Abstract
Growing genetic evidence is converging in favor of common pathogenic mechanisms for autism spectrum disorders (ASD), intellectual disability (ID or mental retardation) and schizophrenia (SCZ), three neurodevelopmental disorders affecting cognition and behavior. Copy number variations and deleterious mutations in synaptic organizing proteins including NRXN1 have been associated with these neurodevelopmental disorders, but no such associations have been reported for NRXN2 or NRXN3. From resequencing the three neurexin genes in individuals affected by ASD (n = 142), SCZ (n = 143) or non-syndromic ID (n = 94), we identified a truncating mutation in NRXN2 in a patient with ASD inherited from a father with severe language delay and family history of SCZ. We also identified a de novo truncating mutation in NRXN1 in a patient with SCZ, and other potential pathogenic ASD mutations. These truncating mutations result in proteins that fail to promote synaptic differentiation in neuron coculture and fail to bind either of the established postsynaptic binding partners LRRTM2 or NLGN2 in cell binding assays. Our findings link NRXN2 disruption to the pathogenesis of ASD for the first time and further strengthen the involvement of NRXN1 in SCZ, supporting the notion of a common genetic mechanism in these disorders.
Introduction
Copy number variations (CNVs) affecting the NRXN1 gene have recently been reported in ASD (Kim et al. 2008; Szatmari et al. 2007), SCZ (Guilmatre et al. 2009; Kirov et al. 2008; Rujescu et al. 2009) and non-syndromic ID (NSID) (Ching et al. 2010; Zahir et al. 2008) patients. NRXN1 generates multiple splice variants of the longer α-neurexin and shorter β-neurexin proteins, all of which function in synaptic adhesion, differentiation, and maturation (Craig and Kang 2007; Sudhof 2008). CNVs are genomic deletions and/or duplications of >1 kb to several megabases in size that may affect one or many contiguous genes or only non-coding regions. Deleterious mutations affecting one or a small number of bases in genes disrupted by a CNV (de novo or transmitted from an affected parent) are also likely to predispose to the same disease. However, there have been few systematic resequencing efforts to detect individual base changes in such genes of neurodevelopmental disorders such as ASD, SCZ and ID, and to our knowledge none has been published on sequencing all the neurexin genes in these three diseases. Furthermore, emphasis has been on NRXN1 with no evidence linking NRXN2 or NRXN3 to neurodevelopmental disorders. We therefore evaluated the role of rare or de novo point mutations in the etiology of ASD, SCZ and NSID by screening for mutations in the neurexin gene family (NRXN1, NRXN2, NRXN3) in 379 subjects affected by ASD, SCZ or NSID, and also screened the same exons/splice sites in 285 unaffected control individuals.
Subjects and methods
Subjects
The ASD, SCZ, NSID and control cohorts and ascertainment strategies were previously well described (Gauthier et al. 2010; Hamdan et al. 2009; Piton et al. 2010). Blood samples were obtained from all members of these cohorts as well as from their parents, except for control individuals whose parent DNAs were not available. Samples were collected through informed consent after approval of each of the studies by the respective institutional ethics committees. Genomic DNA was extracted from blood samples using the Puregene DNA kit and according to the manufacturer's protocol (Gentra System, USA). Paternity and maternity of each individual of all families were confirmed using six highly informative unlinked microsatellite markers.
Sequencing
Coding exons and flanking splice junctions of each gene were sequenced on a 3730XL DNA Analyzer System. Primers were designed using the ExonPrimer program (Supplementary Table 1). Exon numbering (Wilson et al. 2003) and cDNA sequence (Moessner et al. 2007) are as previously described. PolyPhred (version 6.11) and Mutation Surveyor (version 3.10, SoftGenetics Inc., PA, USA) were used for mutation detection analysis. All variations were confirmed by re-amplifying the fragment and resequencing the proband and both parents using reverse and forward primers.
Copy number variation and functional analyses
Details of the materials and methods used for the copy number variation and functional analyses are given in the Supplementary material online.
Results
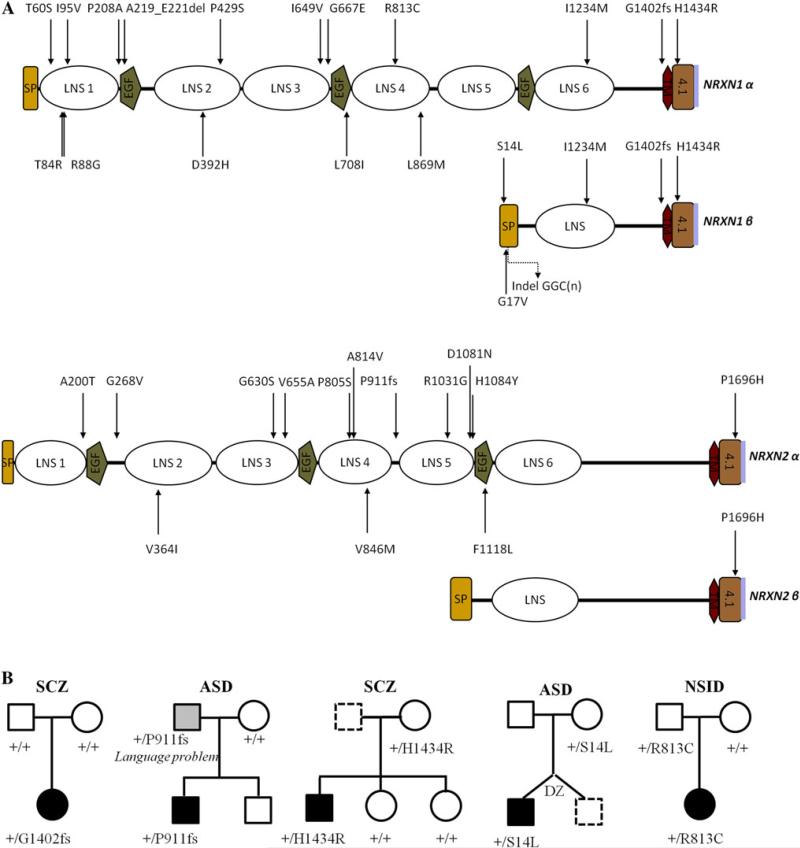
We identified one heterozygous frameshift mutation (c.4205insACGG; NM_004801.4) occurring in exon 22 of NRXN1, affecting both major isoforms (α-NRXN1 and β-NRXN1), in a female diagnosed with disorganized type SCZ. This mutation was absent from blood DNA samples from either of the biological parents, indicating that it was de novo. Neither this mutation nor any other protein-truncating NRXN1 mutations were present in 285 ethnically matched controls. The patient, of Hungarian origin, was born at full-term and without any medical complications. Both her parents are not affected by any psychiatric diseases and there was no family history of mental illness, and developmental milestones were reached within the normal range. This insertion of four nucleotides creates a frameshift at position 1,402 (G1402fs) followed by a premature stop codon, leading to a predicted protein lacking the C-terminal transmembrane and cytoplasmic domains (Fig. 1; Supplementary Table 2).
