Abstract
The development of novel fluorescence methods for the detection of key biomolecules is of great interest, both in basic research and in drug discovery. Particularly relevant and widespread molecules in cells are ADP and GDP, which are the products of a large number of cellular reactions, including reactions catalysed by nucleoside triphosphatases and kinases. Previously, biosensors for ADP were developed in this laboratory, based on fluorophore adducts with the bacterial actin homologue ParM. It is shown in the present study that one of these biosensors, tetramethylrhodamine–ParM, can also monitor GDP. The biosensor can be used to measure micromolar concentrations of GDP on the background of millimolar concentrations of GTP. The fluorescence response of the biosensor is fast, the response time being <0.2 s. Thus the biosensor allows real-time measurements of GTPase and GTP-dependent kinase reactions. Applications of the GDP biosensor are exemplified with two different GTPases, measuring the rates of GTP hydrolysis and nucleotide exchange.
Keywords: fluorescence, GTPase, nucleotide-exchange factor, rhodamine stacking, sensor
Abbreviations: DTT, dithiothreitol; ESI, electrospray ionization; GEF, guanine-nucleotide-exchange factor; hGBP5, human guanylate-binding protein 5; IATR, iodoacetamidotetramethylrhodamine; Sos, Son of Sevenless; Soscat, catalytic domain of Sos; TCEP, tris(2-carboxyethyl)phosphine
INTRODUCTION
GTPases play essential roles in many cellular processes such as signal transduction, cytoskeletal reorganization, vesicle trafficking, regulation of translation, membrane remodelling and pathogen defence [1–3]. Typically the function of GTPases relies on cycling between different protein conformations, determined by the state of the bound nucleotide, either GDP or GTP. Switching between these active and inactive states is tightly controlled by regulatory proteins that affect the rates of exchange of bound nucleotide (GDP to GTP) and GTP hydrolysis to GDP and Pi. Mutations in GTPases or their regulators that interfere with the GTPase cycle cause a number of human diseases, an example being the frequent involvement of Ras-related GTPases in human cancer [4]. Assays to measure the activity of GTPases are therefore of great interest for basic research as well as drug discovery.
Fluorescent reagentless biosensors are widely used tools for the detection and quantitative measurement of biologically important molecules and hence for reporting biological activity. They consist of a biomolecule (protein, DNA and RNA) that specifically binds the analyte and a fluorescent reporter group that couples the recognition of that analyte to a fluorescence change. Protein-based biosensors are a subgroup of reagentless biosensors, where the recognition unit is a fluorescently labelled protein or protein domain. Examples include biosensors for Pi, sugars, amino acids, cations such as Ca2+, nucleotides and single-stranded DNA [5–10]. These types of biosensors offer a number of advantages. First, the signal change can be very fast, limited only by the speed of ligand binding to the protein or the associated conformational change. Hence they can be used for real-time measurements of enzyme activity. Secondly, the protein–ligand interaction can provide high specificity. Finally, a key feature of reagentless biosensors is that only a single component, the labelled protein, is needed for detection, thereby minimizing interference with the system under study.
Fluorescent biosensors for ADP were previously developed by engineering ParM, an actin homologue from bacteria that is involved in plasmid segregation [11,12]. The design of the biosensor relies on a large conformational change of ParM that occurs on nucleotide binding: the two subdomains of the actinfold rotate against each other by 25°, thereby closing the nucleotide-binding site located in a cleft between them [13]. This conformational change was coupled to an optical signal first by introducing cysteine residues at specific locations near the rim of the binding cleft. These cysteine residues provide specific sites to couple with thiol-reactive probes (maleimides or iodoacetamides) [14]. The fluorophores were placed such that their fluorescence responds to the conformation change on ADP binding. Two distinct versions with different properties were produced using either a coumarin, MDCC [11], or a tetramethylrhodamine [12] as the reporter. The coumarin version makes use of the environmental sensitivity of the signal of the single fluorophore.
In contrast, the rhodamine version is achieved by labelling two cysteine residues of a ParM mutant with either 5- or 6-IATR (iodoacetamidotetramethylrhodamine) (Supplementary Figure S1 at http://www.BiochemJ.org/bj/440/bj4400043add.htm). The two labelling sites are located on the two different subdomains, either side of the binding cleft. In the nucleotide-free state the two rhodamines are in close proximity and are likely to form a stacking interaction that largely quenches their fluorescence [15]. Nucleotide binding causes an increase in fluorescence, probably by disturbing the stacking interaction of the rhodamines. In addition to introducing the cysteine residues for label attachment, other mutations were introduced to enhance discrimination against nucleoside triphosphate binding and prevent the ParM polymerizing, so it remains a monomer [11,12]. The final protein used for this rhodamine-labelled biosensor is (His6/K33A/D63C/T174A/T175N/D224C/C287A) ParM [12]. It is demonstrated in the present study that the tetramethylrhodamine-labelled ParM is also suitable for the detection of other nucleoside diphosphates, including GDP, thus providing a novel method to measure the activity of GTPases and other enzymes that produce GDP. The biosensor is characterized for its interaction with guanine and other nucleotides. Example assays demonstrate the use of the biosensor to assay GDP release by enzymes following GTP hydrolysis or due to nucleotide exchange.
