Introduction
Mechanical stresses elicit cellular reactions mediated by chemical signals. Defective responses to forces underlie human medical disorders1–4, such as cardiac failure5 and pulmonary injury6. The actin cytoskeleton’s connectivity enables it to transmit forces rapidly over large distances7, implicating it in these physiological and pathological responses. Here we identify the actin-binding protein, filamin A (FLNa) 8, 9 as a central mechanotransduction element of the cytoskeleton. We reconstituted a minimal system consisting of actin filaments, FLNa and two FLNa-binding partners: the cytoplasmic tail of β-integrin, and FilGAP. Integrins form an essential mechanical linkage between extracellular and intracellular environments, with β integrin tails connecting to the actin cytoskeleton by binding directly to filamin 4. FilGAP is a FLNa-binding GTPase-activating protein specific for Rac, which in vivo regulates cell spreading and bleb formation10. Using Fluorescence Loss After photoConversion (FLAC), a novel high-speed alternative to FRAP11, we demonstrate that both externally-imposed bulk shear and myosin II driven forces differentially regulate the binding of these partners to FLNa. Consistent with structural predictions, strain increases β-integrin binding to FLNa, whereas it causes FilGAP to dissociate from FLNa, providing a direct and specific molecular basis for cellular mechanotransduction. These results identify the first molecular mechanotransduction element within the actin cytoskeleton, revealing that mechanical strain of key proteins regulates the binding of signaling molecules.
Main text
The composite cytoskeleton network in vivo provides dynamic cellular structure and actively generates movement. A physiological reconstituted in vitro network of actin and filamin A (FLNa) creates an elastic gel mechanically dominated by the rod-like actin filaments and crosslinked by flexible FLNa molecules. Applying strain to this network readily deforms FLNa crosslinks (Fig 1a,b), and the specific structure and actin binding of FLNa suggest how these deformations might affect FLNa’s interactions with some of its ~90 other binding partners currently identified9.
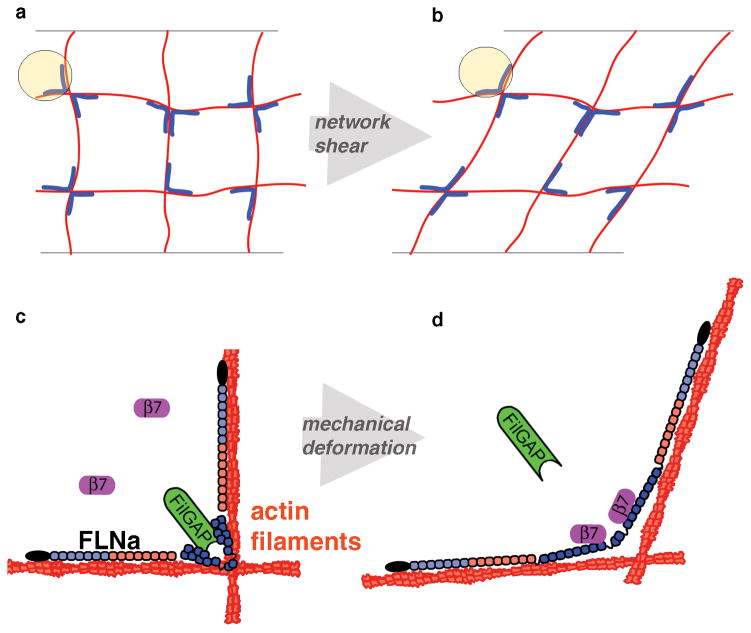
Figure 1. Differential mechanotransduction in FLNa occurs through spatial separation of binding sites and opening cryptic sites.
a) A Filamin (blue) crosslinked actin (red) gel forms an orthogonal network. b) When this network is strained, crosslinks are deformed. c) The actin-binding domain of FLNa is shown in black, followed by repeats 1–7 (light blue) and 8–15 (red), which form the linear rod 1 region. Repeats 16–23 (dark blue) form the compact rod 2 region. FilGAP (green) binds repeats 23 and the cytoplasmic domain of β7 integrin (purple) is unbound. d) When FLNa is mechanically deformed, the cryptic integrin site on repeat 21 is exposed allowing β7 integrin to bind, while repeats 23 are spatially separated, preventing FilGAP from binding both.