Fig. 1.
a Schematic structure of the two major isoforms (alpha and beta) of NRXN1 and NRXN2 proteins (LNS 1–6, laminin neurexin sex hormone binding domains; EGF, epidermal growth factor-like domains; SP, signal peptide; TM, transmembrane region; 4.1, band 4.1 N binding region; purple line, PDZ domain binding site). Arrows indicate non-synonymous variants found during the screening of our diseases (top) and control (bottom) cohorts. b Pedigrees of families showing putative causative variations in NRXN1 or NRXN2 that have been tested functionally
In a second case, diagnosed with autism and of European ancestry (Italian/Greek), we discovered a frameshift mutation (c.2733delT; NM_138732.2), located in exon 12 of NRXN2. This patient is a boy born after a normal full-term pregnancy. The Autism Diagnostic Observation Schedule-Generic (ADOS-G) score is 21 (autism cutoff = 12). This mutation was inherited from the father who had a severe language delay (did not speak until 5 years of age). The maternal aunt of the father has a diagnosis of SCZ (DNA not available). This mutation is predicted to produce a protein lacking the last two laminin/neurexin/sex hormone-binding globulin (LNS) domains, one EGF domain, and the C-terminal transmembrane and cytoplasmic domains (Fig. 1).
Seven rare missense variants (observed once in the cases) and two inframe insertions/deletions were identified in NRXN1 in our disease cohorts (Supplementary Table 2). Of these, two (R813C and H1434R) are predicted to affect protein function by at least two prediction programs (SIFT, Polyphen or Panther; Supplementary Table 2). Co-segregation analysis revealed that variant H1434R is not present in the two unaffected sisters. The R813C missense variant, affecting a highly conserved arginine residue within one LNS domain, was inherited from an unaffected parent. We also identified, in a male of European origin (German/French) diagnosed with ASD, a rare heterozygous maternally inherited S14L missense in NRXN1, a variant now identified in four subjects with ASD and not in over 1,201 controls (Feng et al. 2006; Kim et al. 2008; and this study). Ten other disease-cohort specific missense variants, all inherited from an unaffected parent, were identified in NRXN2 and in NRXN3. No co-segregation was possible for any of these missense variants as they were all from sporadic cases. No de novo or other truncating mutation was detected in NRXN3. The ASD and NSID cohorts were studied for CNV analysis using the Affymetrix 5.0 and Affymetrix 6.0 or array CGH platforms, respectively, and no CNVs were detected in any of the neurexin genes in the 142 ASD and 94 NSID tested.
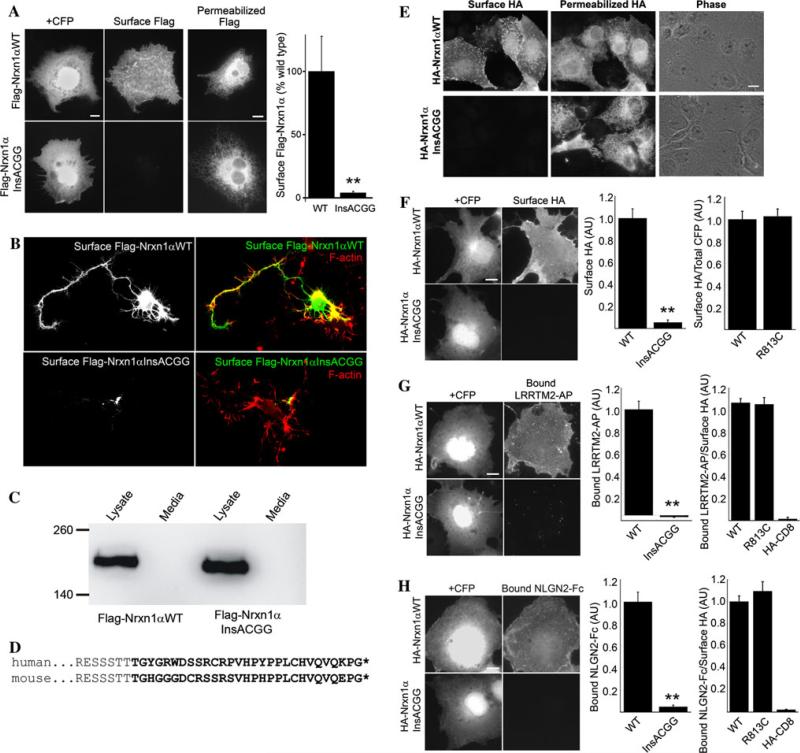
The NRXN1 c.4205insACGG mutant is predicted to generate neurexin1α and 1β truncated just before the transmembrane domain and should thus disrupt cell surface localization. To test this prediction, we expressed the human cDNA for neurexin1α wild type or insACGG mutant with a Flag tag added to the mature extracellular N-terminus. Whereas wild-type Flag-neurexin-1α targeted efficiently to the cell surface, as detected by bright staining of unpermeabilized cells with Flag antibody, little or no insACGG mutant was detected on the cell surface (Fig. 2a). Immunofluorescence of permeabilized cells confirmed similarly high level expression of both constructs indicating that the NRXN1 insACGG mutant is expressed intracellularly, but does not accumulate at the cell surface. We confirmed lack of surface localization of NRXN1 insACGG in transfected primary cultured hippocampal neurons. Whereas wild-type Flag-neurexin-1α was detected on the neuron surface including along the entire axons of relatively immature neurons, we could not detect the insACGG mutant on the neuron surface (Fig. 2b). The truncated protein generated by the insACGG mutation could either be secreted or be trapped in an intracellular domain and never reach the cell surface. To distinguish between these possibilities, we performed Western blots of transfected cell lysates and media. The insACGG mutant Flag-neurexin1α was detected in lysate at similar abundance as wild type, but was not detected in media (Fig. 2c). Thus, the mutant protein does not reach the cell surface.
Fig. 2.