EXPERIMENTAL
ParM expression, purification and labelling with IATR
The ParM construct for rhodamine labelling, ParM (His6/K33A/D63C/T174A/T175N/D224C/C287A), was generated from vector pJSC1 containing wild-type ParM [16] by site-directed mutagenesis (Stratagene) [12]. The identity of the construct was verified by DNA sequencing. The protein construct comprised full-length ParM (amino acids 1–320) plus an additional 10 amino acid peptide (QSGSHHHHHH) at the C-terminus. Expression was performed in BL21-Ai cells (Invitrogen) as described previously [12]. ParM expression cultures were grown at 30°C in 2×TY medium containing 100 μg·ml−1 ampicillin until the attenuance at 600 nm was 0.4–0.6 cm−1, when overexpression was then induced by the addition of 2 mg·l−1 arabinose. Cells were harvested after 16 h of induction, resuspended in lysis buffer {30 mM Tris/HCl (pH 8.0), 25 mM KCl, 1 mM TCEP [tris(2-carboxyethyl)phosphine] and 0.1% Triton X-100} and stored at −80°C. His-tagged ParM was then purified by Ni-chelate chromatography. Cleared cell extracts were supplemented with 500 mM NaCl and filtered through a 0.45 μm syringe filter (Sartorius). The lysate was loaded on to a 5 ml HisTrap HP column (GE Healthcare), equilibrated with buffer A [30 mM Tris/HCl (pH 8.0), 500 mM NaCl and 1 mM TCEP]. The column was washed with buffer A until the absorbance at 280 nm reached the baseline, followed by 50 ml of buffer B [30 mM Tris/HCl (pH 8.0), 25 mM KCl and 1 mM TCEP]. ParM was eluted with a 100 ml linear gradient of 0–250 mM imidazole in buffer B, adjusted to pH 8.0. Fractions containing ParM were pooled and 10 mM DTT (dithiothreitol) was added. Protein solutions were concentrated using a Vivaspin-20 centrifugal concentrator (Vivascience) and then further purified over a HiLoad Superdex 75 column (26/60, GE Healthcare) equilibrated in 30 mM Tris/HCl (pH 7.5), 25 mM KCl, 1 mM EDTA and 5 mM DTT. Fractions containing monomeric ParM were pooled and concentrated again as described above. The concentration of ParM mutants was determined by absorbance measurements using the molar absorption coefficient 34380 M−1·cm−1 at 280 nm, calculated from the primary sequence [17]. Proteins were shock-frozen in liquid nitrogen and stored at −80°C.
The ParM construct contains two exposed cysteine residues (Cys63 and Cys224) that are accessible to modification with thiol-reactive iodoacetamide probes. A third cysteine residue in the construct (Cys100) is buried in the core of the structure and is not readily accessible to labelling (see below). Labelling of ParM with 5- or 6-IATR [18] was performed as described previously [12]. In brief, DTT was removed from ParM solutions using a PD10 column (GE Healthcare) and ParM was labelled with IATR at a 4-fold molar excess for 90 min at room temperature (21°C) in 30 mM Tris/HCl (pH 7.5) and 25 mM KCl. The reaction was stopped by the addition of 2-mercaptoethanesulfonate, which captures the unreacted IATR. Free fluorophore was removed on a PD10 column. Labelled ParM was further purified by anion-exchange chromatography on a HiTrapQ HP column (GE Healthcare) using a gradient of 25–200 mM KCl. The protein was concentrated in an Amicon ultracentrifugal filter device (Millipore), shock-frozen in liquid nitrogen and stored at −80°C. Concentrations of tetramethylrhodamine-labelled ParM mutants were determined from the absorbance spectra using the molar absorption coefficient of a small molecule thiol adduct of 5-IATR at its isosbestic point, ϵ528=52000 M−1·cm−1 [19]. The labelling stoichiometry of two labels per protein molecule was confirmed by ESI (electrospray ionization)-MS.
Nucleotides
All nucleotides were purchased from Sigma–Aldrich at the highest purity available. GTP was purified further by anion-exchange chromatography and its purity was analysed by HPLC as described previously [11]. Nucleotide concentrations were determined from their absorbance spectra in 20 mM Tris/HCl (pH 7.5) using the molar absorption coefficients ϵ253=13700 cm−1 for guanine, ϵ259=15400 cm−1 for adenine, ϵ260=9900 cm−1 for uracil, ϵ260=6100 cm−1 for cytosine and ϵ260=8700 cm−1 for thymidine nucleotides.
Fluorescence titrations
Equilibrium fluorescence measurements were performed using a Cary Eclipse fluorescence spectrophotometer (Varian). 5- or 6-ATR–ParM (0.5 μM) was titrated with different nucleotides and the rhodamine fluorescence was recorded at 577 nm after excitation at 553 nm. Data were analysed by fitting hyperbolic curves using the program GraFit 5.0. Titrations were performed in 30 mM Tris/HCl (pH 7.5), 25 mM KCl, 3 mM MgCl2 and 5 μM BSA at 20°C.
Stopped-flow fluorescence experiments
Stopped-flow measurements were performed using a HiTech SF61 DX2 stopped-flow instrument equipped with a Xe/Hg lamp (TgK Scientific). Association kinetics of 5- and 6-ATR–ParM and GDP were measured under pseudo-first-order conditions with excess of GDP over ParM. Tetramethylrhodamine fluorescence was excited at 546 nm and emission was detected with a photomultiplier after filtering emitted light through a 570 nm long-pass filter. Experimental traces were analysed by single exponential curve fitting using the HiTech software. All measurements were performed at 20°C in 30 mM Tris/HCl (pH 7.5), 25 mM KCl, 3 mM MgCl2 and 5 μM BSA.
GTP hydrolysis assay
Steady-state kinetics of GTP hydrolysis by hGBP5 (human guanylate-binding protein 5) a/b [20] were measured in 50 mM Tris/HCl (pH 8.0), 25 mM KCl, 5 mM MgCl2, 2 mM DTT and 5 μM BSA at 25°C using a Cary Eclipse fluorescence spectrophotometer (Varian). Different concentrations of GTP (0–1 mM) and 5-ATR-ParM (0.25 μM final concentration) were incubated in the cuvette at 25°C and the reaction was started by the addition of hGBP5 (0.22 μM final concentration). Rhodamine fluorescence was excited at 553 nm and detected at 577 nm. The fluorescence response of 5-ATR-ParM was calibrated by titrating ParM with a GDP standard solution in the presence of 0, 0.5 and 1 mM GTP. Data were analysed by linear regression. The slope of the calibration was dependent on the GTP concentration present, albeit the difference was small: approximately 10% difference between 0 and 1 mM GTP. Assuming a linear relationship between the slope of the calibration curve and [GTP], for each [GTP] used in the assay a slope was calculated by linear interpolation. These were used to convert the measured fluorescence time traces into the time course of GDP formation. Initial rates were determined by linear fitting to the data points with less than 10% of the GTP turned over. Michaelis–Menten plots (initial rate against [GTP]) were generated and analysed by hyperbolic curve fitting using GraFit 5.0 [21].