FLNa is an extended homodimer composed of two identical subunits, each having an N-terminal actin-binding domain followed by 24 immunoglobulin (Ig) repeats 12(Fig 1c,d). The actin-binding domains and repeats 1–15 are designated “rod 1”, which forms a linear structure that binds actin filaments. Repeats 16–23 comprising “rod 2”, however, form compact globular clusters that do not interact with actin filaments and contain most of FLNa’s binding partner sites. Strain-dependent reversible straightening of these domains contributes to FLNa-actin network flexibility and may regulate local binding partner affinity (Fig S1). Here we examine the effects of mechanical strain on FLNa’s interactions with two key rod 2 binding partners; cytoplasmic β-tail integrin, which nucleates an extensively characterized signalling13 and adhesion14 complex, and FilGAP, a GTPase specific for Rac, a regulator of cellular activity such as actin assembly10. Mechanical strain may regulate partner binding, and we propose that stretching FLNa crosslinks causes FilGAP to unbind whereas integrin binds more strongly (Figs 1c–d, S1). Neighboring Ig repeats cover integrin binding sites on FLNa repeats 19 and 2115, 16, yet computational simulations suggest that rod 2 of FLNa is highly flexible, and that physiological forces are sufficient to expose these cryptic sites allowing integrin to bind17, 18 (Fig S1a,b). FilGAP binding occurs on each repeat 23, suggesting FilGAP is able to bind repeat 23 on both subunits simultaneously when unstressed, providing sufficient avidity to promote FilGAP association with FLNa (Figs 1c, S1c). Mechanical stretching of FLNa spatially separates repeats 23, preventing FilGAP from binding simultaneously to both 19, thus causing it to dissociate (Figs 1d, S1d).
To test these hypotheses and measure the effect of mechanical stress on binding-partner interactions with FLNa, we reconstituted networks of F-actin and FLNa containing the binding partner FilGAP or β7-integrin. To quantify the strain-dependent kinetics of these partners to FLNa, we developed a novel high-speed analogue to FRAP11, Fluorescence Loss After photoConversion (FLAC), which takes advantage of the rapid photo-activation or conversion of photo-activateable fluorescent proteins (PAFPs). In FLAC, a sample with initially non-fluorescent binding partner is locally pulsed with a 50 ms 405 nm light, rapidly and permanently activating PAFP-conjugated partner fluorescence (Figs S4 & S5). Photoactivation fluorescently marks the sample faster and without the high excitation flux required for conventional photo-bleaching. Post activation, unbound PAFP rapidly diffuses away, decreasing the fluorescent signal, while bound PAFP dissociates more slowly. The time-dependent decay of PAFP intensity reveals the kinetics of the FLNa binding partner, as a slower decay curve indicates slower unbinding, providing a direct high-speed assay of dissociation.
We tested the utility of these PAFP constructs in assaying binding kinetics by reconstituting F-actin, PAFP-labeled binding partners, with different forms of FLNa that have higher or lower affinity for β7 integrin or FilGAP. Consistent with immunoprecipitation data (Fig S3b,c), the fluorescence decay of PA-GFP β7 integrin was faster in wild-type FLNa networks, than in the del41 mutant (movie S1), demonstrating relatively stronger binding in the del41 mutant compared to wild-type. The fluorescence decay of PA-GFP FilGAP was slower in wild-type FLNa networks, than in the M2474E mutant (movie S2), also in agreement with immunoprecipitation data (Fig S3a).