NRXN1 c.4205insACGG results in functional deficiency in surface trafficking and binding to LRRTM2 and NLGN2 in cell binding assays. a COS-7 cells were co-transfected with Flag-neurexin1α human cDNA expression constructs and CFP to mark transfected cells. CFP-positive COS cells cotransfected for wild-type neurexin1α showed variable, often bright, surface Flag immunofluorescence by live cell primary antibody staining. In contrast, little or no surface Flag immunofluorescence was detectable for the insACGG neurexin1α mutant, fluorescence was similar to that of untransfected cells (*p < 0.005 t test, n = 20 CFP-positive cells each group from two experiments). The mutant construct was expressed as shown by bright intracellular Flag staining of permeabilized cells. b Defective surface trafficking of Flag-neurexin1α insACGG mutant was confirmed in primary cultured hippocampal neurons. Whereas neurons expressing wild-type Flag neurexin1α showed bright staining for Flag on the surface including along the axon length, surface Flag staining was not detectable for neurons expressing the insACGG mutant (the small localized signal may represent non-specific permeabilization). Neurons were subsequently stained with phalloidin for F-actin. c Western blot analysis revealed a similar amount of mutant insACGG Flag-neurexin1α as wild type in transfected cell lysate and none in media, indicating that the mutant protein is trapped in the cell. d Insertion of ACGG at the equivalent position in mouse NRXN1 results in aberrant translation of a homologous sequence (bold) and truncation (*) at the equivalent residue. Thus, for further analysis we used a rodent cDNA. e Live cell surface HA immunofluorescence was bright in cells expressing wild-type HA-neurexin1α, but reduced to background level for the insACGG mutant. Permeabilized cell HA immunofluorescence confirmed expression of both constructs. For quantitative assays comparing wild type and insACGG HA-neurexin1α-CFP, we used cotransfected CFP as a marker of expressing cells (since the frameshift resulted in lack of expression of the downstream CFP from the mutant construct, and all other fluorescence channels were needed for antibody staining in the coculture assays); we confirmed that the majority of CFP-positive cells co-labeled for permeabilized HA, equally for wild type (88 ± 2%) and insACGG mutant (86 ± 3%). f Surface expression was strong in CFP-positive cells cotransfected for HA-neurexin1α-CFP wild type but reduced to background in cells cotransfected for the insACGG mutant (**p < 0.0001 t test, n = 20 cells each). The R813C missense variant, and also the H8P and E715K variants (not shown), exhibited surface accumulation similar to wild type (ANOVA p = 0.043, each missense p > 0.05 compared with wild type by Tukey–Kramer post hoc test, n = 40 cells each; also t test p > 0.1 for R813C vs. wild type). g LRRTM2-AP binding was strong in CFP-positive cells cotransfected for HA-neurexin1α-CFP wild type, but reduced to background in cells cotransfected for the insACGG mutant (**p < 0.0001 t test, n = 25 cells each). The R813C missense variant, and also the H8P and E715K variants (not shown), exhibited normal binding of LRRTM2-AP undistinguishable from wild type (ANOVA p > 0.1, n = 30 cells each; also t test p > 0.1 for R813C vs. wild type). HA-CD8 is shown here as a negative control. h NLGN2-Fc binding was strong in CFP-positive cells cotransfected for HA-neurexin1α-CFP wild type but reduced to background in CFP-positive cells cotransfected for the insACGG mutant (**p < 0.0001 t test, n = 15 cells each). The R813C missense variant, and also the H8P and E715K variants (not shown), exhibited normal binding of NLGN2-Fc undistinguishable from wild type (ANOVA p > 0.1, n = 40 cells each; also t test p > 0.1 for R813C vs. wild type). Scale bars, 10 μm
We next tested whether NRXN1 c.4205insACGG disrupts synaptogenic activity. The function of neurexins as synaptic organizing proteins is mediated in part by iso-form-dependent extracellular binding to their postsynaptic partners, the neuroligins (NLGNs) (Craig and Kang 2007; Ichtchenko et al. 1995) and leucine-rich repeat transmembrane neuronal proteins (LRRTMs) (de Wit et al. 2009; Ko et al. 2009; Siddiqui et al. 2010). Neurexin-1a induces clustering of postsynaptic GABAergic proteins via binding NLGN2 (Boucard et al. 2005; Kang et al. 2008), and induces clustering of postsynaptic glutamatergic proteins via binding to LRRTM1 and LRRTM2 (de Wit et al. 2009; Ko et al. 2009; Siddiqui et al. 2010). Such synaptogenic activity is often demonstrated with neuron co-cultures, where neurexin expressed in COS cells induces postsynaptic protein clustering in dendrites of contacting cultured rat hippocampal neurons (Biederer and Scheiffele 2007; Craig et al. 2006). However, our human Flag-neurexin1α construct exhibited poor synaptogenic activity for all markers in comparison with a rodent HA-neurexin-1α-CFP construct in COS cell coculture with rat hippocampal neurons. This difference in activity may have been due to differences in expression vector, presence and position of the tags, and/or species variability (98.6% amino acid identity). To facilitate further analysis, we tested the impact of the NRXN1 insACGG by inserting it at the equivalent position in the rodent HA-neurexin1α-CFP construct predicted to result in aberrant translation of a homologous sequence and truncation at the equivalent amino acid (Fig. 2d). In these experiments we used an isoform lacking the insert at splice site four, important for interaction with LRRTMs (Siddiqui et al. 2010).
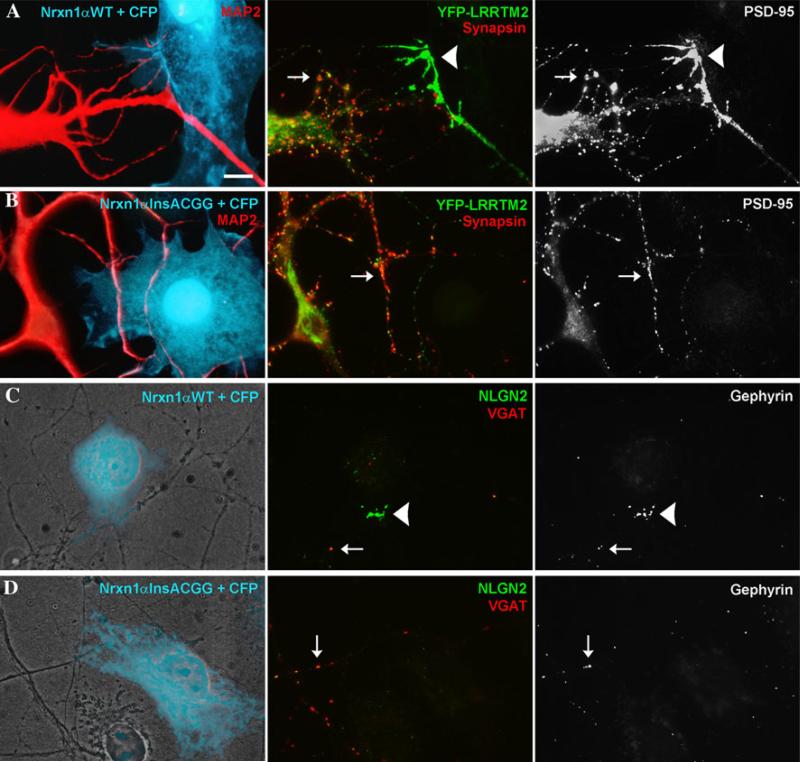
As for the human cDNA, insACGG in rodent HA-neurexin1α-CFP disrupted surface localization in transfected COS cells reducing cell surface immunofluorescence levels to background although intracellular protein was detected (Fig. 2e, f). Neurexin-1α insACGG mutant expressed in COS cells failed to bind partners that bind with high affinity to wild-type neurexin-1α, the ectodomain of LRRTM2 fused to alkaline phosphatase (LRRTM2-AP; Fig. 2g) and the ectodomain of NLGN2 fused to immunoglobulin constant region (NLGN2-Fc; Fig. 2h). Furthermore, HA-neurexin1α insACGG exhibited no synaptogenic activity in the neuron coculture assay. Whereas wild-type HA-neurexin1α induced robust clustering of glutamatergic postsynaptic components YFP-LRRTM2 and endogenous PSD-95 at dendrite contact sites, the insACGG mutant induced no detectable clustering of either glutamatergic component (Fig. 3a, b; Supplementary Figure 1). Similarly, whereas wild-type HA-neurexin1α-CFP induced clear clustering of GABAergic postsynaptic components endogenous NLGN2 and gephyrin at dendrite contact sites, the insACGG mutant induced no detectable clustering of either GABAergic component (Fig. 3c, d; Supplementary Figure 2).