Nucleotide-exchange assays
Nucleotide exchange of H-Ras was measured in 20 mM Hepes (pH 7.4), 150 mM KCl, 5 mM MgCl2, 2 mM DTT and 5 μM BSA at 25°C using a Cary Eclipse fluorescence spectrophotometer (Varian). GDP-bound Ras (Ras–GDP) and 5-ATR-ParM (concentrations given in the legend of Figure 4) were incubated at 25°C in the cuvette and the exchange reaction was started by adding 1 mM GTP and/or the catalytic domain of Sos (Son of Sevenless; residues 564–1049, Soscat) [22]. Fluorescence was recorded at excitation and emission wavelengths of 553 nm and 577 nm respectively. The biosensor response was calibrated as described for the GTPase assay above, but using the calibration in the presence of 1 mM GTP for all data to calculate GDP concentrations from the fluorescence signal. Data points up to maximally 10% turnover were analysed to obtain initial rates by linear regression. Initial rates were plotted as a function of Ras–GDP concentration and hyperbolic curve fitting was performed with GraFit 5.0 [21].
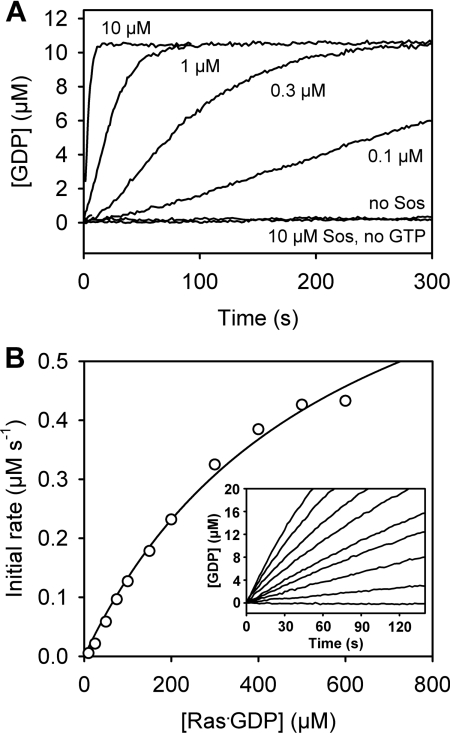
Figure 4. Sos-catalysed nucleotide exchange of Ras.
(A) GDP-to-GTP exchange at different concentrations of Soscat. Ras–GDP (10 μM) and 5-ATR-ParM (0.25 μM) were incubated in the cuvette before GDP dissociation was initiated by the addition of 1 mM GTP and/or Soscat at the concentration indicated in the Figure. The concentration of free GDP was calculated from a calibration as in Figure 3(A). (B) GDP-to-GTP exchange at different concentrations of Ras–GDP. The experiment was performed as described in (A), but with constant 25 nM Soscat and various concentrations of Ras–GDP. The inset shows time courses of GDP dissociation at 0, 25, 50, 75, 100, 150, 200, 300 and 500 μM Ras·GDP. Initial rates were determined by linear regression and plotted against Ras–GDP concentration. The solid line represents the hyperbolic fit to the experimental data. Measurements were performed in 20 mM Hepes (pH 7.4), 150 mM KCl, 5 mM MgCl2, 2 mM DTT and 5 μM BSA at 25°C.
RESULTS AND DISCUSSION
Native ParM, the protein scaffold of the ADP biosensors described previously, binds guanine nucleotides in addition to adenine nucleotides [23]. The coumarin-labelled biosensor MDCC–ParM binds GDP, but does not exhibit a fluorescence change [11], so this version is not suitable for GDP detection. In contrast, both isomers of the tetramethylrhodamine-based biosensor 5- and 6-ATR-ParM (His6/K33A/D63C/T174A/T175N/D224C/C287A) respond to GDP binding with a ~10-fold increase in fluorescence (Figure 1 and Table 1). Before describing the application of the double rhodamine ParM-based biosensor to GDP measurement, the precise identity of this molecule is considered.
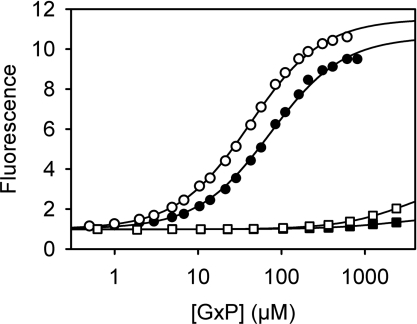
Figure 1. Affinity of the biosensor for guanine nucleotides.
5-ATR–ParM (0.5 μM) was titrated with GDP (circles) or GTP (squares) under two different salt conditions, 25 mM KCl (closed symbols) or 150 mM KCl (open symbols), in 30 mM Tris/HCl (pH 7.5), 3 mM MgCl2 and 5 μM BSA at 20°C. Rhodamine fluorescence was excited at 553 nm and emission was recorded at 577 nm. Dissociation constants for GDP, obtained from hyperbolic curve fits, are summarized in Table 1.
Table 1. Affinity and fluorescence change for the binding of different nucleoside diphosphates to tetramethylrhodamine-labelled ParM.
Equilibrium dissociation constants (Kd) and fluorescence changes (F+/F−) upon nucleotide binding were determined by fluorescence titration as shown in Figure 1. Data were obtained at 20°C in 30 mM Tris/HCl (pH 7.5), 3 mM MgCl2, 5 μM BSA and 25 mM KCl or 150 mM KCl where indicated.
| 5-ATR-ParM | 6-ATR-ParM | |||
|---|---|---|---|---|
| Nucleotide | F+/F− | Kd (μM) | F+/F− | Kd (μM) |
| GDP | 10.6 | 75.0 | 9.5 | 91.2 |
| GDP (150 mM KCl) | 11.7 | 45.8 | 10.1 | 48.5 |
| ADP* | 14.9 | 30.2 | 14.5 | 37.0 |
| ADP (150 mM KCl) * | 15.2 | 15.3 | 12.3 | 15.8 |
| dGDP | 9.0 | 105 | 8.7 | 149 |
| dADP | 13.1 | 27.5 | 12.2 | 41.7 |
| CDP | 9.5 | 826 | 4.8 | 760 |
| dCDP | 10.6 | 707 | 7.4 | >1000 |
| UDP | 11.6 | 101 | 7.2 | 96.4 |
| TDP | 14.1 | 29.0 | 15.1 | 45.0 |
*Values are from [12].