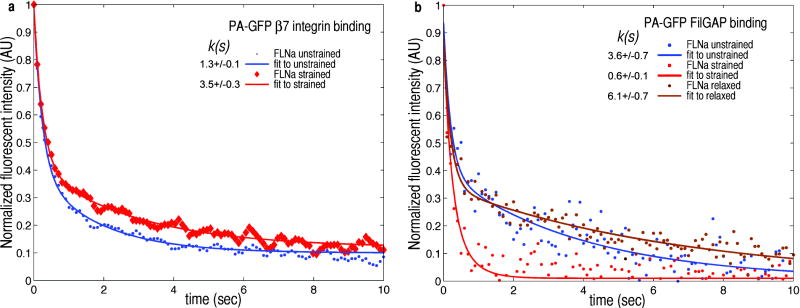
We measured the mechanosensitive aspect of PAFP-binding partner interactions with FLNa. We sheared networks of F-actin and FLNa containing PAFP tagged FilGAP or β7-integrin in a precise and highly controlled fashion using a microscope stage comprised of a stationary coverslip for the bottom of the sample and a piezo-controlled linear actuator at the top. When the FLNa-F-actin network was not strained, β7-integrin had a characteristic exponential decay time of 0.4 +/− 0.1s. The application of a shear strain, γ=0.28, increased this time to 1.0 +/− 0.1s (Fig 2a). The change in fluorescence decay rate describes how the geometric state of FLNa affects dissociation of β7 -integrin; thus, mechanically stretching FLNa molecules enhanced the β7 -integrin binding. In contrast, FilGAP behaved qualitatively oppositely: unstrained networks had a characteristic fluorescence decay time of 2.3 +/− 0.4s, which decreased to 0.3 +/− 0.1 s when a 0.28 shear strain was applied (Fig 2b). FLNa does not permanently cross-link actin, and by unbinding and rebinding on the time-scale of ~6 min (Fig S6), it dynamically allows the network to relax to an unstressed state. After 10 min under strain the network had sufficient time to dissipate internal stress through FLNa remodeling, and the fluorescence decay time increased to 3.4 +/− 0.5 s, demonstrating the reversibility of strain modulated FilGAP binding to FLNa (Fig 2b).
Figure 2. External bulk shear on F-actin-FLNa networks alters FLNa’s binding affinity for β7 integrin and FilGAP.
a) Fluorescence intensity in time of PA-GFP β7 integrin after photoactivation. When unstrained (blue) fluorescence of β7 integrin decays with a characteristic time constant k(s) of 1.3 seconds. Following the application of γ=0.28 shear strain, the time constant increases to 3.5 seconds, as the integrin dissociates more slowly from FLNa (n=18). b), Fluorescence intensity in time of PA-GFP FilGAP after photoactivation. Unstrained (blue) FilGAP’s fluorescence decay time k is 3.6 s. A 4% strain (red) decreased k to 0.6 s from its unstrained decay of 3.6 s. This behavior is reversible, and after allowing the network to relax strain for 10 minutes, k increases to 6.1 s (brown) (n=10).
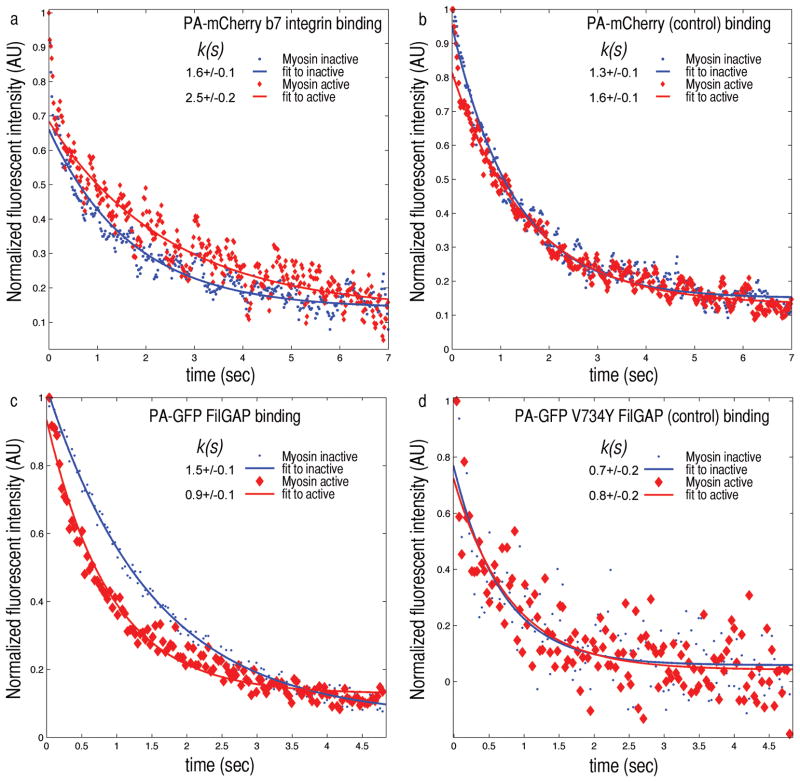
While the application of unidirectional shear revealed the effects of strain on partner binding to FLNa, cells commonly generate internal stresses using molecular motors such as myosin. To examine the effects of cytoskeleton-induced stress, and as a physiological complementary technique to external shear, we included myosin II in the networks to generate contractile stress20 (Fig S9 and movie S3). We allowed the composite network to assemble and come to an unstressed equilibrium state over ~6 hours after the incorporated myosin