Fig. 3.
NRXN1 c.4205insACGG results in functional deficiency in synaptogenic activity in neuron coculture assays. a, b COS cells were transfected with HA-neurexin1α-CFP vectors and CFP to mark the transfected cells and cocultured with rat hippocampal neurons transfected with YFP-LRRTM2. CFP-positive cells cotransfected for HA-neurexin1α-CFP wild type a induced clustering of YFP-LRRTM2 and endogenous PSD-95 at contact sites with transfected neuron dendrites (arrowheads; 87 of 92 contacts positive). Only synapsin-negative clusters were counted as induced (arrowheads); synapsin-positive clusters were considered to represent endogenous synapses (arrows). In contrast, in spite of equal contact of the CFP-positive cotransfected cells with MAP2-positive dendrites, the insACGG mutant b failed to induce clustering of either glutamatergic postsynaptic component (0 of 75 contacts positive). Only synapsin-positive clusters of YFP-LRRTM2 and PSD-95 (arrows) were seen corresponding to endogenous synapses. c CFP-positive cells cotransfected for HA-neurexin1α-CFP wild type and cocultured with hippocampal neurons induced clustering of endogenous NLGN2 and gephyrin at contact sites with neuron processes (arrowheads; 15 positives scored). d In contrast, the insACGG mutant failed to induce clustering of either GABAergic postsynaptic component (0 positives). Only VGAT-positive clusters of NLGN2 and gephyrin (arrows) were seen corresponding to endogenous synapses. Scale bar, 10 μm
We also functionally tested some NRXN1 missense variants that are predicted to be disruptive to protein function, but essentially did not observe differences from wild type. NRXN1 S14L in the signal peptide of neurexin1β identified in multiple patients with ASD as discussed above was assayed in the context of rodent CFP-neurexin1β. However, cell surface trafficking in COS cells was not significantly different from wild type for S14L nor for the common variant G17V (data not shown, n = 45 cells each). NRXN1 R813C identified here in a patient with NSID (Supplementary Table 2), and E715K and R8P (H8P in rat) reported by Yan et al. (Yan et al. 2008) in individuals with ASD were assayed quantitatively in the context of the rodent HA-neurexin1α-CFP homolog. HA-neurexin1α-CFP R813C, E715K, and H8P did not differ from wild type with respect to surface trafficking in COS cells, relative binding affinity to LRRTM2-AP or NLGN2-Fc, or clustering of YFP-LRRTM2 in neuron coculture (Fig. 2e–g and data not shown). We also qualitatively assessed NRXN1 H1434R identified here in a patient with SCZ (Supplementary Table 2), and observed efficient surface trafficking of human Flag-neurexin1α H1434R in COS cells, similar to wild type (data not shown).
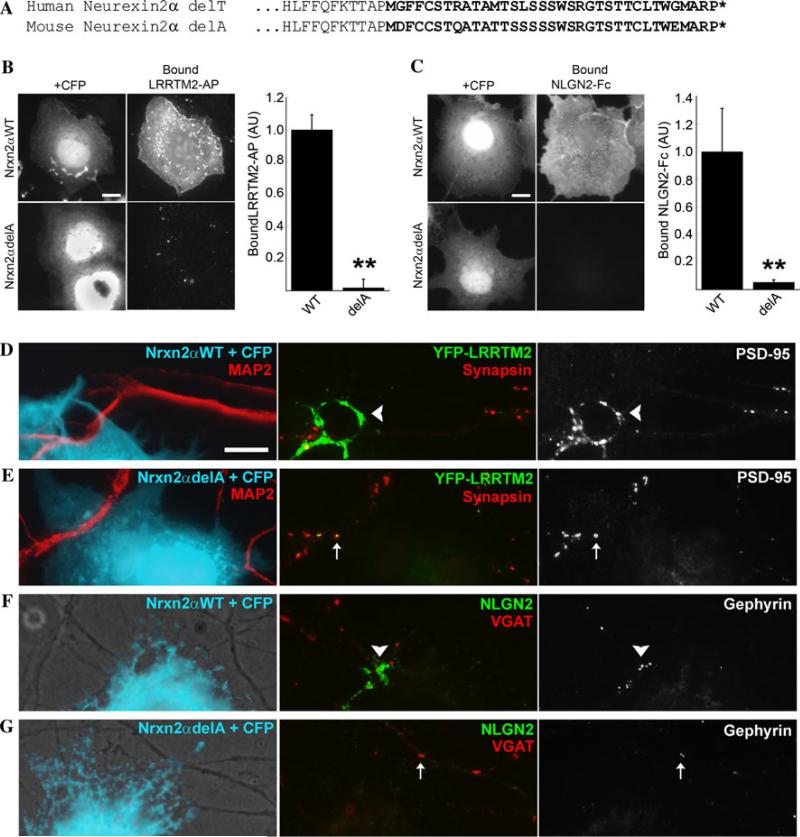
To assess functionally the NRXN2 frameshift c.2733delT/P911fs, we deleted the corresponding residue (delA) in the mouse neurexin2α cDNA, predicting aberrant translation of a homologous sequence and truncation at the equivalent residue (Fig. 4a). This truncation site is at the end of the fourth LNS domain and thus does not affect neurexin2β, but removes nearly half of the neurexin2α protein, including the binding sites for NLGNs and LRRTMs in LNS6, the transmembrane domain, and the intracellular domain. Indeed, unlike the high affinity binding of wild-type neurexin2α-CFP, the delA mutant expressed in COS cells was unable to bind either LRRTM2-AP or NLGN2-Fc (Fig. 4b, c). Furthermore, the neurexin2α delA mutant exhibited a loss of synaptogenic activity in the neuron coculture assay. Whereas wild-type neurexin2α-CFP induced robust clustering of glutamatergic components YFP-LRRTM2 and PSD-95, and GABAergic components NLGN2 and gephyrin at dendrite contact sites, the delA mutant essentially failed to cluster these postsynaptic components (Fig. 4d–g; Supplementary Figures 3–5). In summary, as expected, the frameshifts identified in NRXN1 and NRXN2 showed complete loss of function in binding to recombinant LRRM2 or NLGN2 in cell-based assays and complete loss of synaptogenic activity in neuron coculture.