The substitution reactions of iodoacetates and iodoacetamides are very selective for thiols (such as in cysteine residues) [14,24–26], although they can also react with methionine, tyrosine, histidine and lysine residues. This is, in part, due to the much larger nucleophilicity of thiol over other available groups on a protein that could take part in the nucleophilic substitution reaction [27] (Supplementary Figure S1). Under conditions of neutral pH, ambient temperature and limited excess of iodoacetamide over thiols, the reaction with cysteine is orders of magnitude faster than reaction with any other amino acid [14,24–29]. The ParM construct described here (His6/K33A/D63C/T174A/T175N/D224C/C287A) contains two exposed cysteine residues (Cys63 and Cys224) that, according to the known crystal structures, are accessible to modification; indeed, they were added for that purpose. A third wild-type cysteine in the construct (Cys100) is buried in the core of the structure and so is unlikely to be modified. MS showed that each molecule of this construct is labelled with two acetamidotetramethylrhodamine moieties [12]. In order to confirm that the two labels are on the two exposed cysteine residues, two mutants were constructed with only one exposed cysteine present, ParM (His6/K33A/D63A/T174A/T175N/D224C/C287A) and ParM (His6/K33A/D63C/T174A/T175N/D224A/C287A), as well as one mutant where both exposed cysteine residues were removed, ParM (His6/K33A/D63A/T174A/T175N/D224A/C287A). These proteins were reacted with 5-IATR under the same conditions as the biosensor construct and analysed by ESI-MS (Supplementary Figure S2 at http://www.BiochemJ.org/bj/440/bj4400043add.htm) and absorbance spectroscopy (Supplementary Figure S3 at http://www.BiochemJ.org/bj/440/bj4400043add.htm). Both mutants with only one exposed cysteine residue (63A/224C and 63C/224A) reacted with only one 5-IATR molecule per protein (Supplementary Table S1 at http://www.BiochemJ.org/bj/440/bj4400043add.htm). The mutant that had both cysteine residues removed (63A/224A) showed only a very small fraction of labelling (~4% of the protein, Supplementary Figure S3) and the majority (96%) was unlabelled. Thus the two labels on the biosensor are on Cys63 and Cys224, introduced for this purpose. It is possible that a small fraction (<5%) of the protein has some unspecific labelling on other amino acids, but importantly this is unlikely to impair the biosensor function.
As previously shown for ADP, 5- and 6-ATR–ParM behave very similarly to each other with other nucleoside diphosphates in terms of affinity, kinetics and signal change. Below, characterization of the 5-isomer is described, and then the similarities and differences of 6-ATR–ParM are summarized.
A titration of GDP with 5-ATR–ParM, measuring fluorescence, showed that this nucleotide binds only slightly weaker than ADP (2.5-fold) (Table 1). The dissociation constant was determined as 75 μM in a buffer containing 25 mM KCl. The GDP affinity is approximately 2-fold higher at 150 mM KCl. The biosensor strongly discriminates between GDP and GTP (Figure 1). GTP binds much weaker than GDP and shows only a small fluorescence increase. The sensitivity of ATR–ParM is more than 100-fold higher for GDP than for GTP; whereas 10 μM GDP causes a >2-fold change in fluorescence, >1 mM GTP is needed to produce the same change (Figure 1). These data suggest that 5-ATR–ParM is well suited for sensing GDP in the presence of GTP. At nucleotide concentrations higher than those shown in Figure 2, the fluorescence decreases after reaching a maximum. However, this is outside the concentration range of possible GDP measurements due to saturation of the biosensor and therefore does not affect the biosensor performance.
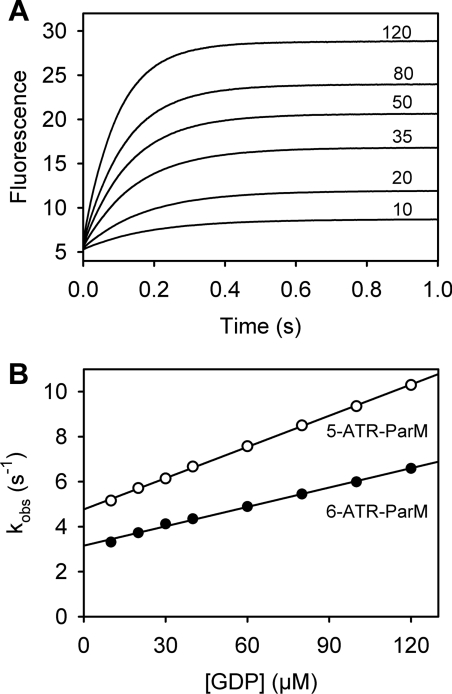
Figure 2. Kinetics of GDP binding to the biosensor.
(A) Fluorescence time courses after mixing 0.1 μM 5-ATR–ParM with increasing concentrations of GDP in a stopped-flow apparatus. Micromolar GDP concentrations are indicated in the Figure. All concentrations are those in the mixing chamber. Fluorescence was excited at 546 nm and emission was detected after a 570 nm long-pass filter. Data were analysed by single exponential curve fitting yielding the observed rate constant kobs. (B) Plot of kobs against GDP concentration for 5-ATR– (open circles) and 6-ATR– (closed circles) ParM. Association (kon) and dissociation (koff) rate constants were obtained from linear regression analysis. For 5-ATR–ParM, kon is 0.047 μM−1·s−1 and koff is 4.8 s−1; for 6-ATR–ParM, kon is 0.029 μM−1·s−1 and koff is 3.2 s−1. Experiments were performed in 30 mM Tris/HCl (pH 7.5), 25 mM KCl, 3 mM MgCl2 and 5 μM BSA at 20°C.
ATR–ParM also responds to other nucleoside diphosphates (dADP, dGDP, CDP, dCDP, TDP and UDP) with a fluorescence increase upon binding that ranges between 9-fold and 14-fold (Table 1). Except for CDP and dCDP, which bind considerably weaker (Kd>700 μM), the dissociation constants of the other nucleotides are in the same range as GDP and ADP (28–105 μM).