II had ceased contracting by enzymatically exhausting the pool of added ATP, and dynamic FLNa remodeling had dissipated internal stresses. For unstressed FLNa, we measured β7 -integrin and FilGAP fluorescence decay times of 1.6 +/− 0.1 s and 1.5 +/− 0.1 s, respectively (Fig 3a,c). Including photolabile ‘caged’ ATP in the sample allowed us to release fresh ATP and restart myosin motor activity21, 22, which contracts the actin network and strains FLNa crosslinks. Myosin stressed FLNa increased the integrin unbinding time to 2.5 +/− 0.2 s, while decreasing the FilGAP unbinding time to 0.9 +/− 0.1 s (Fig 3a,c). The application of either external shear or myosin contraction resulted in increased integrin binding and decreased FilGAP binding, demonstrating the robust yet opposite behaviors of these FLNa binding partners.
Figure 3. Myosin II forces applied to F-actin-FLNa networks changes FLNa’s binding affinity to β7 integrin and FilGAP.
a) When depleted of ATP, myosin is in a rigor state. The FLNa within the network is not stressed and PA-GFP β7 integrin fluorescence decays with a characteristic time constant k(s) of 1.6 seconds (blue). After caged ATP is released myosin reactivates, straining FLNa crosslinks. The decay time constant increases to 2.5 seconds as the integrin dissociates more slowly from FLNa under stress. b) PA-mCherry alone as a control shows no significant difference in the unstrained or strained state. c) Fluorescence intensity in time of PA-GFP FilGAP after photoactivation. In the ATP depleted state FilGAP’s fluorescence decay time k is 1.5 s, and after releasing the caged ATP (red) k decreases to 0.9 s. PA-GFP V734Y FilGAP, a non-FLNa binding mutant as a control, shows no significant difference in the decay times of unstrained (0.7 s) or strained state (0.8 s) (n=20).
The FLNa crosslinked actin cytoskeleton is a large percolated network that readily transmits mechanical signals over long intra-cellular distances due to the filamentous actin structure, yet FLNa is mechanosensitive at nanometer molecular deformations. This is in contrast to focal adhesion mechanosensitivity, which detects local mechanics and is limited to small spatial and strain scales due to their size and connectivity23, 24.
In conclusion, we have developed in vitro systems to determine quantitative protein-protein interactions under mechanical force. Using PAFPs with the FLAC technique provides the advances in time-resolution necessary for measuring transient kinetics, without the harsh intensity or duration of bleaching exposure required for FRAP. The results presented here establish FLNa as the first mechanotransductive substrate within the cytoskeleton, and highlight the utility of in vitro systems combined with the power of the FLAC technique to determine quantitative responses of specific proteins.
