Abstract
Hedgehog (Hh) signaling is essential for multiple aspects of embryogenesis1, 2. In Drosophila, Hh transduction is mediated by a cytoplasmic signaling complex3-5 that includes the putative serinethreonine kinase Fused (Fu) and the kinesin Costal 2 (Cos2), yet Fu does not play a conserved role in Hh signaling in mammals6, 7. Mouse Fu mutants are viable and appear to respond normally to Hh signaling. Here we show that mouse Fu is essential for construction of the central pair (CP) apparatus of motile, 9+2 cilia and offers a novel model of human primary ciliary dyskinesia. We found that mouse Fu physically interacts with Kif27, a mammalian Cos2 ortholog8, and linked Fu to known structural components of the CP apparatus, providing evidence for the first regulatory component involved in CP construction. We also demonstrated that zebrafish Fu is required both for Hh signaling and cilia biogenesis in Kupffer’s vesicle. Mouse Fu rescued both Hh-dependent and independent defects in zebrafish. Our results delineate a novel pathway for CP apparatus assembly, identify common regulators of Hh signaling and motile ciliogenesis, and add insight into evolution of the Hh cascade.
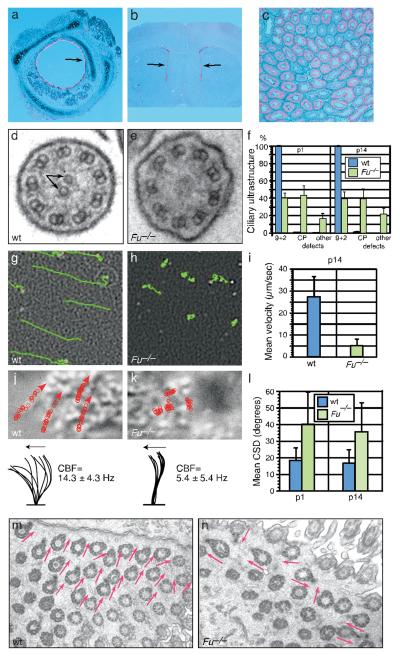
To further investigate the role of Fu in mammalian Hh signaling, we asked whether Hh-dependent Smo localization to the primary cilium is affected in the absence of Fu. Primary cilia, which have a “9+0” arrangement of 9 outer doublet microtubules (MTs), are required for Hh responses and contain several Hh pathway components2, 9. We found that Fu-/- mouse embryonic fibroblasts (MEFs) formed primary cilia normally, trafficked Smo to the primary cilium in response to Hh ligand, and exhibited a typical Gli transcriptional response (Supplementary Fig. 1, 2; data not shown). This suggests that the single mammalian Fu ortholog is dispensable for Hh signaling. To explore the function of Fu in mice, we examined its expression in postnatal tissues by in situ hybridization. Fu transcript was expressed strongly in the respiratory epithelium, the ependymal lining of the ventricles in the brain, and in oviduct and testis (Fig. 1a-c; data not shown). These expression patterns are reminiscent of genes involved in biogenesis of motile cilia, which function in these tissues to propel mucus, fluid and cells. In contrast to the primary cilium, the classical “9+2” motile cilium consists of 9 outer doublet MTs and two singlet central pair (CP) MTs10. The CP apparatus plays a key role in regulating ciliary motility but its formation is poorly understood since the centriole-derived basal body, from which the cilia axoneme extends, does not provide a template for CP outgrowth. Disruption of human motile cilia function leads to primary ciliary dyskinesia (PCD), which is associated with recurrent respiratory infection, hydrocephalus and infertility11-13. To determine whether motile cilia function is compromised in Fu-/- mice, we studied cilia axonemal ultrastructure by transmission electron microscopy. In wild-type animals, over 99% of tracheal and ependymal motile cilia displayed a typical “9+2” configuration (Fig. 1d, f; Supplementary Fig. 3, 4). By contrast, approximately 60% of Fu-/- cilia have abnormal ciliary ultrastructure; two-thirds of these lack the CP apparatus (Fig. 1e, f; Supplementary Fig. 3, 4). CP defects in Fu mutants are apparent at the basal plate region characterized by an electron-dense thick band, where the CP originates in wild-type cilia14 (Supplementary Fig. 4). Our findings indicate that mammalian Fu is dispensable for Hh signaling and specifically participates in generation of the CP apparatus in motile cilia axonemes.