In order to assess whether the biosensor is suitable for real-time monitoring of GDP formation, the kinetics of GDP binding were determined using stopped-flow (Figure 2). Binding kinetics were measured under pseudo-first-order conditions with excess GDP over 5-ATR–ParM. Time courses show that GDP binding is fast, completed in <1 s even at the lowest concentration of GDP (Figure 2A). The fluorescence traces are well described by single exponentials. The observed rate constants show a linear dependence on the concentration of GDP, indicating that the speed of the fluorescence response in the concentration range measured is controlled by the bimolecular binding step rather than a conformational change (Figure 2B). Association (kon) and dissociation (koff) rate constants obtained from linear regression were 0.047 μM−1·s−1 and 4.8 s−1 respectively. The slightly lower affinity of the biosensor for GDP in comparison with ADP [12] was mainly due to a lower kon value, whereas the koff value was very similar. The dissociation constant obtained from the kinetics (Kd=koff/kon) was 103 μM, which is in reasonable agreement with the value from equilibrium titrations. Given that the fluorescence response of the biosensor to GDP occurs with rate constant koff+kon[GDP], this response is always greater than koff (4.8 s−1). This corresponded to a response time of <200 ms (1/koff). Thus the biosensor is well suited for real-time measurements of GDP in steady-state or slow transient kinetic assays using manual mixing. In order to observe ‘pure’ GDP release rate constants using fast kinetic techniques (e.g. stopped-flow), this rate constant needs to be ~10-fold slower than the biosensor response, corresponding to release rate constants of <0.5 s−1.
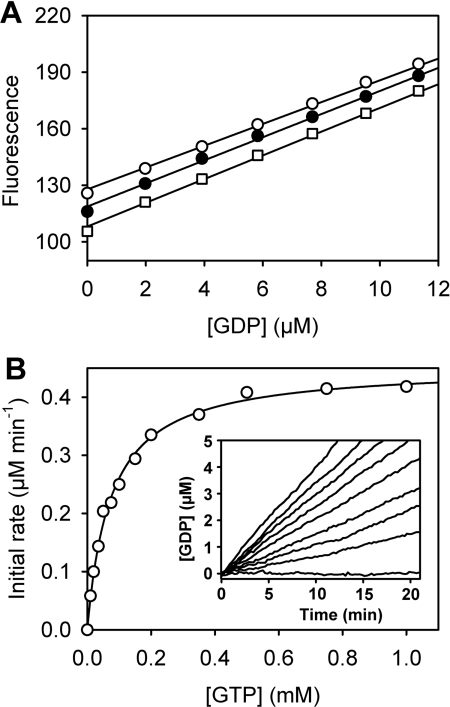
The performance of the GDP biosensor was tested in real-time assays by measuring the steady-state GTPase activity of hGBP5. hGBP5 is a mammalian dynamin-related GTPase that is induced by interferons, shows tumour-associated expression and has a role in innate immunity [3,30]. GTP hydrolysis by hGBP5 was measured at different GTP concentrations by monitoring GDP formation using 5-ATR–ParM (Figure 3). To calibrate the fluorescence response, 5-ATRParM was titrated with GDP in the presence of different concentrations of GTP under the assay conditions (Figure 3A). The fluorescence response was linearly dependent on GDP up to ~20 μM. The presence of GTP had only a small effect on the slope of the calibration: <10% difference was observed between 0 and 1 mM GTP. The calibration accounted for this as described in the Experimental section. Fluorescence time courses were converted into GDP concentrations and initial rates were determined by linear regression (Figure 3B, inset). Michaelis–Menten plots of initial rates against GTP concentration gave the parameters KM=79 μM and Vmax=0.467 μM·min−1 (Figure 3B). The catalytic activity (kcat) of 2.1 min−1, determined here, was in good agreement with a previous study, where the activity of hGBP5 was determined as 1.7 min−1 in an HPLC-based assay under very similar solution conditions [20].
Figure 3. Assay of the GTPase activity of hGBP5.
(A) Calibration of the fluorescence signal of the biosensor. 5-ATR-ParM (0.25 μM) was titrated with GDP in the absence of GTP (□), with 0.5 mM GTP (●) or with 1 mM GTP (○), and fluorescence emission was monitored as described in Figure 1. The slopes from linear regression analysis are 6.3 μM−1, 6.1 μM−1 and 5.8 μM−1 at 0, 0.5 and 1 mM GTP respectively. (B) Kinetics of GTP hydrolysis catalysed by hGBP5 were measured under steady-state conditions with 0.22 μM hGBP1, different concentrations of GTP and 0.25 μM 5-ATR-ParM. The inset shows time courses of GDP formation at 0, 10, 20, 35, 50, 100, 150, 200 and 1000 μM GTP. Initial rates were determined by linear fits to data points below 10% turnover and are plotted against GTP concentration. A hyperbolic curve fit yields the Michaelis–Menten parameters KM=79 μM and Vmax=0.467 μM·min−1. Measurements were performed in 50 mM Tris/HCl (pH 8.0), 25 mM KCl, 5 mM MgCl2, 2 mM DTT and 5 μM BSA at 25°C.