Fig. 4.
NRXN2 c.2733delT (delA in mouse) results in functional deficiency in binding to LRRTM2 and NLGN2 in cell binding assays, and deficiency in synaptogenic activity in neuron coculture assays. a Compared with delT in human, delA at the equivalent position in mouse NRXN2 results in aberrant translation of a homologous sequence (bold) and truncation (*) at the equivalent residue. Thus for further analysis we used a mouse cDNA. b LRRTM2-AP binding was strong in CFP-positive cells cotransfected for neurexin2α-CFP wild type, but reduced to background in cells cotransfected for the delA mutant (**p < 0.0001 t test, n = 15 cells each). c NLGN2-Fc binding was strong in CFP-positive cells cotransfected for neurexin2α-CFP wild type, but reduced to background in cells cotransfected for the delA mutant (**p < 0.0001 t test, n = 15 cells each). d, e COS cells were transfected with neurexin2α-CFP vectors and CFP to mark the transfected cells and cocultured with rat hippocampal neurons transfected with YFP-LRRTM2. CFP-positive cells cotransfected for neurexin2α-CFP wild type d induced clustering of YFP-LRRTM2 and endogenous PSD-95 at contact sites with transfected neuron dendrites (arrowheads; 65 of 71 contacts positive). In contrast, despite equal contact with MAP2-positive dendrites, the delA mutant e essentially failed to induce clustering of either glutamatergic postsynaptic component (2 of 88 contacts positive). Only synapsin-positive clusters of YFP-LRRTM2 and PSD-95 (arrows) were seen corresponding to endogenous synapses. f, g CFP-positive cells cotransfected for neurexin2α-CFP wild type f and cocultured with hippocampal neurons induced clustering of endogenous NLGN2 and gephyrin at contact sites with neuron processes (arrowheads; 13 positives scored). In contrast, the delA mutant g failed to induce clustering of either GABAergic postsynaptic component (0 positive). Only VGAT-positive clusters of NLGN2 and gephyrin (arrows) were seen corresponding to endogenous synapses. Scale bars, 10 μm
Discussion
We report here a novel de novo truncating mutation in NRXN1 in a patient with SCZ, and, for the first time, a truncating mutation in NRXN2 in a patient with ASD. Both truncations severely disrupt the major characterized functions of neurexins as synaptic organizing proteins mediating glutamatergic and GABAergic synaptic differentiation. Upon their initial discovery (Ushkaryov et al. 1992), neurexins were appreciated to be neuron-specific and broadly expressed in brain as thousands of potential splice variants (Missler and Sudhof 1998). Recent studies indicate that neurexins embedded in the presynaptic membrane interact in an isoform-specific code with multiple key postsynaptic surface proteins: NLGNs (Craig and Kang 2007; Sudhof 2008), LRRTMs (de Wit et al. 2009; Ko et al. 2009; Siddiqui et al. 2010), and Cbln-GluRδ (Uemura et al. 2010). Thus, neurexins are emerging as a very important family of synaptic organizing proteins.
Our findings emphasize the importance of neurexin molecular pathways in the pathogenesis of neurodevelopmental disorders ASD, SCZ, and ID. Other genes involved in this pathway are good candidates for these diseases. Indeed, mutations in multiple neuroligins NLGN1, NLGN3, and NLGN4 have been previously reported in ASD and ID (Glessner et al. 2009; Jamain et al. 2003; Laumonnier et al. 2004; Marshall et al. 2008; Sudhof 2008; Zhang et al. 2009), and variants in LRRTM1 and LRRTM4 in SCZ and ASD (Francks et al. 2007; Pinto et al. 2010). As is the case for NRXN truncating variants in our study, while these variants occur in only a small proportion of ASD, SCZ, and ID patients, they often segregate with disease phenotype and often alter protein function, and thus are thought to be causally involved.
Both the NRXN1 insertion and NRXN2 deletion reported herein lead to a protein that does not anchor into the membrane due to the lack of the C-terminal transmembrane and cytoplasmic domains. The NRXN2 deletion also removes LNS6, the binding site for NLGNs and LRRTMs (Boucard et al. 2005; Kang et al. 2008; Ko et al. 2009; Siddiqui et al. 2010). In the NRXN1 insertion mutant, LNS6 is expressed but non-functional due to the lack of membrane anchor and lack of cell surface trafficking. Thus, neither the NRXN1 insertion nor the NRXN2 deletion variant exhibited synaptogenic function in neuron coculture. In the context of an individual with one wild-type allele, these frameshift mutants may be similar to a null allele, or potentially more deleterious. Particularly for the NRXN1 insACGG mutant, truncated protein binding to NLGNs and LRRTMs might inhibit binding of these ligands to the wild-type neurexin. Such nuances are difficult to model in our culture systems. The functional assays here suggest the mutants act as null alleles at least, with deficiencies resulting from dosage effects, but it may be important to consider potential additional interference of the truncated protein with the function of remaining wild-type protein.
The missense variants assessed here did not detectably alter surface trafficking in COS cells or binding to ligands LRRTM2 or NLGN2. These missense variants might affect neuron-specific trafficking or interactions with other neurexin partners. α-neurexins also bind dystroglycan (Sugita et al. 2001) and must have key interacting partners as yet unidentified to mediate their coupling to presynaptic calcium channels (Missler et al. 2003). Interestingly, H1434R is located in the region with which neurexins interact with band 4.1 N, an association thought to be important for linking neurexins to presynaptic F-actin and stabilizing its interaction with CASK (Biederer and Sudhof 2001).
