE (to J.A. Engelman), the Ellison Foundation Scholar Award (to J.A. Engelman), the American Lung Cancer Association Lung Cancer Discover Award (to J.A. Engelman), Uniting Against Lung Cancer Foundation (to K.-K. Wong), and the Susan Spooner Research Fund (to K.-K. Wong). H. Ebi was supported by a fellowship from the Kanae Foundation for the Promotion of Medical Science. We also thank Mike Waring and Andrew Cosgrove (Massachusetts General Hospital) for FACS analysis and Anna Kreshock for assistance with IHC. We thank the GI tumor bank at Brigham and Women’s Hospital for assistance in obtaining samples for analyses.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(11):4311–4321. doi:10.1172/JCI57909.
References
- 1.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. . N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janne PA, Gray N, Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8(9):709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 5.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 6.McDermott U, et al. Genomic alterations of anaplastic lymphoma kinase may sensitize tumors to anaplastic lymphoma kinase inhibitors. Cancer Res. 2008;68(9):3389–3395. doi: 10.1158/0008-5472.CAN-07-6186. [DOI] [PubMed] [Google Scholar]
- 7.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 9.Faber AC, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A. 2009;106(46):19503–19508. doi: 10.1073/pnas.0905056106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yakes FM, Chinratanalab W, Ritter CA, King W, Seelig S, Arteaga CL. Herceptin-induced inhibition of phosphatidylinositol-3 kinase and Akt Is required for antibody-mediated effects on p27, cyclin D1, and antitumor action. Cancer Res. 2002;62(14):4132–4141. [PubMed] [Google Scholar]
- 11.Sleijfer S, Wiemer E, Verweij J. Drug Insight: gastrointestinal stromal tumors (GIST) — the solid tumor model for cancer-specific treatment. Nat Clin Pract Oncol. 2008;5(2):102–111. doi: 10.1038/ncponc1037. [DOI] [PubMed] [Google Scholar]
- 12.Lievre A, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 13.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. . N Engl J Med. 2008;359(17):1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 14.Benvenuti S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67(6):2643–2648. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 15.De Roock W, et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 2008;19(3):508–515. doi: 10.1093/annonc/mdm496. [DOI] [PubMed] [Google Scholar]
- 16.Lievre A, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26(3):374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 17.Khambata-Ford S, et al. Expression of epiregulin and amphiregulin and K-ras mutation status predict disease control in metastatic colorectal cancer patients treated with cetuximab. J Clin Oncol. 2007;25(22):3230–3237. doi: 10.1200/JCO.2006.10.5437. [DOI] [PubMed] [Google Scholar]
- 18.Bokemeyer C, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27(5):663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 19.McCormick F. Success and failure on the ras pathway. Cancer Biol Ther. 2007;6(10):1654–1659. doi: 10.4161/cbt.6.10.5153. [DOI] [PubMed] [Google Scholar]
- 20.Young A, Lyons J, Miller AL, Phan VT, Alarcon IR, McCormick F. Ras signaling and therapies. Adv Cancer Res. 2009;102:1–17. doi: 10.1016/S0065-230X(09)02001-6. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez-Viciana P, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370(6490):527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 22.Zhang XF, et al. Normal and oncogenic p21ras proteins bind to the amino-terminal regulatory domain of c-Raf-1. Nature. 1993;364(6435):308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 23.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 24.Der CJ, Van Dyke T. Stopping ras in its tracks. Cell. 2007;129(5):855–857. doi: 10.1016/j.cell.2007.05.031. [DOI] [PubMed] [Google Scholar]