A second application of the GDP biosensor is nucleotide exchange of the small GTPase Ras, a key signalling molecule in the regulation of cell growth, proliferation, differentiation and apoptosis [31]. Ras is activated by GEFs (guanine-nucleotide-exchange factors) that convert GDP-bound Ras into GTP-bound Ras by accelerating nucleotide dissociation. Sos is one of these Ras-specific exchange factors. Nucleotide exchange from Ras–GDP to Ras–GTP was measured using 5-ATR–ParM to monitor released GDP (Figure 4). A large excess of free GTP over Ras–GDP was used to get complete exchange of GTP for GDP, ensuring quasi-irreversible dissociation of GDP. GDP release from 10 μM Ras–GDP was very slow in the absence of nucleotide-exchange factor and strongly accelerated by addition of the catalytic domain of Sos (Soscat) (Figure 4A). If there was no GTP present to replace the dissociating GDP, a net release of GDP was not detectable. (Figure 4A). This finding indicates that Sos alone cannot displace GDP from Ras. In other words, the nucleotide-free complex of Ras and exchange factor (Sos–Ras) is not formed in significant amounts (<5%) under these conditions. The apparent half-times of the exchange reaction were 240 s, 80 s, 25 s and 2.5 s at 0.1, 0.3, 1 and 10 μM Soscat respectively. Under single-turnover conditions (10 μM Soscat, 10 μM Ras–GDP) the apparent rate constant obtained from single exponential curve fitting was 0.28 s−1. This is >10-fold faster than the single-turnover rate constant reported previously at the same concentration of Soscat but lower Ras–GDP (1 μM) [32,33]. The large difference could be explained by the allosteric feedback mechanism governing Sos-catalysed nucleotide exchange [34]. Beside the catalytic Ras binding site, Soscat possesses a second interaction site, where Ras–GTP and, to a lesser extent, Ras–GDP binding causes an allosteric activation of Sos catalytic activity [34]. A higher concentration of Ras might lead to a higher occupation of this allosteric site and thus faster nucleotide exchange. In addition, the use of fluorescently labelled mantGDP in the studies of Ford et al. [32,33] might also contribute to the observed difference in rate constants, because nucleotide dissociation from Ras is slower for mant-nucleotides than for unlabelled nucleotides [35].
Nucleotide exchange of the same system was measured under steady-state conditions using a constant low concentration of Soscat and increasing concentrations of Ras–GDP (Figure 4B). The initial rate of GDP dissociation increased with Ras–GDP concentration with approximately hyperbolic dependence. Although the data do not allow an accurate determination of the Michaelis–Menten parameters since saturation is not reached, lower limits can be estimated for KM>400 μM and kcat>20 s−1. Such a high apparent KM value has also been observed for the related Ras GEF, Cdc25Mm (KM=386 μM) [36]. The intrinsic exchange rate for Ras–GDP, determined with the GDP biosensor, was 3.6×10−5 s−1 (results not shown) in agreement with previous studies [33,36]. Thus nucleotide exchange is accelerated by Soscat by more than five orders of magnitude, as found for many GEFs of small GTPases [37].
So far, the 5-isomer of ATR–ParM has been described for its use as a biosensor for nucleoside diphosphates, in particular GDP. Generally the ParM version, labelled with the 6-IATR, behaves very similarly to 5-ATR-ParM with respect to NDP and dNDP binding (Table 1, and Figures 2 and 3). 6-ATR–ParM shows a similar fluorescence response as 5-ATR–ParM to the different nucleoside diphosphates, with slightly lower affinity for NDPs, including GDP, and similar binding kinetics. Hence, 6-ATR–ParM can be used in the same way as 5-ATR–ParM to measure nucleoside diphosphates.
In summary, 5-ATR–ParM and 6-ATR–ParM function as biosensors for GDP that can be used to measure rates of GTP hydrolysis and nucleotide exchange of GTPases. This is a wide ranging group of enzymes, including small G-proteins, heterotrimeric G-proteins, GTPases involved in transcription and translation, and large dynamin-related GTPases. GTP-dependent kinases are another group of enzymes that could be studied with these biosensors, albeit they are fewer in number than GTPases. These kinases, for example phosphoenolpyruvate carboxykinase, transfer the γ-phosphate of GTP on to a substrate, thereby releasing GDP. In addition there are other metabolic enzymes that produce GDP, such as a variety of enzymes that handle GDP sugars, for example transferases involved in carbohydrate biosynthetic pathways, as well as specific examples, GDP-L-galactose phosphorylase and GDP-mannose glycosyl hydrolase.
ATR–ParM shows a fast fluorescence response (<200 ms response time) allowing real-time measurements of GDP concentration changes on the time scale of seconds. The biosensor has micromolar sensitivity for GDP and discriminates well against GTP. The dynamic range for GDP detection, estimated from the concentrations at 10% and 90% saturation, was 5–500 μM in Tris buffer (pH 7.5) and 150 mM KCl. These upper and lower limits depend on the exact buffer conditions and will be shifted, e.g by ionic strength, pH value and temperature, according to the GDP dissociation constant. The fluorescence response of the biosensor is approximately linear at concentrations well below the Kd value, whereas at higher concentrations a hyperbolic equation can be used for calibration. The contribution of GTP binding to the signal is negligible (<5%) at concentrations up to ~800 μM (Figure 1), but can be corrected by appropriate calibrations if high accuracy or higher concentrations of GTP are needed. In practice, the biosensor is able to detect down to 2 μM concentrations of GDP in the presence of mM GTP, as demonstrated in the GTPase and nucleotide exchange assays (Figure 3 and 4). The broad specificity for nucleoside diphosphates and deoxynucleoside diphosphates enables the biosensor to be used for measuring reactions that release other diphosphates, in addition to ADP and GDP. On the other hand, due to the similar affinities of the biosensor for the NDPs, the presence of any other diphosphate in addition to the one to be studied would interfere with the detection, unless its concentration is much lower than its Kd for the biosensor. Further engineering of the active site by site-directed mutagenesis could potentially create new variant biosensors with higher specificity for particular diphosphates. To our knowledge there is only one earlier described reagentless biosensor for GDP, consisting of a coumarin-labelled nucleoside diphosphate kinase [38]. That biosensor is also not specific for the base and can be used to detect all NDPs and dNDPs. However, it does not report the absolute concentration of NDP, but the ratio of diphosphate to triphosphate. The reason is that the fluorescence reports not on nucleotide binding, but on the phosphorylation state of nucleoside diphosphate kinase, which is controlled by the NTP/NDP ratio. An antibody-based assay has been described, aimed at high-throughput assays and using fluorescence polarization [39]. Other commonly used methods for measurement in vitro of nucleotide binding and GTP hydrolysis employ either radioactive nucleotides, HPLC analysis of labelled or unlabelled nucleotides, or measurement of released Pi from the GTPase. For Pi measurement, there are colorimetric assays, e.g. Malachite Green [40,41], coupled enzyme assays, for example using purine nucleoside phosphorylase [42,43] and phosphate biosensors [5,44,45]. However, only the coupled enzyme and biosensor assays can be used in a real-time format, but they are not applicable to measuring guanine-nucleotide exchange. The other methods require stopping the reaction at different time points to analyse individual samples, a process that can be laborious and time consuming, especially if long separation protocols are required. Although assays based on absorbance detection can have comparable sensitivity with the GDP biosensor (low micromolar concentrations), radioactive assays are generally much more sensitive, but they require special equipment and safety precautions. A large number of fluorescence-based assays rely on fluorescently labelled nucleotides, for example [36,46–48], and there are also studies where labelled GTPases or regulatory proteins are used, for example [49,50]. These methods are sensitive (possibly down to tens of nanomolar) and are particularly useful for transient kinetic studies. However, one drawback is that they require modification of the natural nucleotide or protein, and thus potentially disturb the system under study. In addition, these assays are not truly generic and, in particular with labelled GTPases, an assay has to be developed for each GTPase.