Some mutations in ASD patients have been modeled in genetically targeted mice. For example, NLGN4 knockout (KO) mice, mimicking X-linked NLGN4 truncations, exhibit selective deficits in social interaction and ultrasonic vocalization (Jamain et al. 2008), phenocopying some aspects of ASD. The neurexin-2α KO heterozygous mice with unaffected 2β (Missler et al. 2003) might be a reasonable model for the NRXN2 delT mutation reported here in ASD. Newborn neurexin-2α homozygous KOs show reduced spontaneous synaptic transmission in brainstem, a milder reduction than the other α-neurexin KOs (Dudanova et al. 2007; Missler et al. 2003), but extensive analysis including behavior has not been reported. While the ideal model for the NRXN1 insACGG mutant in SCZ might be a heterozygous neurexin-1α and 1β KO, interestingly the neurexin-1α KO with unaffected 1β shows a decrease in prepulse inhibition (Etherton et al. 2009). Prepulse inhibition, a measure of sensorimotor gating, correlates quantitatively with symptom severity in SCZ patients and has reasonable validity in animal models (Braff and Geyer 1990; Swerdlow et al. 1994). These single gene mutants generally alter synapse composition and function, but not synapse number. Even the phenotypically severe perinatal lethal triple α-neurexin KO mice have normal numbers of excitatory synapses (Dudanova et al. 2007; Missler et al. 2003) consistent with neuropathological observations from SCZ as well as ASD brains that neurons and synapses are still intact (Belmonte et al. 2004; Harrison 1999). Our findings here, particularly implicating NRXN2 for the first time in ASD emphasize the importance for future studies to define the precise contributions of specific neurexin iso-forms and pathways to the development of synapses important for cognition and language processing.
Finally, deleterious chromosomal rearrangements, CNVs, and point mutations should all be considered as potential genetic cause of neurodevelopmental disorders. As shown here, genes present in CNVs or any other chromosomal rearrangements reported in patients, but not in healthy individuals should be considered as strong candidate genes and warrant sequencing in large cohorts. Taken together, our results suggest that the neurexin family is very important in the etiology of neurodevelopmental disorders.
Supplementary Material
Acknowledgments
We thank all families who kindly participated in this study. We are also thankful to all other S2D members. This work was supported by Genome Canada and Génome Québec, GRSNC of the Fonds de Recherche en Santé du Québec, and received co-funding from Université de Montréal for the ‘Synapse to Disease’ (S2D) project as well as funding from the Canadian Foundation for Innovation. The NSID cohort was recruited with funding from the Réseau de Médecine Génétique Appliquée (RMGA). Functional analysis was supported by Canadian Institutes of Health Research grant MOP84241 and National Institutes of Health grant MH70860 (to A.M.C.). G.A.R. holds the Canada Research Chair in Genetics of the Nervous System; A.M.C. holds the Canada Research Chair in Neurobiology; P.D. holds the Canada Research Chair in Neuroscience; T.J.S. holds a Michael Smith Foundation for Health Research Postdoctoral Fellowship. A portion of the Schizophrenia cohort was collected through the Collaborative Network for Family Study in Psychiatry (“Réseau d'étude familiale en Psychiatry”, REFAPSY), supported by the Fondation Pierre Deniker. We also acknowledge the efforts of the members and Génome Québec Innovation Centre Sequencing (Pierre Lepage, Sébastien Brunet and Hao Fan Yam) and Bioinformatic (Louis Létourneau and Louis Dumond Joseph) groups, and thank Xiling Zhou for preparation of neuron cultures.
Footnotes
J. Gauthier, T. J. Siddiqui contributed equally to this work.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-011-0975-z) contains supplementary material, which is available to authorized users.
Conflict of interest None.
Contributor Information
Julie Gauthier, Centre of Excellence in Neuromics of Université de Montréal, Centre Hospitalier de l’Université de Montréal, Montreal, Canada; Department of Medicine, Université of Montréal, Montreal, Canada.
Tabrez J. Siddiqui, Department of Psychiatry, Brain Research Centre, University of British Columbia, 2211 Wesbrook Mall, Vancouver, BC V6T 2B5, Canada
Peng Huashan, McGill University Department of Neurology, Centre for Research in Neuroscience, Montreal General Hospital, McGill University Health Centre, 1650 Cedar Avenue, Montreal, QC H3G 1A4, Canada.
Daisaku Yokomaku, Department of Psychiatry, Brain Research Centre, University of British Columbia, 2211 Wesbrook Mall, Vancouver, BC V6T 2B5, Canada.
Fadi F. Hamdan, Centre of Excellence in Neuromics of Université de Montréal, CHU Sainte-Justine Research Center, 3175, Côte Ste-Catherine, 6th Floor, Block 7, Montreal, QC H3T 1C5, Canada
Nathalie Champagne, Department of Pathology and Cell Biology, Le Groupe de Recherche sur le Système Nerveux Central, Université de Montréal, Montreal, Canada.
Mathieu Lapointe, Department of Pathology and Cell Biology, Le Groupe de Recherche sur le Système Nerveux Central, Université de Montréal, Montreal, Canada.
Dan Spiegelman, Centre of Excellence in Neuromics of Université de Montréal, Centre Hospitalier de l’Université de Montréal, Montreal, Canada; Department of Medicine, Université of Montréal, Montreal, Canada.
Anne Noreau, Centre of Excellence in Neuromics of Université de Montréal, Centre Hospitalier de l’Université de Montréal, Montreal, Canada; Department of Medicine, Université of Montréal, Montreal, Canada.
Ronald G. Lafrenière, Centre of Excellence in Neuromics of Université de Montréal, Centre Hospitalier de l’Université de Montréal, Montreal, Canada Department of Medicine, Université of Montréal, Montreal, Canada.
Ferid Fathalli, Department of Psychiatry, Douglas Mental Health University Institute, McGill University, Pavilion Frank B. Common Rm. F-2142, 6875 LaSalle Blvd, Verdun, Montreal, QC H4H 1R3, Canada.
Ridha Joober, Department of Psychiatry, Douglas Mental Health University Institute, McGill University, Pavilion Frank B. Common Rm. F-2142, 6875 LaSalle Blvd, Verdun, Montreal, QC H4H 1R3, Canada.
Marie-Odile Krebs, Laboratory of Pathophysiology of Psychiatric Diseases, University Paris Descartes, U894, Sainte-Anne Hospital, 7 rue Cabanis, 75014 Paris, France.
Lynn E. DeLisi, VA Boston Healthcare Service, Harvard Medical School, Brockton, MA, USA
Laurent Mottron, Pervasive Developmental Disorders Specialized Clinic, Rivière-des-Prairies Hospital, University of Montreal, Montreal, QC H1E 1A4, Canada.
Éric Fombonne, Department of Psychiatry, Montreal Children's Hospital, Montreal, QC H3Z 1P2, Canada.