Figure 1. Mouse Fu is required for central pair (CP) apparatus construction.
a-c, Expression of Fu (pink signal) in the mouse tracheal epithelium (a), ependyma of the lateral ventricles (b), and testis (c) at postnatal (p) day 14 by section in situ hybridization to Fu. Arrows indicate sites of Fu expression. d, e, Transmission electron micrographs (TEM) of motile cilia from wild-type (wt) and Fu-/- tracheae. Arrows denote the CP microtubules. f, Quantification of ultrastructural defects from p1 (wt, n = 4 animals, mean 88 cilia/animal analyzed; Fu-/-, n = 4 animals, mean 64 cilia/animal analyzed) and p14 (wt, n = 7, mean 113 cilia/animal analyzed; Fu-/-, n = 6 animals, mean 93 cilia/animal analyzed) tracheae. Error bars indicate standard deviation (s.d.). g, h, Traces of fluorescent bead movement over tracheal explants. i, Mean particle velocity in p14 wt (n = 5) and Fu-/- (n = 5) tracheae. Error bars indicate s.d. j, k, Traces of cilia beat path overlaid on still DIC images of tracheal cilia (upper), and lateral traces of cilia waveform (lower). Mean ciliary beat frequency (CBF) was calculated from 30 cilia (n = 3 animals for wt and Fu-/-). Arrows indicate directions of the forward effective strokes. l, Quantification of circular standard deviation (CSD) of basal feet from p1 (wt, n = 24 cells from 4 animals; Fu-/-, n = 38 cells from 4 animals; P < 3.4 × 10−6; unpaired Student’s t-test) and p14 (wt, n = 31 cells from 4 animals; Fu-/-, n = 36 cells from 3 animals; P < 2.6 × 10−7; unpaired Student’s t-test). Error bars indicate s.d. m, n, Representative TEM images of basal foot polarity (arrows) in p14 wt and Fu-/- tracheae.
Mice lacking functional Fu are born with no obvious phenotype, but fail to thrive in comparison with wild-type or heterozygous littermates and die before postnatal (p) day 216, 7. To determine the functional consequences of CP apparatus loss in Fu-/- animals, we examined fluid flow in tracheal explants. Analysis of fluorescent bead movement revealed strong distal-proximal directional flow in wild-type explants, whereas beads overlaid on Fu-/- tracheae exhibited severely impaired velocity and little to no directional movement (Fig. 1g-i; Supplementary Movie 1, 2). We next asked if elimination of the CP apparatus in Fu-/- animals disrupted cilia motion. In wild-type tracheae, cilia beat in a linear path with a quick forward power stroke and a slower recovery stroke15 (Fig. 1j; Supplementary Movie 3, 5). The majority of Fu-/- cilia moved stiffly and had a significantly reduced stroke amplitude; a subset were either immotile or beat in a slow, circular motion (Fig. 1k; Supplementary Movie 4, 6). In contrast to wild-type motile cilia, which beat coordinately to produce a metachronic wave16, cilia in Fu-/- animals which did beat appeared disoriented with respect to their neighbors (Fig. 1k; Supplementary Movie 4, 6). This prompted us to investigate if cilia orientation, specified by a basal body accessory structure known as the basal foot, was perturbed17, 18. In wild-type tracheae, basal feet were properly aligned with each other (Fig. 1m). In Fu-/- mutants, basal feet were disoriented and frequently pointed at right angles or antiparallel to one another (Fig. 1n), and the circular standard deviation (CSD) of cilia orientation within a given cell was significantly higher (Fig. 1l). Loss of the CP apparatus in Fu-/- mice thus eliminated directional fluid flow, resulting from unccordinated ciliary beating and global disorganization of cilia polarity.
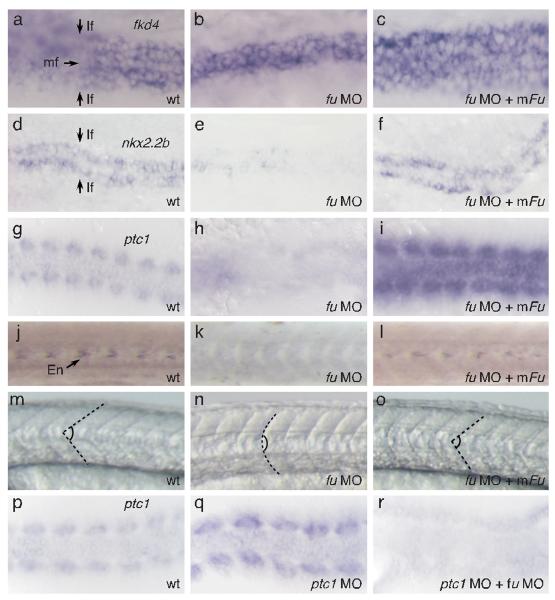
We hypothesized that Fu in different metazoan species might participate in both Hh signaling and ciliogenesis. We examined the role of Fu in zebrafish since fu morphants exhibit mild Hh-dependent somite phenotypes19. By delivering a higher concentration of fu morpholino, we observed stronger Hh phenotypes including cyclopia and loss of lateral floor plate (Fig. 2a, b, d, e; Supplementary Fig. 5; data not shown), similar to smo mutants20. Knockdown of zebrafish fu activity greatly reduced patched1 (ptc1) expression in somites, suggesting disruption of Hh responses (Fig. 2g, h). The Hh-dependent muscle pioneer population, marked by expression of engrailed1 (en1), was lost (Fig. 2j, k) and fu morphants developed U-shaped instead of chevron-shaped somites (Fig. 2m, n). In ptc1 morphants, Hh target genes are up-regulated cell autonomously (Fig. 2p, q; Supplementary Fig. 10)19. Up-regulation of Hh target genes is abolished in ptc1; fu double morphants (Fig. 2r), indicating that fu functions cell autonomously in Hh-responsive cells to control Hh signaling. Taken together, these results provide convincing evidence for an integral role of Fu in the zebrafish Hh pathway. We then asked if mouse Fu compensated for loss of zebrafish Fu. Surprisingly, co-injection of mouse Fu mRNA and zebrafish fu morpholino rescued all Hh phenotypes including restoration of ptc1 expression, lateral floor plate formation, muscle pioneer differentiation and somite shape (Fig. 2c, f, i, l, o; Supplementary Fig. 5; data not shown). By contrast, co-injection of Drosophila fu mRNA and the zebrafish fu morpholino failed to rescue Hh phenotypes (data not shown). Thus, mouse Fu retains the necessary information to participate in the fish Hh pathway, implying that a common mechanism underlies critical aspects of Hh signaling and motile ciliogenesis.