In conclusion, the biosensor for GDP described in the present study provides a novel generic method to measure hydrolysis activity and guanine-nucleotide exchange of GTPases, or the activity of GTP-dependent kinases. This method can be used for functional and mechanistic studies on such systems, but may also find application in high-throughput screening of inhibitors for this important protein class.
Online data
AUTHOR CONTRIBUTION
Martin Webb and Simone Kunzelmann conceived the study and wrote the paper. Simone Kunzelmann performed the experiments.
ACKNOWLEDGEMENTS
We thank Mark Wehner, Dr Daniel Filchtinski and Dr Christian Herrmann (Ruhr-University Bochum, Germany) for the gifts of purified hGBP5 and H-Ras, and Dr Nicolas Nassar (Stony Brook University, New York, U.S.A.) for the purified protein Soscat. We also thank Gordon Reid (NIMR, London, U.K.) for technical assistance.
FUNDING
This work was supported by Medical Research Council Technology and by the Medical Research Council, UK [grant number U117512742].
References
- 1.Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125–132. doi: 10.1038/348125a0. [DOI] [PubMed] [Google Scholar]
- 2.Praefcke G. J., McMahon H. T. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat. Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 3.Shenoy A. R., Kim B. H., Choi H. P., Matsuzawa T., Tiwari S., MacMicking J. D. Emerging themes in IFN-γ-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbacid M. Ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 5.Brune M., Hunter J. L., Corrie J. E., Webb M. R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- 6.de Lorimier R. M., Smith J. J., Dwyer M. A., Looger L. L., Sali K. M., Paavola C. D., Rizk S. S., Sadigov S., Conrad D. W., Loew L., Hellinga H. W. Construction of a fluorescent biosensor family. Protein Sci. 2002;11:2655–2675. doi: 10.1110/ps.021860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou J., Hofer A. M., Lurtz M. M., Gadda G., Ellis A. L., Chen N., Huang Y., Holder A., Ye Y., Louis C. F., et al. Developing sensors for real-time measurement of high Ca2+ concentrations. Biochemistry. 2007;46:12275–12288. doi: 10.1021/bi7007307. [DOI] [PubMed] [Google Scholar]
- 8.Berg J., Hung Y. P., Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat. Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamura H., Nhat K. P., Togawa H., Saito K., Iino R., Kato-Yamada Y., Nagai T., Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc. Natl. Acad. Sci. U.S.A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillingham M. S., Tibbles K. L., Hunter J. L., Bell J. C., Kowalczykowski S. C., Webb M. R. Fluorescent single-stranded DNA binding protein as a probe for sensitive, real-time assays of helicase activity. Biophys. J. 2008;95:3330–3339. doi: 10.1529/biophysj.108.133512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunzelmann S., Webb M. R. A biosensor for fluorescent determination of ADP with high time resolution. J. Biol. Chem. 2009;284:33130–33138. doi: 10.1074/jbc.M109.047118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunzelmann S., Webb M. R. A fluorescent, reagentless biosensor for ADP based on tetramethylrhodamine-labeled ParM. ACS Chem. Biol. 2010;5:415–425. doi: 10.1021/cb9003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den Ent F., Moller-Jensen J., Amos L. A., Gerdes K., Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson I., Spence M. T. Z. The Molecular Probes Handbook. Carlsbad: Life Technologies Corporation; 2010. [Google Scholar]
- 15.Selwyn J. E., Steinfeld J. I. Aggregation of equilibriums of xanthene dyes. J. Phys. Chem. 1972;76:762–774. [Google Scholar]
- 16.Salje J., Lowe J. Bacterial actin: architecture of the ParMRC plasmid DNA partitioning complex. EMBO J. 2008;27:2230–2238. doi: 10.1038/emboj.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pace C. N., Vajdos F., Fee L., Grimsley G., Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munasinghe V. R. N., Corrie J. E. T. Optimised synthesis of 6-iodoacetamidotetramethylrhodamine. ARKIVOC. 2006;ii:143–149. [Google Scholar]
- 19.Corrie J. E. T., Craik J. S. Synthesis and characterisation of pure isomers of iodoacetamidotetramethylrhodamine. J. Chem. Soc. Perkin Trans. 1994;I:2967–2973. [Google Scholar]
- 20.Wehner M., Herrmann C. Biochemical properties of the human guanylate binding protein 5 and a tumor-specific truncated splice variant. FEBS J. 2010;277:1597–1605. doi: 10.1111/j.1742-4658.2010.07586.x. [DOI] [PubMed] [Google Scholar]
- 21.Leatherbarrow R. J. Grafit Version 5. Horley, U.K.: Erithacus Software Ltd; 2001. [Google Scholar]
- 22.Ford B., Hornak V., Kleinman H., Nassar N. Structure of a transient intermediate for GTP hydrolysis by ras. Structure. 2006;14:427–436. doi: 10.1016/j.str.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Popp D., Narita A., Oda T., Fujisawa T., Matsuo H., Nitanai Y., Iwasa M., Maeda K., Onishi H., Maeda Y. Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability. EMBO J. 2008;27:570–579. doi: 10.1038/sj.emboj.7601978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crestfield A. M., Moore S., Stein W. H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J. Biol. Chem. 1963;238:622–627. [PubMed] [Google Scholar]
- 25.Glazer A. N. Chemical modification of proteins. In: Neurath H., editor. The Proteins. New York: Academic Press; 1976. pp. 1–103. Vol. II. [Google Scholar]
- 26.Gurd F. R. N. Carboxymethylation. Methods Enzymol. 1967;11:532–541. [Google Scholar]
