Abstract
Detection of specific chromosomal abnormalities by FISH and metaphase cytogenetics allows risk stratification in multiple myeloma; however, gene expression profiling (GEP) based signatures may enable more specific risk categorization. We examined the utility of 2 GEP-based risk stratification systems among patients undergoing initial therapy with lenalidomide in the context of a phase 3 trial. Among 45 patients studied at baseline, 7 (16%) and 10 (22%), respectively, were high-risk using the GEP70 and GEP15 signatures. The median overall survival for the GEP70 high-risk group was 19 months versus not reached for the rest (hazard ratio = 14.1). Although the medians were not reached, the GEP15 also predicted a poor outcome among the high-risk patients. The C-statistic for the GEP70, GEP15, and FISH based risk stratification systems was 0.74, 0.7, and 0.7, respectively. Here we demonstrate the prognostic value for GEP risk stratification in a group of patients primarily treated with novel agents. This trial was registered at www.clinicaltrials.gov as #NCT00098475.
Introduction
Multiple myeloma (MM) is characterized by significant heterogeneity in outcome that is primarily driven by the underlying genetic abnormalities.1,2 Routine use of metaphase cytogenetics and FISH has allowed a better understanding of the spectrum of genetic abnormalities and to identify abnormalities associated with a poor outcome.3–6 These include translocations involving the immunoglobulin heavy chain (IgH) locus on chromosome 14 and chromosomes 4, 16, and 20, deletion of 17p, and deletions involving chromosome 13 seen on metaphase cytogenetics.1,7 However, these abnormalities alone do not account for the heterogeneity and led to evaluation of other approaches, such as gene expression profiling (GEP) of tumor cells to risk stratify patients.8–13 Several GEP signatures have been proposed by different groups, primarily in the context of autologous stem cell transplantation (ASCT).14–16 However, there are limited data regarding their utility in the context of patients primarily treated with novel agents, such as lenalidomide. We undertook the current study to examine the prognostic value of the GEP70 classification system that was developed by researchers at University of Arkansas and has since been extensively validated, in the setting of a phase 3 trial of lenalidomide and dexamethasone in newly diagnosed MM.14 In addition, we also examined the GEP15 system that was proposed by the Intergroupe Francophone du Myelome investigators.15
Methods
The E4A03 clinical trial randomized patients with previously untreated MM to lenalidomide and either standard-dose dexamethasone (40 mg days 1-4, 9-12, and 17-21) or low-dose dexamethasone (40 mg weekly).17 After the first 4 cycles of therapy, patients could discontinue therapy to pursue ASCT or continue therapy on study until progression. Overall, 445 patients were randomized: 222 patients to the low-dose arm and 223 to the high-dose arm. The results have been published previously and demonstrated improved overall survival (OS) for patients receiving low-dose dexamethasone.17 All patients provided written informed consent before entering the trial in accordance with the Declaration of Helsinki. Institutional Review Boards at all participating Eastern Cooperative Oncology Group institutions approved the study.
Baseline bone marrow samples were obtained from consenting patients and shipped to a central Eastern Cooperative Oncology Group laboratory. The marrow aspirates were subjected to a fully automated ROBOSEP cell separation system that uses immunomagnetic technology to positively select for CD 138+ cells, which then were stored in RNAlater for subsequent analysis. The purity of the sorting was confirmed by 3-color immunofluorescent slide-based assessment on the sorted cells. The plasma cell gene expression profiles were analyzed using high-density oligonucleotide microarrays containing probes for 50 000 transcripts and variants, including 14 500 known genes (U133 Plus Version 2.0 array; Affymetrix) as per the manufacturer's recommendations.10,18 All samples were run individually with no pooling. The GEP70 signature was determined as previously described, using log2-transformed raw MAS Version 5.0 signals.14 A cut-off of 0.66 was used for separating the high-risk GEP signature from standard risk. The GEP15 classification was performed as previously described, with the patients in highest quartile for the risk score being considered as high risk.15 FISH was performed on these samples as previously described.10,19 All microarray data are available for viewing in the Gene Expression Omnibus under accession number GSE31504.