Figure 2. Mouse Fu is capable of rescuing Hh-related phenotypes in zebrafish fu morphants.
a, Whole-mount in situ hybridization to fkd4 (purple signal) in both medial and lateral floor plate of wt zebrafish embryos at 24 hours post fertilization (hpf). b, c, fkd4 expression is lost in the lateral floor plate of fu morphants (b) and is restored when mouse Fu is expressed (c). d, Whole-mount in situ hybridization to nkx2.2b (purple signal) in lateral floor plate of wt zebrafish embryos at 24 hpf. e, f, nkx2.2b expression is lost in the lateral floor plate of fu morphants (d) and is restored when mouse Fu is expressed (e). View is dorsal. mf, medial floor plate; lf, lateral floor plate. g, Whole-mount in situ hybridization to ptc1 (purple signal) in somites of wt zebrafish embryos at 10-somite stage. View is dorsal. h, i, ptc1 expression is greatly reduced in somites of fu morphants (h) and is restored when mouse Fu is expressed (i). j, Immunohistochemistry against En (arrow), which labels the muscle pioneer population in wt zebrafish somites at 24 hpf. View is lateral. k, l, Rescue of En expression in fu morphant somites (k) by co-injection with mouse Fu (l). m, Lateral view of chevron-shaped somites in wt zebrafish embryos at 24 hpf. n, o, Rescue of U-shaped somites in fu morphants (n) by co-injection with mouse Fu (o). Dotted lines delineate the boundaries of somites. p, Whole-mount in situ hybridization to ptc1 (purple signal) in somites of wt zebrafish embryos at 10-somite stage. View is dorsal. q, r, Upregulation of ptc1 expression in ptc1 morphants (q) is abolished by knocking down fu (r).
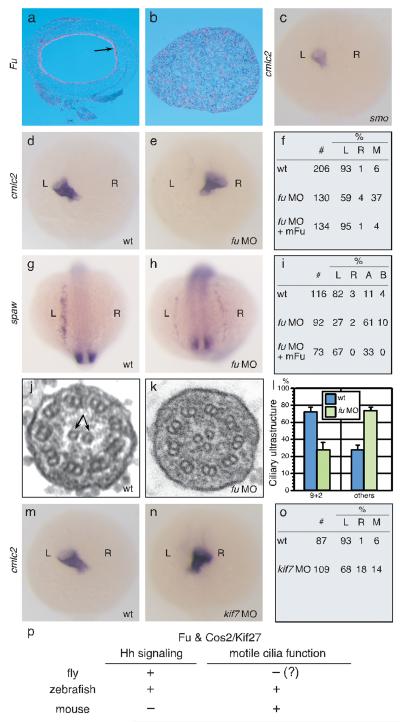
Fu may have an ancient, conserved role in regulating microtubule or motile cilia function since the genomes of many organisms, including plants and flagellated unicellular eukaryotes, contain genes encoding a highly conserved Fu kinase domain21 (Supplementary Fig. 8). To test this idea, we examined Fu expression in different species by in situ hybridization and found strong expression in the chick tracheal epithelium and the oviduct and testis of Xenopus tropicalis (Fig. 3a, b; data not shown), in a pattern similar to mouse Fu. We then focused on zebrafish, which utilize 9+2 motile cilia on the surface of Kupffer’s vesicle (KV) to generate a counterclockwise flow essential for establishment of left-right (L-R) asymmetry22. If zebrafish Fu also participates in 9+2 cilia biogenesis, we reasoned that L-R asymmetry would be disrupted. We examined the positioning of the heart and visceral organs by cardiac myosin light chain2 (cmlc2/myl7) and fork head domain protein FKD2 (fkd2/foxa3) expression, respectively. In contrast to zebrafish smo mutants, in which disrupted Hh signaling does not perturb L-R asymmetry20 (Fig. 3c), 41% of fu morphants displayed reversed or midline hearts (Fig. 3d, e, f), while 30% of injected embryos had abnormal positioning of the gut, liver and pancreas (Supplementary Fig. 11). To investigate if Fu is required for early establishment of asymmetric gene expression in the left lateral plate mesoderm (LPM), we studied the expression pattern of southpaw (spaw) and paired-like homeodomain transcription factor 2 (pitx2) in fu morphants. In 73% of fu morphants, spaw was found to be on the right side, bilateral, or absent in the LPM (Fig. 3g, h, i). Similarly, 71% of fu morphants had significantly reduced or absent pitx2 staining in the LPM (data not shown). Co-injection of mouse Fu, but not Drosophila fu, with fu morpholino was sufficient to restore L-R asymmetry (Fig. 3f, i; data not shown).