- 27.Way J. C. Covalent modification as a strategy to block protein-protein interactions with small-molecule drugs. Curr. Opin. Chem. Biol. 2000;4:40–46. doi: 10.1016/s1367-5931(99)00049-6. [DOI] [PubMed] [Google Scholar]
- 28.Means G. E., Feeny R. E. Chemical Modification of Proteins. San Francisco: Holden-Day; 1971. [Google Scholar]
- 29.Sippel T. O. New fluorochromes for thiols: maleimide and iodoacetamide derivatives of a 3-phenylcoumarin fluorophore. J. Histochem. Cytochem. 1981;29:314–316. doi: 10.1177/29.2.7019305. [DOI] [PubMed] [Google Scholar]
- 30.Fellenberg F., Hartmann T. B., Dummer R., Usener D., Schadendorf D., Eichmuller S. GBP-5 splicing variants: new guanylate-binding proteins with tumor-associated expression and antigenicity. J. Invest. Dermatol. 2004;122:1510–1517. doi: 10.1111/j.0022-202X.2004.22613.x. [DOI] [PubMed] [Google Scholar]
- 31.Vojtek A. B., Der C. J. Increasing complexity of the Ras signaling pathway. J. Biol. Chem. 1998;273:19925–19928. doi: 10.1074/jbc.273.32.19925. [DOI] [PubMed] [Google Scholar]
- 32.Ford B., Skowronek K., Boykevisch S., Bar-Sagi D., Nassar N. Structure of the G60A mutant of Ras: implications for the dominant negative effect. J. Biol. Chem. 2005;280:25697–25705. doi: 10.1074/jbc.M502240200. [DOI] [PubMed] [Google Scholar]
- 33.Ford B., Boykevisch S., Zhao C., Kunzelmann S., Bar-Sagi D., Herrmann C., Nassar N. Characterization of a Ras mutant with identical GDP- and GTP-bound structures. Biochemistry. 2009;48:11449–11457. doi: 10.1021/bi901479b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Margarit S. M., Sondermann H., Hall B. E., Nagar B., Hoelz A., Pirruccello M., Bar-Sagi D., Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 35.Neal S. E., Eccleston J. F., Webb M. R. Hydrolysis of GTP by p21NRAS, the NRAS protooncogene product, is accompanied by a conformational change in the wild-type protein: use of a single fluorescent probe at the catalytic site. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3562–3565. doi: 10.1073/pnas.87.9.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenzen C., Cool R. H., Prinz H., Kuhlmann J., Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 37.Vetter I. R., Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 38.Brune M., Corrie J. E., Webb M. R. A fluorescent sensor of the phosphorylation state of nucleoside diphosphate kinase and its use to monitor nucleoside diphosphate concentrations in real time. Biochemistry. 2001;40:5087–5094. doi: 10.1021/bi002484h. [DOI] [PubMed] [Google Scholar]
- 39.Zielinski T., Kimple A. J., Hutsell S. Q., Koeff M. D., Siderovski D. P., Lowery R. G. Two Gαi1 rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J. Biomol. Screen. 2009;14:1195–1206. doi: 10.1177/1087057109347473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itaya K., Ui M. A new micromethod for the colorimetric determination of inorganic phosphate. Clin. Chim. Acta. 1966;14:361–366. doi: 10.1016/0009-8981(66)90114-8. [DOI] [PubMed] [Google Scholar]
- 41.Quan A., Robinson P. J. Rapid purification of native dynamin I and colorimetric GTPase assay. Methods Enzymol. 2005;404:556–569. doi: 10.1016/S0076-6879(05)04049-8. [DOI] [PubMed] [Google Scholar]
- 42.Webb M. R. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. U.S.A. 1992;89:4884–4887. doi: 10.1073/pnas.89.11.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb M. R., Hunter J. L. Interaction of GTPase-activating protein with p21ras, measured using a continuous assay for inorganic phosphate release. Biochem. J. 1992;287:555–559. doi: 10.1042/bj2870555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nixon A. E., Brune M., Lowe P. N., Webb M. R. Kinetics of inorganic phosphate release during the interaction of p21ras with the GTPase-activating proteins, p120-GAP and neurofibromin. Biochemistry. 1995;34:15592–15598. doi: 10.1021/bi00047a026. [DOI] [PubMed] [Google Scholar]
- 45.Binns D. D., Helms M. K., Barylko B., Davis C. T., Jameson D. M., Albanesi J. P., Eccleston J. F. The mechanism of GTP hydrolysis by dynamin II: a transient kinetic study. Biochemistry. 2000;39:7188–7196. doi: 10.1021/bi000033r. [DOI] [PubMed] [Google Scholar]
- 46.Eccleston J. F., Gratton E., Jameson D. M. Interaction of a fluorescent analogue of GDP with elongation factor Tu: steady-state and time-resolved fluorescence studies. Biochemistry. 1987;26:3902–3907. doi: 10.1021/bi00387a024. [DOI] [PubMed] [Google Scholar]
- 47.John J., Sohmen R., Feuerstein J., Linke R., Wittinghofer A., Goody R. S. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29:6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 48.Kunzelmann S., Praefcke G. J., Herrmann C. Nucleotide binding and self-stimulated GTPase activity of human guanylate-binding protein 1 (hGBP1) Methods Enzymol. 2005;404:512–527. doi: 10.1016/S0076-6879(05)04045-0. [DOI] [PubMed] [Google Scholar]
- 49.Kraemer A., Brinkmann T., Plettner I., Goody R., Wittinghofer A. Fluorescently labelled guanine nucleotide binding proteins to analyse elementary steps of GAP-catalysed reactions. J. Mol. Biol. 2002;324:763–774. doi: 10.1016/s0022-2836(02)01136-1. [DOI] [PubMed] [Google Scholar]
- 50.Newcombe A. R., Stockley R. W., Hunter J. L., Webb M. R. The interaction between rac1 and its guanine nucleotide dissociation inhibitor (GDI), monitored by a single fluorescent coumarin attached to GDI. Biochemistry. 1999;38:6879–6886. doi: 10.1021/bi9829837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.