Two-sided Fisher exact tests were used to test for differences between categorical variables. Two-sided Wilcoxon rank-sum tests were used to compare continuous variables. Survival analysis was done using the method described by Kaplan and Meier. Differences between survival curves were tested for statistical significance using the 2-sided log-rank test. C-statistic was used to determine the predictive value of the GEP score.20
Results and discussion
Forty-five patients had adequate sample for successful RNA extraction and GEP studies; the baseline characteristics are provided in Table 1. The baseline characteristics and the survival outcomes were similar between the 45 patients and the remaining 400 patients, other than patients in the GEP subset being slightly younger, reflecting lack of selection bias.
Table 1.
Baseline characteristics for the 45 patients, also stratified according to the GEP or FISH risk groups
| Variable | Category | Total (n = 45) | GEP70 (n = 45) |
FISH (n = 44) |
GEP15 (n = 45) |
|||
|---|---|---|---|---|---|---|---|---|
| Standard-risk (n = 38) | High-risk (n = 7) | Standard-risk (n = 34) | High-risk (n = 10) | Standard-risk (n = 35) | High-risk (n = 10) | |||
| Age, y | Median (Q1, Q3) | 61.0 (43, 77) | 59.5 (43, 77) | 61.0 (53, 71) | 60.3 (43, 77) | 61.2 (55, 67) | 61.0 (53.0, 68.0) | 60.0 (56.0, 66.5) |
| Sex | Male | 29 (64) | 27 (71) | 2 (29) | 23 (67) | 6 (60) | 23 (65.7) | 6 (60.0) |
| ISS (n = 43) | Stage I | 13 (30) | 12 (32) | 1 (17) | 11 (33) | 2 (22) | 9 (26.5) | 4 (44.4) |
| Stage II | 21 (49) | 19 (51) | 2 (33) | 16 (49) | 5 (56) | 19 (55.9) | 2 (22.2) | |
| Stage III | 9 (21) | 6 (16) | 3 (50) | 6 (18) | 2 (22) | 6 (17.6) | 3 (33.3) | |
| ECOG PS | 0 | 18 (40) | 17 (45) | 1 (14) | 14 (41) | 4 (40) | 14 (40.0) | 4 (40.0) |
| 1 | 21 (47) | 16 (42) | 5 (71) | 16 (47) | 4 (40) | 16 (45.7) | 5 (50.0) | |
| 2 | 6 (13) | 5 (13) | 1 (14) | 4 (12) | 2 (20) | 5 (14.3) | 1 (10.0) | |
| Bone disease | Yes | 29 (64) | 23 (61) | 6 (86) | 23 (67) | 5 (50) | 23 (65.7) | 6 (60.0) |
| FISH (n = 44)* | Standard-risk | 34 (77) | 32 (84) | 2 (33) | — | — | 30 (88.2) | 4 (40.0) |
| High-risk | 10 (23) | 6 (16) | 4 (67) | — | — | 4 (11.8) | 6 (60.0) | |
| β2-microglobulin, μg/mL | Median (Q1, Q3) | 3.6 (2.5, 4.8) | 3.6 (2.3, 4.5) | 5.1 (2.8, 7.1) | 3.5 (2.3, 4.8) | 3.6 (2.8, 4.3) | 3.6 (2.5, 4.7) | 3.1 (2.3, 5.6) |
| C-reactive protein, mg/dL | Median (Q1, Q3) | 0.5 (0.1, 1.1) | 0.5 (0.1, 0.8) | 2.0 (0.7, 2.9) | 0.5 (0.1, 0.9) | 0.5 (0.4, 1.9) | 0.4 (0.1, 0.9) | 0.6 (0.5, 1.8) |
| Lactate dehydrogenase, U/L | Median (Q1, Q3) | 147.0 (121, 188) | 147.0 (120, 176) | 192.0 (142, 270) | 147.0 (121, 168) | 172.5 (124, 219) | 147.0 (122.0, 180.0) | 147.0 (97.0, 191.0) |
| Serum M spike, g/dL | Median (Q1, Q3) | 3.4 (2.5, 5.1) | 3.3 (2.5, 4.9) | 4.5 (3.2, 5.4) | 3.2 (2.3, 5.2) | 4.8 (3.5, 5.1) | 3.2 (2.2, 5.2) | 4.6 (3.6, 4.9) |
| Albumin, g/dL† | Median (Q1, Q3) | 3.6 (3.0, 4.0) | 3.6 (3.1, 4.0) | 3.3 (2.8, 3.7) | 3.7 (3.2, 4.0) | 3.1 (2.8, 3.3) | 3.7 (3.1, 4.0) | 3.4 (2.7, 3.9) |
| Creatinine, mg/dL | Median (Q1, Q3) | 1.1 (0.9, 1.3) | 1.1 (0.9, 1.3) | 1.3 (1.0, 1.8) | 1.1 (0.9, 1.2) | 1.2 (0.8, 1.4) | 1.1 (0.9, 1.3) | 1.1 (0.9, 1.4) |
| Hemoglobin, g/dL | Median (Q1, Q3) | 10.8 (9.7, 11.9) | 11.0 (9.8, 12.0) | 9.9 (9.3, 10.9) | 11.1 (10.1, 12.2) | 9.9 (9.2, 11.5) | 10.8 (9.6, 11.9) | 10.6 (9.8, 11.9) |
— indicates not applicable.