Figure 3. Zebrafish fu has a Hh-independent role in L-R asymmetry and generation of 9+2 cilia.
a, b, Section in situ hybridization to Fu (pink signal) in chick trachea (a) and X. tropicalis testis (b). Arrow indicates sites of Fu expression. c, Whole mount in situ hybridization to cmlc2 (purple signal) in smohi1640Tg fish embryos at 24 hpf. View is dorsal. d, e, Whole-mount in situ hybridization to cmlc2 in wt (d) and fu morphants at 24 hpf. View is dorsal. (e). f, Summary of cardiac laterality defects in wt (n = 206), fu morphants (n = 130), and fu morphants rescued with mouse Fu (n = 134). L, left; R, right; M, medial. g, h, Whole mount in situ hybridization to spaw at 15-somite stage. View is dorsal. i, Summary of spaw expression in the lateral plate mesoderm (LPM) in wt (n = 116), fu morphants (n = 92), and fu morphants rescued with mouse Fu (n = 73). L, left; R, right; A, absent; B, bilateral. j, k, Electron micrograph of Kupffer’s vesicle (KV) cilia from wt (j) and fu morphant (k). l, Quantification of ultrastructural defects in KV cilia from wt and fu morphants. Error bars indicate standard deviation (s.d.). m, n, Whole mount in situ hybridization to cmlc2 in wt and kif7 morphants at 24 hpf. View is dorsal. o, Summary of cardiac laterality defects in wt (n = 87) and kif7 morphants (n = 109). p, Summary of essential Fu and Cos2/Kif27 functions in metazoan model organisms.
To confirm a direct role of Fu in regulating KV function, we injected fluorescein-labeled fu morpholino into dorsal forerunner cells (DFCs)23, which migrate at the leading edge of the embryonic shield to produce KV. 44% of embryos with strong fluorescent signal in the DFCs developed cardiac laterality but not somite defects (data not shown), indicating that knockdown of fu in KV accounts for the L-R asymmetry defects. KV cilia in fu morphants displayed disorganized axonemal structures, including loss and acquisition of extra CP microtubules (Fig. 3j, k; data not shown), indicating a conserved role of vertebrate Fu in CP construction. Loss of fu affected cilia motility as revealed by injecting rhodamine-conjugated dextran beads into KV of fu morphants at the 8-somite stage (Supplementary Movies 7 and 8). Defects in establishing a counterclockwise flow in fu morphants were rescued by mouse Fu (Supplementary Movie 9). Taken together, the data strongly support a conserved, Hh-independent role of Fu in vertebrate 9+2 cilia biogenesis.
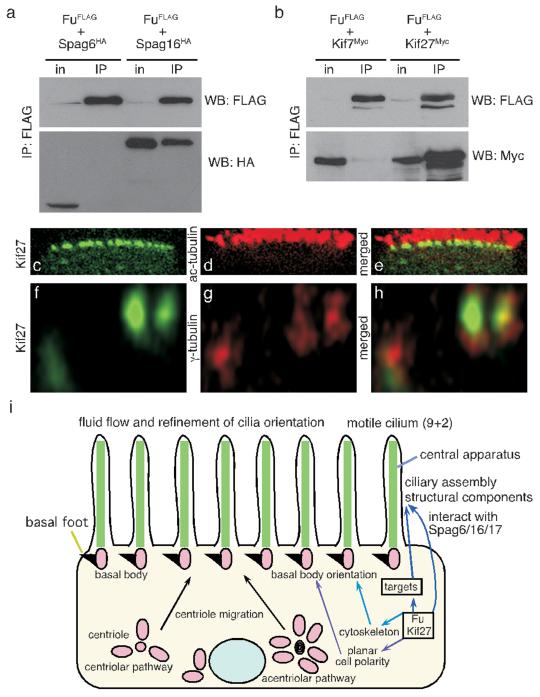
The process of CP construction is poorly characterized and Fu is the first regulatory component known to control its assembly. To determine how Fu might control this process, we tested the ability of Fu to interact with SPAG6/PF16 and SPAG16L/PF20, evolutionarily conserved components of the CP apparatus24, 25. When expressed in HEK 293T cells, Fu-FLAG efficiently co-immunoprecipitated SPAG16L-HA, but not SPAG6-HA (Fig. 4a). Interestingly, SPAG16L localizes to the sperm CP apparatus26, and its Chlamydomonas ortholog PF20 decorates the C2 microtubule along the intermicrotubule bridges between CP MTs27. This suggests a direct role for Fu in assembly or maintenance of the CP apparatus.