Jacques L. Michaud, Centre of Excellence in Neuromics of Université de Montréal, CHU Sainte-Justine Research Center, 3175, Côte Ste-Catherine, 6th Floor, Block 7, Montreal, QC H3T 1C5, Canada
Pierre Drapeau, Department of Pathology and Cell Biology, Le Groupe de Recherche sur le Système Nerveux Central, Université de Montréal, Montreal, Canada.
Salvatore Carbonetto, McGill University Department of Neurology, Centre for Research in Neuroscience, Montreal General Hospital, McGill University Health Centre, 1650 Cedar Avenue, Montreal, QC H3G 1A4, Canada.
Ann Marie Craig, Department of Psychiatry, Brain Research Centre, University of British Columbia, 2211 Wesbrook Mall, Vancouver, BC V6T 2B5, Canada.
Guy A. Rouleau, Centre of Excellence in Neuromics of Université de Montréal, Centre Hospitalier de l’Université de Montréal, Montreal, Canada Department of Medicine, Université of Montréal, Montreal, Canada; CHUM Research Centre, 2099, Alexandre De-Seve Street, Room Y-3633, Montreal, QC H2L 2W5, Canada.
References
- Belmonte MK, Cook EH, Jr, Anderson GM, Rubenstein JL, Greenough WT, Beckel-Mitchener A, Courchesne E, Boulanger LM, Powell SB, Levitt PR, Perry EK, Jiang YH, DeLorey TM, Tierney E. Autism as a disorder of neural information processing: directions for research and targets for therapy. Mol Psychiatry. 2004;9:646–663. doi: 10.1038/sj.mp.4001499. [DOI] [PubMed] [Google Scholar]
- Biederer T, Scheiffele P. Mixed-culture assays for analyzing neuronal synapse formation. Nat Protoc. 2007;2:670–676. doi: 10.1038/nprot.2007.92. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sudhof TC. CASK and protein 4.1 support F-actin nucleation on neurexins. J Biol Chem. 2001;276:47869–47876. doi: 10.1074/jbc.M105287200. [DOI] [PubMed] [Google Scholar]
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Sudhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to alpha- and beta-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Braff DL, Geyer MA. Sensorimotor gating and schizophrenia. Human and animal model studies. Arch Gen Psychiatry. 1990;47:181–188. doi: 10.1001/archpsyc.1990.01810140081011. [DOI] [PubMed] [Google Scholar]
- Ching MS, Shen Y, Tan WH, Jeste SS, Morrow EM, Chen X, Mukaddes NM, Yoo SY, Hanson E, Hundley R, Austin C, Becker RE, Berry GT, Driscoll K, Engle EC, Friedman S, Gusella JF, Hisama FM, Irons MB, Lafiosca T, LeClair E, Miller DT, Neessen M, Picker JD, Rappaport L, Rooney CM, Sarco DP, Stoler JM, Walsh CA, Wolff RR, Zhang T, Nasir RH, Wu BL. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:937–947. doi: 10.1002/ajmg.b.31063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Kang Y. Neurexin-neuroligin signaling in synapse development. Curr Opin Neurobiol. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AM, Graf ER, Linhoff MW. How to build a central synapse: clues from cell culture. Trends Neurosci. 2006;29:8–20. doi: 10.1016/j.tins.2005.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Sylwestrak E, O'Sullivan ML, Otto S, Tiglio K, Savas JN, Yates JR, 3rd, Comoletti D, Taylor P, Ghosh A. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64:799–806. doi: 10.1016/j.neuron.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudanova I, Tabuchi K, Rohlmann A, Sudhof TC, Missler M. Deletion of alpha-neurexins does not cause a major impairment of axonal pathfinding or synapse formation. J Comp Neurol. 2007;502:261–274. doi: 10.1002/cne.21305. [DOI] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proc Natl Acad Sci USA. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Francks C, Maegawa S, Lauren J, Abrahams BS, Velayos-Baeza A, Medland SE, Colella S, Groszer M, McAuley EZ, Caffrey TM, Timmusk T, Pruunsild P, Koppel I, Lind PA, Matsumoto-Itaba N, Nicod J, Xiong L, Joober R, Enard W, Krinsky B, Nanba E, Richardson AJ, Riley BP, Martin NG, Strittmatter SM, Moller HJ, Rujescu D, St Clair D, Muglia P, Roos JL, Fisher SE, Wade-Martins R, Rouleau GA, Stein JF, Karayiorgou M, Geschwind DH, Ragoussis J, Kendler KS, Airaksinen MS, Oshimura M, DeLisi LE, Monaco AP. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol Psychiatry. 2007;12:1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, Champagne N, Lafreniere RG, Xiong L, Spiegelman D, Brustein E, Lapointe M, Peng H, Cote M, Noreau A, Hamdan FF, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Joober R, Fathalli F, Mouaffak F, Haghighi AP, Neri C, Dube MP, Samuels ME, Marineau C, Stone EA, Awadalla P, Barker PA, Carbonetto S, Drapeau P, Rouleau GA. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proc Natl Acad Sci USA. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, Wood S, Zhang H, Estes A, Brune CW, Bradfield JP, Imielinski M, Frackelton EC, Reichert J, Crawford EL, Munson J, Sleiman PM, Chiavacci R, Annaiah K, Thomas K, Hou C, Glaberson W, Flory J, Otieno F, Garris M, Soorya L, Klei L, Piven J, Meyer KJ, Anagnostou E, Sakurai T, Game RM, Rudd DS, Zurawiecki D, McDougle CJ, Davis LK, Miller J, Posey DJ, Michaels S, Kolevzon A, Silverman JM, Bernier R, Levy SE, Schultz RT, Dawson G, Owley T, McMahon WM, Wassink TH, Sweeney JA, Nurnberger JI, Coon H, Sutcliffe JS, Minshew NJ, Grant SF, Bucan M, Cook EH, Buxbaum JD, Devlin B, Schellenberg GD, Hakonarson H. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Dubourg C, Mosca AL, Legallic S, Goldenberg A, Drouin-Garraud V, Layet V, Rosier A, Briault S, Bonnet-Brilhault F, Laumonnier F, Odent S, Le Vacon G, Joly-Helas G, David V, Bendavid C, Pinoit JM, Henry C, Impallomeni C, Germano E, Tortorella G, Di Rosa G, Barthelemy C, Andres C, Faivre L, Frebourg T, Saugier Veber P, Campion D. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66:947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Cote M, Perreault-Linck E, Carmant L, D'Anjou G, Fombonne E, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Mouaffak F, Joober R, Mottron L, Drapeau P, Marineau C, Lafreniere RG, Lacaille JC, Rouleau GA, Michaud JL. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ. The neuropathology of schizophrenia. A critical review of the data and their interpretation. Brain. 