Statistically significant for comparison with GEP70 (P = .018) and for comparison with GEP15 (P = .004).
Statistically significant for FISH high- versus low-risk (P = .015).
Overall, 7 patients (15.6%) were considered high-risk by GEP70, a proportion similar to that described previously.14,21 In contrast, 10 (22.7%) of 44 patients with FISH data available were considered high-risk by the presence of t(4;14), t(14;16), t(14;20), or del 17p. Six of the FISH high-risk patients and 2 of the standard-risk patients were reclassified into the low- and high-risk categories by GEP70, respectively (Table 1). The distributions of the clinical and laboratory features between the GEP70 high and standard-risk are given in Table 1, with no clear difference between the risk groups. This is probably a reflection of the smaller numbers as previous studies have suggested an enrichment of International Staging System stage 3 patients and higher serum lactate dehydrogenase in the GEP70 high-risk patients.14
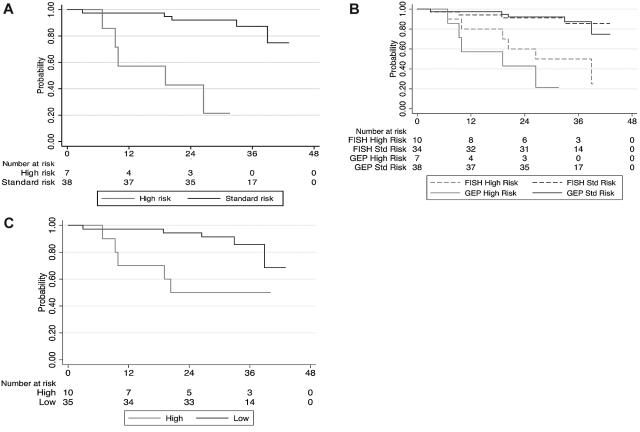
The OS was significantly shorter for the GEP70 high-risk group compared with the remaining patients; hazard ratio (high vs standard) estimate was 14.1 (95% confidence interval [CI], 3.3-60.5, P < .001). The median OS was 19 months for the GEP70 high-risk group and did not reach the median for the standard-risk group (Figure 1A). Furthermore, the predictive ability of the gene score was assessed using the C-statistic. The estimated C-statistic for GEP70 score (when used dichotomously) was 0.74 (95% CI, 0.61-0.88), a value conventionally considered as reflecting a prediction model with good discriminatory ability. In comparison, the C-statistic for FISH-based risk stratification was 0.70 (95% CI, 0.55-0.84). The median OS for the 10 patients considered to be high risk by FISH was 39 months and did not reach median for the rest (Figure 1B); the hazard ratio (high vs standard) was 5.8 (95% CI, 1.62-20.5, P = .007). The median time to progression for the patients with GEP70 high risk was 9 months compared with the rest (23 months; P = .3). In comparison, the median time to progression for the FISH high-risk group was 16 months versus the rest (23 months; P = .4).
Figure 1.
Impact of risk stratification on overall survival. (A) Kaplan-Meier curves comparing the OS from diagnosis among the high- and standard-risk GEP (GEP70) patients receiving initial therapy with lenalidomide and dexamethasone. (B) Kaplan-Meier curves comparing the OS from diagnosis among the high- and standard-risk patients based on GEP (GEP70) and FISH abnormalities, receiving initial therapy with lenalidomide and dexamethasone. (C) Kaplan-Meier curves comparing the OS from diagnosis among the high- and standard-risk patients based on GEP15 score, receiving initial therapy with lenalidomide and dexamethasone.