Figure 4. Mouse Fu interacts with the CP protein SPAG16L and the Cos2 ortholog Kif27.
a, Western blot of immunoprecipitated mouse Fu-FLAG to detect its physical interaction with mouse SPAG6-HA or SPAG16L-HA from HEK 293T lysates. WB, western blot; in, input; IP, immunoprecipitation. b, Western blot of immunoprecipitated mouse Fu-FLAG to determine its physical association with mouse Kif7-myc or Kif27-myc from HEK 293T lysates. c-h, Confocal images of fully differentiated mouse tracheal epithelial cells (MTECs) to visualize localization of Kif27-GFP to the basal body (marked by anti-γ-tubulin) of motile cilia labeled with acetylated tubulin (ac-tubulin). j, Model of Fu, Kif27, and SPAG16 function in motile cilia construction.
In fly, Fu binds to the kinesin Cos2 to transduce the Hh signal downstream of Smo. We examined if mouse Fu bound to the mouse Cos2 orthologs Kif7 and Kif27. When expressed in HEK 293T cells and mouse tracheal epithelial cells (MTECs), Fu-FLAG bound strongly to Kif27-myc, but not Kif7-myc (Fig. 4b; data not shown), implicating Kif27 in generation or regulation of 9+2 cilia. We expressed Kif27-GFP in MTECs via lentiviral infection and assessed its localization throughout MTEC differentiation induced by the creation of an air-liquid interface (ALI). During this process, hundreds of centrioles migrate to the apical surface of the cell, dock with the membrane to form basal bodies, and serve as templates for the outgrowth of the outer MT doublets of the ciliary axoneme28. At ALI days 0 and 5, Kif27-GFP punctae were associated with centrioles as determined by γ-tubulin staining (Supplementary Fig. 6). Kif27-GFP associated with the base of the cilium after axoneme outgrowth (Fig. 4c, d, e, f, g, h). Fu-mCherry was broadly distributed in the cytoplasm of MTECs throughout differentiation, overlapping with Kif27 (Supplementary Fig. 6). Fu and Kif27 expression are upregulated during MTEC differentiation, consistent with their essential roles in motile ciliogenesis (Supplementary Fig. 7). Efforts to demonstrate Fu kinase activity in vitro have not been successful, suggesting the requirement of a special microenvironment for its activity. We speculate that Fu has multiple substrates, some of which could reside in the cytoplasm and control CP assembly indirectly (Fig. 4i). Our data favor a model in which Kif27 and/or SPAG16L directs the localization or activity of Fu for CP construction (Fig. 4i).
Despite the non-essential role of Fu in mammalian Hh signaling, the protein retains an interaction with the Cos2 ortholog Kif27. Analysis of Cos2, Kif7 and Kif27 sequences indicates that the Kif7/27 genes may have arisen through a duplication event (Supplementary Fig. 9). The four fish species examined do not contain an obvious Kif27 ortholog, suggesting either that Kif27 was lost after gene duplication or that the duplication event occurred after divergence of the fish and amphibian lineages. Supporting the latter, morpholino knockdown of kif7 in zebrafish (z) resulted in both Hh-specific phenotypes and disruption of L-R asymmetry (Fig. 3m, n, o; data not shown), indicating a dual role for Kif7 in Hh signaling29 and motile ciliogenesis, similar to zFu which co-immunoprecipitates with zKif7 (Supplementary Fig. 12). There are conflicting reports on the role of Kif7 and Kif27 in vertebrate Hh signaling29, 30; based on the data here, we predict that Kif27 does not play a significant role in mammalian Hh signal transduction and mice lacking functional Kif27 may exhibit phenotypes similar to Fu. We speculate that Fu has evolved or retained its function in CP assembly in vertebrates, and that duplication of ancestral Cos2 in the vertebrate lineage led to partition of functions for Kif7/27, while Kif27 retained its partnership with Fu. Although the requirement of Fu-like activity in mammalian Hh signaling is unproven, if it exists it is likely compensated for by an unrelated kinase (Supplementary Fig. 8). Alternatively, the involvement of the primary cilium as a scaffold for Hh pathway components in mammals could circumvent the need for a Fu-kinesin complex. Consistent with the notion of evolutionary changes in Hh pathway design in different species, Su(fu), a component of the cytoplasmic signaling complex, is dispensable for fly viability, plays a minor role in zebrafish Hh signaling and becomes a major negative regulator in mice2. Further analysis of Fu/Kif27 function in ciliogenesis and Hh signaling in diverse species will provide additional insight into the evolution of this critical signaling pathway.
Methods Summary
Transmission electron microscopy (TEM)
Mouse tissue was fixed in 3% glutaraldehyde /1% paraformaldehyde /0.1 M sodium cacodylate, pH 7.4 at 4°C overnight. Fish embryos were fixed in 2% paraformaldehyde /2% glutaradehyde (EM grade) at RT for 2 hr. Standard processing, embedding and sectioning procedures were followed. Samples were examined on a JEOL 100CX or JEM-1230 transmission electron microscope.
Basal foot polarity
The orientation and circular standard deviation (CSD) of basal feet in EM micrographs was calculated as described18. Circular statistics were calculated using Oriana 2.0 (Kovachs Computing Services).