Mechanotransduction in vitro provides the biological specificity necessary for understanding how these complex regulatory signals may operate in vivo. Cellular mechanotransduction has been shown to induce rapid biochemical activity over long distances 25. Since mechanical stimuli induces relatively large local deformations that decrease in magnitude with distance from application site, FLNa mechanotransduction in vivo likely provides a rapid, distance-sensitive biphasic response by binding or unbinding integrins or FilGAP, respectively, due to the transmitted strain. Physiologically, the localization and binding of these proteins determine their activity. Strain induced binding of integrin to FLNa may compete with talin binding to integrin 26, thus providing a mechanosensitive switch for integrin activation and adhesion. FLNa’s homodimer structure may induce clustering of integrin, thereby reinforcing adhesion and concentrating signaling molecules at a specific location. FilGAP, when unbound from FLNa, relocates to the plasma membrane where it inactivates Rac 10. Active Rac levels profoundly impact cell movement27 and increased Rac activity in FLNa deficient cells correlates with increased apoptosis 28. Moreover, our measurements are consistent with in vivo studies demonstrating that Rac activity and expression appear to be force-regulated by FilGAP-FLNa interactions, since inhibiting FLNa or FilGAP increases Rac levels, yet applying local forces to wild-type cells causes FilGAP to decrease Rac expression 28. Since FLNa does not change FilGAP’s catalytic activity, mechanically-induced redistribution alone might explain its regulation in vivo. Force-dependent conformational changes in structure required for mechanical-regulation have been observed in many proteins, including FLNa in vivo29, 30. By identifying FLNa as the first mechanosensitive element within the cytoskeleton, we have clarified how Rac and integrin activity may be regulated by a specific molecular mechanotransduction pathway. Identifying mechanotransduction elements may direct unique therapeutic approaches by correcting or modulating mechanosensitive binding.
Methods summary
PAFP fluorophore synthesis
PAFP fluorophore cDNA was inserted into binding partners, creating PAFP labeled s7 integrin and FilGAP. Solubility and correct binding of labeled partners was confirmed using western blots (Fig S3).
FLAC methodology
An external 405 nm laser was coupled into a Leica SP5 confocal microscope and used to illuminate a central ~2μm spot for 50 ms, converting the PAFP from its dark to fluorescent state (Figs S4 & S5). The decay in fluorescence intensity, I(t), of the activated fluorophores was monitored and fit with the exponential:
where k is the time constant of characteristic dissociation. Given k values represent best fits +/− 95% confidence intervals.
Sample cell composition
Shear cell samples consisted of 24 μM actin, 0.12 μM FLNa, 1xFB, 2μM Alexa 546 phalloidin, and either PAGFP FilGAP or β7 integrin, and were sheared in a piezo-driven shear cell (see Supplemental information). Sheared FLAC measurements for strained networks were acquired approximately 5–10 s after shear. Myosin samples included 24 μM actin, 0.12 μM FLNa, 1μM myosin II, 1xAB, 2 μM caged ATP, and 2μM Alexa 546 phalloidin, and PAGFP FilGAP, or 2μM Alexa 488 phalloidin and PA-mCherry β7 integrin. Samples were allowed to polymerize and consume available ATP over 6 hours. FLAC measurements were then performed on the ATP-free unstressed network. Subsequently, the caged ATP (Sigma) was released by a 4 s exposure to a diffuse 50 mW 365 nm LED light (Prizmatix Israel), and within 3 s the network could be seen to homogenize under myosin contraction (Fig S9 and movie S3). FLAC measurements were then repeated in this active myosin stressed network to quantify the strain dependent binding activity.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Harvard Materials Research and Engineering Center (DMR-0820484) for confocal imaging, Drs Ginsberg, Lippincott-Schwartz, and Verkhusha for providing cDNA for PA-GFP and PA-mCherry constructs. We thank T. Collins for technical assistance, L. Jawerth, and V. Zaburdaev, for helpful discussions, and J. Wilking and K. Guenthner for help with the manuscript. This work was supported by NIH R01 HL19429 (TPS), NIH T32 HL07680 (AJE), Harvard University Science and Engineering Committee Seed Fund for Interdisciplinary Science (DAW, TPS, FN).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author contributions
The project was conceived by TPS, FN, and DAW. Experiments were designed by AJE TPS, JHH, and FN. Proteins and materials were synthesized and purified by FN and AJE. FLAC experiments were performed by AJE and binding assays by FN. Data was analyzed by AJE. All authors discussed data and aided in preparing the manuscript. AJE and FN contributed equally to this project.
The authors declare no competing financial interests.