1999;122(Pt 4):593–624. doi: 10.1093/brain/122.4.593. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, Ramanantsoa N, Gallego J, Ronnenberg A, Winter D, Frahm J, Fischer J, Bourgeron T, Ehrenreich H, Brose N. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci USA. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Zhang X, Dobie F, Wu H, Craig AM. Induction of GABAergic postsynaptic differentiation by alpha-neurexins. J Biol Chem. 2008;283:2323–2334. doi: 10.1074/jbc.M703957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, Descartes M, Holt L, Braddock S, Troxell R, Kaplan L, Volkmar F, Klin A, Tsatsanis K, Harris DJ, Noens I, Pauls DL, Daly MJ, MacDonald ME, Morton CC, Quade BJ, Gusella JF. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Gumus D, Chen W, Norton N, Georgieva L, Sari M, O'Donovan MC, Erdogan F, Owen MJ, Ropers HH, Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64:791–798. doi: 10.1016/j.neuron.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns JP, Ropers HH, Hamel BC, Andres C, Barthelemy C, Moraine C, Briault S. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, Thiruvahindrapduram B, Fiebig A, Schreiber S, Friedman J, Ketelaars CE, Vos YJ, Ficicioglu C, Kirkpatrick S, Nicolson R, Sloman L, Summers A, Gibbons CA, Teebi A, Chitayat D, Weksberg R, Thompson A, Vardy C, Crosbie V, Luscombe S, Baatjes R, Zwaigenbaum L, Roberts W, Fernandez B, Szatmari P, Scherer SW. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- Missler M, Zhang W, Rohlmann A, Kattenstroth G, Hammer RE, Gottmann K, Sudhof TC. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature. 2003;424:939–948. doi: 10.1038/nature01755. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, Zwaigenbaum L, Fernandez B, Roberts W, Szatmari P, Scherer SW. Contribution of SHANK3 mutations to autism spectrum disorder. Am J Hum Genet. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piton A, Gauthier J, Hamdan FF, Lafreniere RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, Spiegelman D, Kuku F, Duguay J, Destroismaisons L, Jolivet P, Cote M, Lachapelle K, Diallo O, Raymond A, Marineau C, Champagne N, Xiong L, Gaspar C, Riviere JB, Tarabeux J, Cossette P, Krebs MO, Rapoport JL, Addington A, Delisi LE, Mottron L, Joober R, Fombonne E, Drapeau P, Rouleau GA. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2010 doi: 10.1038/mp.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, Toulopoulou T, Picchioni M, Vassos E, Ettinger U, Bramon E, Murray R, Ruggeri M, Tosato S, Bonetto C, Steinberg S, Sigurdsson E, Sigmundsson T, Petursson H, Gylfason A, Olason PI, Hardarsson G, Jonsdottir GA, Gustafsson O, Fossdal R, Giegling I, Moller HJ, Hartmann AM, Hoffmann P, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Djurovic S, Melle I, Andreassen OA, Hansen T, Werge T, Kiemeney LA, Franke B, Veltman J, Buizer-Voskamp JE, Sabatti C, Ophoff RA, Rietschel M, Nothen MM, Stefansson K, Peltonen L, St Clair D, Stefansson H, Collier DA. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui TJ, Pancaroglu R, Kang Y, Rooyakkers A, Craig AM. LRRTMs and neuroligins bind neurexins with a differential code to cooperate in glutamate synapse development. J Neurosci. 2010;30:7495–7506. doi: 10.1523/JNEUROSCI.0470-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S, Saito F, Tang J, Satz J, Campbell K, Sudhof TC. A stoichiometric complex of neurexins and dystroglycan in brain. J Cell Biol. 2001;154:435–445. doi: 10.1083/jcb.200105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow NR, Braff DL, Taaid N, Geyer MA. Assessing the validity of an animal model of deficient sensorimotor gating in schizophrenic patients. Arch Gen Psychiatry. 1994;51:139–154. doi: 10.1001/archpsyc.1994.03950020063007. [DOI] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, Vincent JB, Skaug JL, Thompson AP, Senman L, Feuk L, Qian C, Bryson SE, Jones MB, Marshall CR, Scherer SW, Vieland VJ, Bartlett C, Mangin LV, Goedken R, Segre A, Pericak-Vance MA, Cuccaro ML, Gilbert JR, Wright HH, Abramson RK, Betancur C, Bourgeron T, Gillberg C, Leboyer M, Buxbaum JD, Davis KL, Hollander E, Silverman JM, Hallmayer J, Lotspeich L, Sutcliffe JS, Haines JL, Folstein SE, Piven J, Wassink TH, Sheffield V, Geschwind DH, Bucan M, Brown WT, Cantor RM, Constantino JN, Gilliam TC, Herbert M, Lajonchere C, Ledbetter DH, Lese-Martin C, Miller J, Nelson S, Samango-Sprouse CA, Spence S, State M, Tanzi RE, Coon H, Dawson G, Devlin B, Estes A, Flodman P, Klei L, McMahon WM, Minshew N, Munson J, Korvatska E, Rodier PM, Schellenberg GD, Smith M, Spence MA, Stodgell C, Tepper PG, Wijsman EM, Yu CE, Roge B, Mantoulan C, Wittemeyer K, Poustka A, Felder B, Klauck SM, Schuster C, Poustka F, Bolte S, Feineis-Matthews S, Herbrecht E, Schmotzer G, Tsiantis J, Papanikolaou K, Maestrini E, Bacchelli E, Blasi F, Carone S, Toma C, Van Engeland H, de Jonge M, Kemner C, Koop F, Langemeijer M, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Sudhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Wilson HL, Wong AC, Shaw SR, Tse WY, Stapleton GA, Phelan MC, Hu S, Marshall J, McDermid HE. Molecular characterisation of the 22q13 deletion syndrome supports the role of haploinsufficiency of SHANK3/PROSAP2 in the major neurological symptoms. J Med Genet. 2003;40:575–584. doi: 10.1136/jmg.40.8.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Noltner K, Feng J, Li W, Schroer R, Skinner C, Zeng W, Schwartz CE, Sommer SS. Neurexin 1alpha structural variants associated with autism. Neurosci Lett. 2008;438:368–370. doi: 10.1016/j.neulet.2008.04.074. [DOI] [PubMed] [Google Scholar]
- Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, Marra MA, Friedman JM. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- Zhang C, Milunsky JM, Newton S, Ko J, Zhao G, Maher TA, Tager-Flusberg H, Bolliger MF, Carter AS, Boucard AA, Powell CM, Sudhof TC. A neuroligin-4 missense mutation associated with autism impairs neuroligin-4 folding and endoplasmic reticulum export. J Neurosci. 2009;29:10843–10854. doi: 10.1523/JNEUROSCI.1248-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.