Tracheal flow assays
Tracheae from postnatal (p) day 14 wild-type and Fu-/- mice were excised, cleaned of muscle and vasculature, opened longitudinally, and placed in a drop of PBS on a glass slide. 5 μl of a 0.01% solution of Fluospheres (Invitrogen) were added on top of a single trachea to visualize the direction of ciliary flow. Images were acquired using a SPOT 2.3 camera connected to a Nikon E1000 epifluorescence microscope. Images were captured at a rate of 26 frames per second (fps) over a 50 μm by 50 μm area and were saved as .tiff stacks. Movies were examined in NIH Image J using the enhancing feature of the SpotTracker plugin (Daniel Sage and Susan Gasser) to optimize sphere intensity, and the MtrackJ plugin (Erik Meijering, Biomedical Imaging Group, University Medical Center, Rotterdam) to trace the direction and path length of the sphere. Average velocity was taken to be the straight-line distance a particle traveled from its originating point divided by time, and was calculated in Microsoft Excel.
Full Methods
Animal husbandry
Fu+/− mice were maintained as described6. Wild-type AB fish were used and raised as described31. The slow muscle omitted/smoothened allele20 used in this study is smohi1640Tg.
Molecular biology
Standard molecular biology techniques, including molecular cloning, genomic DNA preparation, RNA isolation, PCR, RT-PCR, and Southern analysis were performed as described32, 33. Fu-FLAG, Fu-4×FLAG, Fu-mCherry, Kif7-3xmyc, Kif27-3xmyc, Kif27-GFP, SPAG16L-3×HA, and SPAG6-3×HA were cloned into pCAGGS (for immunoprecipitation and immunofluorescence in mammalian cells), pCS2+ (for expression in zebrafish), pcDNA3 (for immunoprecipitation and immunofluorescence in mammalian cells), or FuPw (for lentiviral expression) vectors. Detailed methods and maps are available on request.
FuPw vector (courtesy of Kit Wong, Bourne lab, UCSF) contains the HIV-1 flap sequence, the human polyubiquitin C promoter, a multiple cloning site, and the woodchuck hepatitis virus post-transcriptional regulatory element. Flanking this cassette are 5′ and 3′ self-inactivating long-terminal repeats. Expression constructs were co-transfected with the HIV packaging vector pCMVΔ8,9 and the envelope glycoprotein vector pVSV-G into HEK293T cells using Lipofectamine 2000 (Invitrogen).
Morpholino (MO) Injections
Wild-type zebrafish embryos were injected with 1.6–4 ng fu or 8–12 ng kif7 or 0.2 ng ptc1 MO at the one- to two- cell stage. Fluorescein-tagged fu MO (4 ng) was injected into the yolk of 128-cell stage embryos to target dorsal forerunner cells (DFCs). A p53 morpholino was co-injected with fu or kif7 morpholino at the same concentration to block non-specific cell death34. In rescue experiments, 400 pg of mouse Fu mRNA was co-injected with fu MO. In testing genetic epistasis, 0.2 ng of ptc1 and 2ng of fu MO were co-injected. The fu (5′ TGGTACTGATCCATCTCCAGCGACG 3′), kif7 (5′ GCCGACTCCTTTTGGAGACATAGCT 3′) and ptc1 MO (5′ CATAGTCCAAACGGGAGGCAGAAGA 3′) were described previously19.
In situ hybridization
Histological analysis and section in situ hybridization using 33P-labeled riboprobes were performed as described6. Probes for chick, zebrafish and X. tropicalis Fu were amplified by PCR using partial or full-length cDNAs (Open Biosystems) as templates. Zebrafish embryos were raised in medium treated with 0.2 mM 1-phenyl-1-2-thiourea to maintain optical transparency. Whole mount in situ hybridization was performed as described35; probes used were cmlc2 (myl7), fkd2 (foxa3), fkd4 (foxa), nkx2.2b, fused, shh, ptc1, spaw, and pitx2.
Ciliary beat frequency (CBF) and waveform measurements
Tracheae were dissected out from p10-p14 wild-type and Fu-/- animals, and were cut into rings or strips. Tracheae were washed briefly in PBS and placed in DMEM supplemented with 10% FBS, penicillin-streptomycin and L-glutamate. Tissue was placed in a few drops of medium in a 35 mm glass bottom microwell dish (MatTek). Cilia beating was observed using differential interference contrast (DIC) microscopy on a Nikon TE2000E inverted microscope equipped with Perfect Focus, a 60x water immersion objective, 1.5x zoom adapter and an In Vivo Scientific incubator set at 37°C and 5% CO2. A Photometrics Coolsnap HQ2 camera and NIS Elements 2.3 software were used to acquire videos of beating cilia at frame rates of 60-70 fps, depending on the size of the defined region of interest (ROI). CBF was measured by defining an ROI in the upper third of the ciliary shaft, and plotting the changes in pixel intensity over time in the obtained image series. This data was subsequently Fourier transformed in order to obtain the frequency using MatLab. Waveform was analyzed by tracing of cilia from individual movie frames in Adobe Illustrator, or by manual tracking using the MtrackJ plugin (Erik Meijering, Biomedical Imaging Group, University Medical Center, Rotterdam) in NIH ImageJ.
