To the editor:
Genome-wide association studies (GWASs) of sickle cell disease (SCD) patients are a promising tool for identifying genetic modifiers of clinically relevant traits.1,2 In such a study, Solovieff et al2 have uncovered a set of SNPs associated with fetal hemoglobin (HbF) concentration, the major modulator of the clinical course of this disease.
After the introduction of the SCD mutations into the Americas by African slaves, African descendants mixed with individuals of Native American and European origin to various extents across the continent. In Latin America, individuals tend to be more admixed than in African-American populations.3,4 Studying different phenotypes, Solovieff et al2 did not find an association between fetal hemoglobin concentration and European ancestry, while Creary et al5 have reported an association between European ancestry and the proportion of erythrocytes containing HbF.
The level of admixture of SCD patients has implications for the design of association studies aimed at identifying new genetic modifiers of SCD clinical manifestations, particularly if these outcomes are associated with ancestry. If both genetic variants and clinical manifestations are associated with ancestry, there are 2 important issues to address. First, an observed association may be spurious if ancestry is not controlled for.6 Second, the admixture mapping strategy7 may be used: Thousands of ancestry-informative markers may be genotyped across the genome, and the local ancestry along chromosomes can be inferred. The genetic modifier alleles are expected to be located in genomic regions with an excess of ancestry of the population associated with the more common clinical outcome.
The risk of spurious association and the power of admixture mapping increase both with the level of admixture. However, estimates of ancestry are rare in SCD patients. Excluding GWASs, association studies between SNPs and SCD subphenotypes seldom control for ancestry.
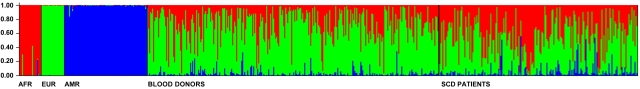
We estimated admixture in 200 patients with SCD and in 291 healthy blood donors; all from the State of Minas Gerais (approximately 20 million inhabitants in South Eastern Brazil). We genotyped 54 SNPs/INDELs validated for admixture studies.3,8,9 The ancestry of the healthy blood donors, who represent the general population reasonably well, was 33.8% African, 57.7% European and 3.5% Amerindian; whereas SCD patients showed 47.3%, 39.7% and 13.0% of African, European and Amerindian ancestry, respectively (estimated following Dupanloup and Bertorelle,10 see supplemental Table 1, available on the Blood Web site; see the supplemental Materials link at the top of the online article). Considering individual admixture (Figure 1), only 11.05% of SCD patients had > 85% African ancestry. Most of the patients (73.37%) had intermediate levels of admixture (15%-85%), and interestingly, 13.8% had predominant European ancestry (> 85%, and for 13.0%, the lower limit of the 90% credibility interval of European ancestry was > 0.60). Therefore, the prevalence of European ancestry is high, and the individual admixture is very heterogeneous in SCD patients from Brazil. Our results suggest that: (1) despite the association of SCD with African ancestry, the label “ethnic/racial” disease seems inappropriate in this population, (2) in association studies with SCD patients, controlling for ancestry is important to avoid spurious association, and (3) Latin American populations of SCD patients are promising targets for admixture mapping of genetic modifiers of ancestry-associated SCD clinical manifestations.
Figure 1.
Individual admixture in healthy blood donors and Sickle Cell Disease patients from Minas Gerais. Admixture was estimated using the method by Pritchard et al implemented in the software Structure.11 Each vertical bar represents an individual and his/her admixture proportions based on the parental populations on the left. Additional methodologic information and results are available as supplemental Methods. Structure was run using the following conditions and parameters: K = 3 (number of parental populations), burn-in period = 100 000, MCMC cycles after burn-in = 100 000, we used a priori information for the individuals from parental populations to assist the clustering (USEPOPINFO = 1), model = ADMIXTURE for the admixed individuals, α parameter was inferred for each population, GENSBACK = 2, MIGRPRIOR = 0.05, allele frequencies was assumed to be correlated.
Authorship
Acknowledgments: The authors thank the personnel from the Hemominas Foundation for their collaboration in sample collection. The study was approved by Ethical Committees from the Universidad Federal de Minas Gerais and the Hemominas Foundation. This research was funded by Brazilian Federal Agencies CNPq and CAPES, the Minas Gerais State Agency FAPEMIG and by the NCI-Fogarty 1R01TW007894-01 GRIP grant.
Contribution: M.C.F.S., M.L.M. and E.T.S. conceived the study; M.C.F.S., Z.M.V., M.L.M. organized samples collection; M.C.F.S., L.W.Z., F.C. and G.B.S.S. performed experiments or analyzed the data; S.D.J.P. provided resources for the INDELS panel; and M.C.F.S. and E.T.S. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Eduardo Tarazona-Santos, PhD, Universidade Federal de Minas Gerais Av Antonio Carlos 6627 Pampulha CP 486 UFMG-ICB- Departamento de Biologia Geral Belo Horizonte, MG 31270-901, Brazil; e-mail: edutars@icb.ufmg.br.
References
- 1.Sebastiani P, Solovieff N, Hartley SW, et al. Genetic modifiers of the severity of sickle cell anemia identified through a genome-wide association study. Am J Hematol. 2010;85(1):29–35. doi: 10.1002/ajh.21572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Solovieff N, Milton JN, Hartley SW, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5′ olfactory receptor gene cluster. Blood. 2010;115(9):1815–1822. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena SD, Di Pietro G, Fuchshuber-Moraes M, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryc K, Velez C, Karafet T, et al. Colloquium paper: genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8954–8961. doi: 10.1073/pnas.0914618107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Creary LE, McKenzie CA, Menzel S, et al. Ethnic differences in F cell levels in Jamaica: a potential tool for identifying new genetic loci controlling fetal haemoglobin. Br J Haematol. 2009;144(9):954–960. doi: 10.1111/j.1365-2141.2008.07532.x. [DOI] [PubMed] [Google Scholar]
- 6.Tarazona-Santos E, Raimondi S, Fuselli S. Controlling the effects of population stratification by admixture in pharmacogenetics. In: Suarez-Kurtz G, editor. Pharmacogenomics in Admixed populations. Austin: Landes Bioscience; 2007. pp. 12–27. [Google Scholar]
- 7.Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annu Rev Genomics Hum Genet. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastos-Rodrigues L, Pimenta JR, Pena SD. The genetic structure of human populations studied through short insertion-deletion polymorphisms. Ann Hum Genet. 2006;70(5):658–665. doi: 10.1111/j.1469-1809.2006.00287.x. [DOI] [PubMed] [Google Scholar]
- 9.Silva MC, Zuccherato LW, Soares-Souza GB, et al. Development of two multiplex mini-sequencing panels of ancestry informative SNPs for studies in Latin Americans: an application to populations of the State of Minas Gerais (Brazil). Genet Mol Res. 2010;9(4):2069–2085. doi: 10.4238/vol9-4gmr911. [DOI] [PubMed] [Google Scholar]
- 10.Dupanloup I, Bertorelle G. Inferring admixture proportions from molecular data: extension to any number of parental populations. Mol Biol Evol. 2001;18(4):672–675. doi: 10.1093/oxfordjournals.molbev.a003847. [DOI] [PubMed] [Google Scholar]
- 11.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]