- 25.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sos ML, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci U S A. 2009;106(43):18351–18356. doi: 10.1073/pnas.0907325106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.She QB, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halilovic E, et al. PIK3CA mutation uncouples tumor growth and cyclin D1 regulation from MEK/ERK and mutant KRAS signaling. Cancer Res. 2010;70(17):6804–6814. doi: 10.1158/0008-5472.CAN-10-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carretero J, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17(6):547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pao W, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103(6):931–943. doi: 10.1016/S0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 32.Yang Y, et al. Phosphatidylinositol 3-kinase mediates bronchioalveolar stem cell expansion in mouse models of oncogenic K-ras-induced lung cancer. PLoS One. 2008;3(5):e2220. doi: 10.1371/journal.pone.0002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta S, et al. Binding of ras to phosphoinositide 3-kinase p110alpha is required for ras-driven tumorigenesis in mice. Cell. 2007;129(5):957–968. doi: 10.1016/j.cell.2007.03.051. [DOI] [PubMed] [Google Scholar]
- 34.Fisher GH, et al. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 2001;15(24):3249–3262. doi: 10.1101/gad.947701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66(3):1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Songyang Z, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72(5):767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 37.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3′-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273(46):30199–30203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 38.Engelman JA, et al. ErbB-3 mediates phosphoinositide 3-kinase activity in gefitinib-sensitive non-small cell lung cancer cell lines. Proc Natl Acad Sci U S A. 2005;102(10):3788–3793. doi: 10.1073/pnas.0409773102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316(5827):1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 40.Guix M, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118(7):2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mondino A, Giordano S, Comoglio PM. Defective posttranslational processing activates the tyrosine kinase encoded by the MET proto-oncogene (hepatocyte growth factor receptor). Mol Cell Biol. 1991;11(12):6084–6092. doi: 10.1128/mcb.11.12.6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garcia-Echeverria C, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell. 2004;5(3):231–239. doi: 10.1016/s1535-6108(04)00051-0. [DOI] [PubMed] [Google Scholar]
- 43.Gong Y, et al. High expression levels of total IGF-1R and sensitivity of NSCLC cells in vitro to an anti-IGF-1R antibody (R1507). PLoS One. 2009;4(10):e7273. doi: 10.1371/journal.pone.0007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brachmann SM, et al. Specific apoptosis induction by the dual PI3K/mTor inhibitor NVP-BEZ235 in HER2 amplified and PIK3CA mutant breast cancer cells. Proc Natl Acad Sci U S A. 2009;106(52):22299–22304. doi: 10.1073/pnas.0905152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurent-Puig P, Lievre A, Blons H. Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res. 2009;15(4):1133–1139. doi: 10.1158/1078-0432.CCR-08-0905. [DOI] [PubMed] [Google Scholar]
- 46.Lim KH, Counter CM. Reduction in the requirement of oncogenic Ras signaling to activation of PI3K/AKT pathway during tumor maintenance. Cancer Cell. 2005;8(5):381–392. doi: 10.1016/j.ccr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 47.Campbell PM, Groehler AL, Lee KM, Ouellette MM, Khazak V, Der CJ. K-Ras promotes growth transformation and invasion of immortalized human pancreatic cells by Raf and phosphatidylinositol 3-kinase signaling. Cancer Res. 2007;67(5):2098–2106. doi: 10.1158/0008-5472.CAN-06-3752. [DOI] [PubMed] [Google Scholar]
- 48.Murphy GA, et al. Involvement of phosphatidylinositol 3-kinase, but not RalGDS, in TC21/R-Ras2-mediated transformation. J Biol Chem. 2002;277(12):9966–9975. doi: 10.1074/jbc.M109059200. [DOI] [PubMed] [Google Scholar]
- 49.Hu YP, et al. Heterogeneity of receptor function in colon carcinoma cells determined by cross-talk between type I insulin-like growth factor receptor and epidermal growth factor receptor. Cancer Res. 2008;68(19):8004–8013. doi: 10.1158/0008-5472.CAN-08-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim KH, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7(6):533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 51.Hamad NM, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16(16):2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15(6):489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villanueva J, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wee S, et al. PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci U S A. 2008;105(35):13057–13062. doi: 10.1073/pnas.0802655105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.