Cell culture, transfections, and immunoprecipitation
HEK 293T cells were maintained in DMEM supplemented with 10% FBS, penicillin-streptomycin, and L-glutamate. Cells were transfected with Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. 48 hr post-transfection, cells were harvested and lysed in lysis buffer (1% Triton X-100, 150 mM NaCl, 50 mM Tris-Cl pH 7.5, 1 mM EDTA, 0.5 mM PMSF, 2 μg/mL pepstatin A, 10 μg/mL leupeptin, 5 μg/mL aprotinin). Lysates were sheared with a 20-gauge needle and remained on ice for 30 min. Lysates were then clarified by centrifugation at 20817 × g for 20 min at 4°C. The supernatant was removed and bound to 50 μl of anti-FLAG M2 agarose beads (Sigma) for 4 hr at 4°C with constant nutation. Beads were washed five times with lysis buffer prior to addition of sample buffer. Immunoprecipitated proteins were analyzed by 7.5% SDS-PAGE and transferred to PVDF for immunoblotting. Antibodies used were rabbit anti-FLAG (Sigma, 1:2000), rabbit anti-myc (Sigma, 1:2000), and rabbit anti-HA (Sigma, 1:1000).
Primary MTEC culture and viral transduction
Primary mouse tracheal epithelial cells (MTECs) were derived from postnatal day 10 to 21 mice and cultured as described36. Lentivirus was produced by co-transfecting cDNAs cloned into the FuPw vector with pCMVΔ8,9 and pVSV-G into HEK 293T cells as described above. Supernatant was harvested 72-96 hr post-transfection, filtered through a 0.45μ PES membrane syringe filter unit (Nalgene), and concentrated ten-fold using a Centriprep Ultracel YM-10 device (Millipore). Infection of MTECs was performed as described37.
Immunofluorescence and microscopy
Cells were fixed in 4% paraformaldehyde for most applications, or in ice-cold methanol for visualization of basal bodies. Standard procedures were used for immunostaining. Primary antibodies used were mouse anti-acetylated-α-tubulin (Sigma, 1:2000) and mouse anti-γ tubulin (Sigma, 1:2000). Secondary antibodies and conjugates used were donkey anti-mouse AlexaFluor 594 (Molecular Probes, 1:2000), donkey anti-mouse FITC (Molecular Probes, 1:2000), and rhodamine-conjugated phalloidin (Sigma, 1:200). Fluorescent confocal images were acquired using a Nikon TE2000U inverted microscope with a Yogokawa CSU22 spinning disk confocal (Solamere Technology Group), a Photometrics Cascade II Camera, and MicroManager software (Vale lab, University of California-San Francisco). Images were acquired with a 100x oil-immersion lens and a 1.5x zoom adapter (Nikon) using two laser lines (488 nm and 568 nm). Confocal stacks were collected using a 0.25 μm step size along the z-axis. Stacks were analyzed and xy, xz, and yz projections generated using ImageJ and the VolumeViewer plugin (Kai Uwe Barthel, Internationale Medieninformatik, Berlin, Germany). Deconvolution was performed with the Iterative Deconvolve 3D plugin (Robert Dougherty, OptiNav, Inc.).
Immunohistochemistry staining
Immunohistochemistry staining using anti-Engrailed (4D9, Developmental Studies Hybridoma Bank) at 1:100 dilution and anti-acetylated tubulin (Sigma, MO) at 1:200 dilution was conducted as described (ref?). Confocal images were acquired with an LSM510 confocal microscope (Zeiss, Germany).
Fluorescent bead injection
Fluorescent beads diluted 1:100 in 1x PBS were injected into Kupffer’s vesicle (KV) at the 8- to 10-somite stage23. Embryos were imaged on a Zeiss Axioplan 2 microscope using a 63x water immersion lens (Zeiss, Germany).
Supplementary Material
Acknowledgements
We thank H. Bourne, C.C. Hui, Z. Zhang, J. Strauss III, and W. Hwang for constructs, antibodies and sharing of unpublished results; R. Harland and T. Mikawa for X. tropicalis and chicken tissue; K. Thorn and S. Dandekar for assistance with microscopy and CBF analysis; Mei-Leng Cheong and Yoko Nozawa for technical assistance; and D. Casso, S. Coughlin, T. Kornberg, W. Marshall, T. Mikawa, K. Wemmer, and members of the Chen and Chuang laboratory for discussion and critical reading of the manuscript. Some data for this study were acquired at the Nikon Imaging Center at UCSF/QB3. This work was supported by grants from the National Institutes of Health to J.-N. C. and P.-T. C., and a Career Investigator Award from the American Lung Association to P.-T. C.
Footnotes
Author information The authors declare no competing financial interests.