References Cited
- 1.Ingber DE. Mechanobiology and diseases of mechanotransduction. Ann Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- 2.Discher DE, Janmey P, Wang YL. Tissue cells feel and respond to the stiffness of their substrate. Science (New York, N Y. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 3.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Developmental cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. S1534-5807(10)00346-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartz MA. Integrins and extracellular matrix in mechanotransduction. Cold Spring Harb Perspect Biol. 2010;2:a005066. doi: 10.1101/cshperspect.a005066. cshperspect.a005066 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruger M, Linke WA. Titin-based mechanical signalling in normal and failing myocardium. J Mol Cell Cardiol. 2009;46:490–498. doi: 10.1016/j.yjmcc.2009.01.004. S0022-2828(09)00021-2 [pii] [DOI] [PubMed] [Google Scholar]
- 6.Birukov KG. Small GTPases in mechanosensitive regulation of endothelial barrier. Microvasc Res. 2009;77:46–52. doi: 10.1016/j.mvr.2008.09.006. S0026-2862(08)00142-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. nrm2594 [pii] [DOI] [PubMed] [Google Scholar]
- 8.Stossel TP, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2:138–145. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura F, Stossel TP, Hartwig JH. The filamins: Organizers of cell structure and function. Cell Adh Migr. 2011;5:14401, pii. doi: 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nature cell biology. 2006;8:803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 11.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. S0006-3495(04)74392-1 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura F, Osborn TM, Hartemink CA, Hartwig JH, Stossel TP. Structural basis of filamin A functions. J Cell Biol. 2007;179:1011–1025. doi: 10.1083/jcb.200707073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 14.Calderwood DA, et al. Increased filamin binding to beta-integrin cytoplasmic domains inhibits cell migration. Nat Cell Biol. 2001;3:1060–1068. doi: 10.1038/ncb1201-1060. [DOI] [PubMed] [Google Scholar]
- 15.Lad Y, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. Embo J. 2007;26:3993–4004. doi: 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heikkinen OK, et al. Atomic structures of two novel immunoglobulin-like domain pairs in the actin cross-linking protein filamin. J Biol Chem. 2009;284:25450–25458. doi: 10.1074/jbc.M109.019661. M109.019661 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pentikainen U, Ylanne J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. Journal of molecular biology. 2009;393:644–657. doi: 10.1016/j.jmb.2009.08.035. S0022-2836(09)01032-8 [pii] [DOI] [PubMed] [Google Scholar]
- 18.Chen HS, Kolahi KS, Mofrad MR. Phosphorylation facilitates the integrin binding of filamin under force. Biophysical journal. 2009;97:3095–3104. doi: 10.1016/j.bpj.2009.08.059. S0006-3495(09)01514-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura F, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS ONE. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koenderink GH, et al. An active biopolymer network controlled by molecular motors. Proc Natl Acad Sci U S A. 2009;106:15192–15197. doi: 10.1073/pnas.0903974106. 0903974106 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphrey D, Duggan C, Saha D, Smith D, Kas J. Active fluidization of polymer networks through molecular motors. Nature. 2002;416:413–416. doi: 10.1038/416413a. [DOI] [PubMed] [Google Scholar]
- 22.Smith DM, et al. Molecular motor-induced instabilities and crosslinkers determine biopolymer organization. Biophys J. 2007 doi: 10.1529/biophysj.106.095919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. nature09621 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grashoff C, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. nature09198 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Na S, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci U S A. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. 0711704105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiema T, et al. The molecular basis of filamin binding to integrins and competition with talin. Mol Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Sanz-Moreno V, et al. Rac activation and inactivation control plasticity of tumor cell movement. Cell. 2008;135:510–523. doi: 10.1016/j.cell.2008.09.043. S0092-8674(08)01236-1 [pii] [DOI] [PubMed] [Google Scholar]
- 28.Shifrin Y, Arora PD, Ohta Y, Calderwood DA, McCulloch CA. The role of FilGAP-filamin A interactions in mechanoprotection. Mol Biol Cell. 2009;20:1269–1279. doi: 10.1091/mbc.E08-08-0872. E08-08-0872 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson CP, Tang HY, Carag C, Speicher DW, Discher DE. Forced unfolding of proteins within cells. Science. 2007;317:663–666. doi: 10.1126/science.1139857. 317/5838/663 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krieger CC, et al. Cysteine shotgun-mass spectrometry (CS-MS) reveals dynamic sequence of protein structure changes within mutant and stressed cells. Proc Natl Acad Sci U S A. 2011;108:8269–8274. doi: 10.1073/pnas.1018887108. 1018887108 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.