We then reclassified the patients using the GEP15 risk score; 10 patients were considered high risk. Four each of the FISH high-risk patients and standard-risk patients were reclassified into the low- and high-risk categories by GEP15 (Table 1). The baseline features were not significantly different between the 10 high-risk patients and the remaining 35 patients as shown in Table 1. There was a significant difference in the OS between GEP15-based risk strata; the hazard ratio estimate was 4.3 (95% CI, 1.23-14.5, P = .023). Both high- and low-risk groups did not reach median OS. The estimated C-statistic was 0.70 (95% CI, 0.56-0.85). The median time to progression for the patients with GEP15 high-risk was 16 months compared with the rest (23 months; P = .3).
Previous studies have suggested that GEP signatures may provide a more specific method for identifying patients with high-risk myeloma, further refining the risk stratification obtained through FISH and cytogenetics.14,15 Much of the prognostic value is derived from the proliferation signature contained in the high-risk signature as well as the reflection of chromosome 1 abnormalities.14 Use of bortezomib improves the outcome of patients with t(4;14) or del13 and may also have some benefit for patients with del17p.22,23 In contrast, the group of patients with a high-risk GEP70 signature still continues to have a dismal outcome despite intensive approaches, such as total therapy regimens.21 Hence, GEP is an attractive tool for developing risk-adapted trials for these poor-risk patients. However, the generalizability of the previous studies has been limited by the uniform use of ASCT in those groups.14,15 This study is the first to demonstrate the value of GEP scoring system outside the context of ASCT, in a cohort of patients treated primarily with novel agents. Moreover, we have evaluated 2 different scoring systems demonstrating prognostic value for both, strengthening the rationale for a GEP-based risk stratification approach. Whether one system is better than the other is not the objective of the current study and cannot be answered with the number of patients studied here and has been addressed in the prior studies.15 This is valuable confirmation supporting the use of GEP-based risk stratification approaches and its routine incorporation into clinical trials designed to test different treatment strategies for high-risk and standard-risk myeloma. In this context, this group of patients is more representative of the myeloma population in general because this trial did not specify transplant eligibility, and the group studied here included 18 (40%) patients older than 65 years.17,24 We have previously shown the prognostic value of FISH-based risk stratification in a lenalidomide-treated population.25 The results presented here confirm the value of the GEP scoring and highlight its value in the setting of initial therapy with lenalidomide and dexamethasone, one of the most commonly used treatments for myeloma, as well as its value in older patients. The small number of patients in this study, however, prevents a comparison of the GEP and FISH-based risk stratification, and an assessment of the incremental value of GEP over FISH-based risk stratification or viceversa.
Acknowledgments
This work was coordinated by the Eastern Cooperative Oncology Group (Dr Robert L. Comis, Chair) and supported by the National Cancer Institute, National Institutes of Health, and the Department of Health and Human Services (Public Health Service grants CA23318, CA66636, CA21115, CA13650, and CA93842) and in part by the National Institutes of Health (CA90628, S.K.K.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: S.K.K. and S.V.R. planned the study, analyzed the data, and wrote the manuscript; H.U. and S.J.J. analyzed the data and wrote the manuscript; J.L.H. performed all the GEP studies and reviewed the manuscript; K.J.H., G.J.A., and S.A.V.W. performed sample processing and reviewed the manuscript; and R.F., D.S.S., P.R.G., and N.S.C. were involved in design of the study and critically reviewed the manuscript.
Conflict-of-interest disclosure: S.K.K. is involved with sponsored research from Celgene. The remaining authors declare no competing financial interests.
Correspondence: Shaji K. Kumar, Division of Hematology, Mayo Clinic, 200 First St SW, Rochester, MN 55905; e-mail: kumar.shaji@mayo.edu.