References
- 1.McMahon AP, Ingham PW, Tabin CJ. Developmental roles and clinical significance of hedgehog signaling. Curr Top Dev Biol. 2003;53:1–114. doi: 10.1016/s0070-2153(03)53002-2. [DOI] [PubMed] [Google Scholar]
- 2.Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- 3.Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–45. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 4.Robbins DJ, et al. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–34. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- 5.Lum L, et al. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12:1261–74. doi: 10.1016/s1097-2765(03)00426-x. [DOI] [PubMed] [Google Scholar]
- 6.Chen MH, Gao N, Kawakami T, Chuang PT. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25:7042–53. doi: 10.1128/MCB.25.16.7042-7053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merchant M, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25:7054–68. doi: 10.1128/MCB.25.16.7054-7068.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katoh Y, Katoh M. KIF27 is one of orthologs for Drosophila Costal-2. Int J Oncol. 2004;25:1875–80. [PubMed] [Google Scholar]
- 9.Davenport JR, Yoder BK. An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am J Physiol Renal Physiol. 2005;289:F1159–69. doi: 10.1152/ajprenal.00118.2005. [DOI] [PubMed] [Google Scholar]
- 10.Davis EE, Brueckner M, Katsanis N. The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle. Dev Cell. 2006;11:9–19. doi: 10.1016/j.devcel.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Afzelius BA. Cilia-related diseases. J Pathol. 2004;204:470–7. doi: 10.1002/path.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marshall WF, Kintner C. Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol. 2008;20:48–52. doi: 10.1016/j.ceb.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zariwala MA, Knowles MR, Omran H. Genetic defects in ciliary structure and function. Annu Rev Physiol. 2007;69:423–50. doi: 10.1146/annurev.physiol.69.040705.141301. [DOI] [PubMed] [Google Scholar]
- 14.McKean PG, Baines A, Vaughan S, Gull K. Gamma-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr Biol. 2003;13:598–602. doi: 10.1016/s0960-9822(03)00174-x. [DOI] [PubMed] [Google Scholar]
- 15.Chilvers MA, Rutman A, O’Callaghan C. Ciliary beat pattern is associated with specific ultrastructural defects in primary ciliary dyskinesia. J Allergy Clin Immunol. 2003;112:518–24. doi: 10.1016/S0091-6749(03)01799-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Dillon RH, Fauci LJ. An integrative computational model of multiciliary beating. Bull Math Biol. 2008;70:1192–215. doi: 10.1007/s11538-008-9296-3. [DOI] [PubMed] [Google Scholar]
- 17.Frisch D, Farbman AI. Development of order during ciliogenesis. Anat Rec. 1968;162:221–32. doi: 10.1002/ar.1091620209. [DOI] [PubMed] [Google Scholar]
- 18.Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- 19.Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13:1169–81. doi: 10.1016/s0960-9822(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 20.Chen W, Burgess S, Hopkins N. Analysis of the zebrafish smoothened mutant reveals conserved and divergent functions of hedgehog activity. Development. 2001;128:2385–96. doi: 10.1242/dev.128.12.2385. [DOI] [PubMed] [Google Scholar]
- 21.Oh SA, et al. A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr Biol. 2005;15:2107–11. doi: 10.1016/j.cub.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 22.Kramer-Zucker AG, et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–21. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 23.Shu X, et al. Na,K-ATPase alpha2 and Ncx4a regulate zebrafish left-right patterning. Development. 2007;134:1921–30. doi: 10.1242/dev.02851. [DOI] [PubMed] [Google Scholar]
- 24.Neilson LI, et al. cDNA cloning and characterization of a human sperm antigen (SPAG6) with homology to the product of the Chlamydomonas PF16 locus. Genomics. 1999;60:272–80. doi: 10.1006/geno.1999.5914. [DOI] [PubMed] [Google Scholar]
- 25.Sapiro R, et al. Sperm antigen 6 is the murine homologue of the Chlamydomonas reinhardtii central apparatus protein encoded by the PF16 locus. Biol Reprod. 2000;62:511–8. doi: 10.1095/biolreprod62.3.511. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, et al. A sperm-associated WD repeat protein orthologous to Chlamydomonas PF20 associates with Spag6, the mammalian orthologue of Chlamydomonas PF16. Mol Cell Biol. 2002;22:7993–8004. doi: 10.1128/MCB.22.22.7993-8004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith EF, Lefebvre PA. PF20 gene product contains WD repeats and localizes to the intermicrotubule bridges in Chlamydomonas flagella. Mol Biol Cell. 1997;8:455–67. doi: 10.1091/mbc.8.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 29.Tay SY, Ingham PW, Roy S. A homologue of the Drosophila kinesin-like protein Costal2 regulates Hedgehog signal transduction in the vertebrate embryo. Development. 2005;132:625–34. doi: 10.1242/dev.01606. [DOI] [PubMed] [Google Scholar]
- 30.Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–86. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- 32.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Third Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2003. [Google Scholar]
- 33.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- 34.Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–43. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- 35.Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–16. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- 36.You Y, Richer EJ, Huang T, Brody SL. Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am J Physiol Lung Cell Mol Physiol. 2002;283:L1315–21. doi: 10.1152/ajplung.00169.2002. [DOI] [PubMed] [Google Scholar]
- 37.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.