References
- 1.Kumar SK, Mikhael JR, Buadi FK, et al. Management of newly diagnosed symptomatic multiple myeloma: updated mayo stratification of myeloma and risk-adapted therapy (mSMART) consensus guidelines. Mayo Clin Proc. 2009;84(12):1095–1110. doi: 10.4065/mcp.2009.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fonseca R. Many and multiple myeloma(s). Leukemia. 2003;17(10):1943–1944. doi: 10.1038/sj.leu.2403090. [DOI] [PubMed] [Google Scholar]
- 3.Fonseca R, Barlogie B, Bataille R, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64(4):1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 4.Avet-Loiseau H. Role of genetics in prognostication in myeloma. Best Pract Res Clin Haematol. 2007;20(4):625–635. doi: 10.1016/j.beha.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009;23(12):2210–2221. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avet-Loiseau H, Attal M, Moreau P, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myelome. Blood. 2007;109(8):3489–3495. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 7.Stewart AK, Bergsagel PL, Greipp PR, et al. A practical guide to defining high-risk myeloma for clinical trials, patient counseling and choice of therapy. Leukemia. 2007;21(3):529–534. doi: 10.1038/sj.leu.2404516. [DOI] [PubMed] [Google Scholar]
- 8.Magrangeas F, Nasser V, Avet-Loiseau H, et al. Gene expression profiling of multiple myeloma reveals molecular portraits in relation to the pathogenesis of the disease. Blood. 2003;101(12):4998–5006. doi: 10.1182/blood-2002-11-3385. [DOI] [PubMed] [Google Scholar]
- 9.Hose D, Reme T, Hielscher T, et al. Proliferation is a central independent prognostic factor and target for personalized and risk adapted treatment in multiple myeloma. Haematologica. 2011;96(1):87–95. doi: 10.3324/haematol.2010.030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67(7):2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 11.Carrasco DR, Tonon G, Huang Y, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9(4):313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 12.Agnelli L, Bicciato S, Mattioli M, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23(29):7296–7306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 13.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108(6):2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaughnessy JD, Jr, Zhan F, Burington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109(6):2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 15.Decaux O, Lode L, Magrangeas F, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the Intergroupe Francophone du Myelome. J Clin Oncol. 2008;26(29):4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 16.Zhan F, Barlogie B, Mulligan G, Shaughnessy JD, Jr, Bryant B. High-risk myeloma: a gene expression based risk-stratification model for newly diagnosed multiple myeloma treated with high-dose therapy is predictive of outcome in relapsed disease treated with single-agent bortezomib or high-dose dexamethasone. Blood. 2008;111(2):968–969. doi: 10.1182/blood-2007-10-119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rajkumar SV, Jacobus S, Callander NS, et al. Lenalidomide plus high-dose dexamethasone versus lenalidomide plus low-dose dexamethasone as initial therapy for newly diagnosed multiple myeloma: an open-label randomised controlled trial. Lancet Oncol. 2010;11(1):29–37. doi: 10.1016/S1470-2045(09)70284-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12(2):131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101(11):4569–4575. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 20.Pencina MJ, D'Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 21.Nair B, van Rhee F, Shaughnessy JD, Jr, et al. Superior results of Total Therapy 3 (2003-33) in gene expression profiling-defined low-risk multiple myeloma confirmed in subsequent trial 2006-66 with VRD maintenance. Blood. 2010;115(21):4168–4173. doi: 10.1182/blood-2009-11-255620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.San Miguel JF, Schlag R, Khuageva NK, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359(9):906–917. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 23.Jagannath S, Richardson PG, Sonneveld P, et al. Bortezomib appears to overcome the poor prognosis conferred by chromosome 13 deletion in phase 2 and 3 trials. Leukemia. 2007;21(1):151–157. doi: 10.1038/sj.leu.2404442. [DOI] [PubMed] [Google Scholar]
- 24.Kyle RA, Gertz MA, Witzig TE, et al. Review of 1027 patients with newly diagnosed multiple myeloma. Mayo Clin Proc. 2003;78(1):21–33. doi: 10.4065/78.1.21. [DOI] [PubMed] [Google Scholar]
- 25.Kapoor P, Fonseca R, Rajkumar SV, et al. Evidence for cytogenetic and fluorescence in situ hybridization risk stratification of newly diagnosed multiple myeloma in the era of novel therapie. Mayo Clin Proc. 2010;85(6):532–537. doi: 10.4065/mcp.2009.0677. [DOI] [PMC free article] [PubMed] [Google Scholar]