Abstract
PLD catalyzes the conversion of the membrane phospholipid phosphatidylcholine to choline and phosphatidic acid (PA). PLD's mission in the cell is two-fold: phospholipid turnover with maintenance of the structural integrity of cellular/intracellular membranes and cell signaling through PA and its metabolites. Precisely, through its product of the reaction, PA, PLD has been implicated in a variety of physiological cellular functions, such as intracellular protein trafficking, cytoskeletal dynamics, chemotaxis of leukocytes and cell proliferation. The catalytic (HKD) and regulatory (PH and PX) domains were studied in detail in the PLD1 isoform, but PLD2 was traditionally studied in lesser detail and much less was known about its regulation. Our laboratory has been focusing on the study of PLD2 regulation in mammalian cells. Over the past few years, we have reported, in regards to the catalytic action of PLD, that PA is a chemoattractant agent that binds to and signals inside the cell through the ribosomal S6 kinases (S6K). Regarding the regulatory domains of PLD2, we have reported the discovery of the PLD2 interaction with Grb2 via Y169 in the PX domain, and further association to Sos, which results in an increase of de novo DNA synthesis and an interaction (also with Grb2) via the adjacent residue Y179, leading to the regulation of cell ruffling, chemotaxis and phagocytosis of leukocytes. We also review the complex regulation by tyrosine phosphorylation by epidermal growth factor receptor (EGF-R), Janus Kinase 3 (JAK3) and Src and the role of phosphatases. Recently, there is evidence supporting a new level of regulation of PLD2 at the PH domain, by the discovery of CRIB domains and a Rac2-PLD2 interaction that leads to a dual (positive and negative) effect on its enzymatic activity. Lastly, we review the surprising finding of PLD2 acting as a GEF. A phospholipase such as PLD that exists already in the cell membrane that acts directly on Rac allows a quick response of the cell without intermediary signaling molecules. This provides only the latest level of PLD2 regulation in a field that promises newer and exciting advances in the next few years.
1. Introduction
PLD catalyzes the conversion of the membrane phospholipid phosphatidylcholine to choline and phosphatidic acid (PA) [1]. PLD's mission in the cell is two-fold: phospholipid turnover with maintenance of the structural integrity of cellular/intracellular membranes [2] and cell signaling through PA and its metabolites (FIGURE 1). For the latter, PLD has been implicated in a variety of physiological cellular functions, such as intracellular protein trafficking, cytoskeletal dynamics, chemotaxis of leukocytes and cell proliferation. Since the late 1980's, PLD activity was found in many diverse mammalian cells and tissues. PLD enzymatic activity was enhanced by a number of cell agonists, such as growth factors like EGF, PDGF, (as well as PMA) and by stimulants of G-protein associated receptors. The study of PLD involves the product of its enzymatic reaction (PA), and its role in signal transduction in the last two decades has been the subject of excellent reviews [3-13]. Regulation of PLD was recognized as involving small GTPases such as RhoA and ARF [14, 15] in conjunction with PKC [16]. The interaction sites on the small G protein RhoA for PLD [17] and PKC [18] have been identified. It was also recognized that PLD can also be regulated by tyrosine kinases. Once activated, PLD can also modulate downstream molecules during cell signaling, chiefly mTOR, S6K and Ras [19]. In this review, we will present first the salient points of the PLD gene and protein structure and the enzyme's physiological and pathological roles, and then, we will concentrate on the data from our own lab that concerns itself with regulation of mammalian PLD2.
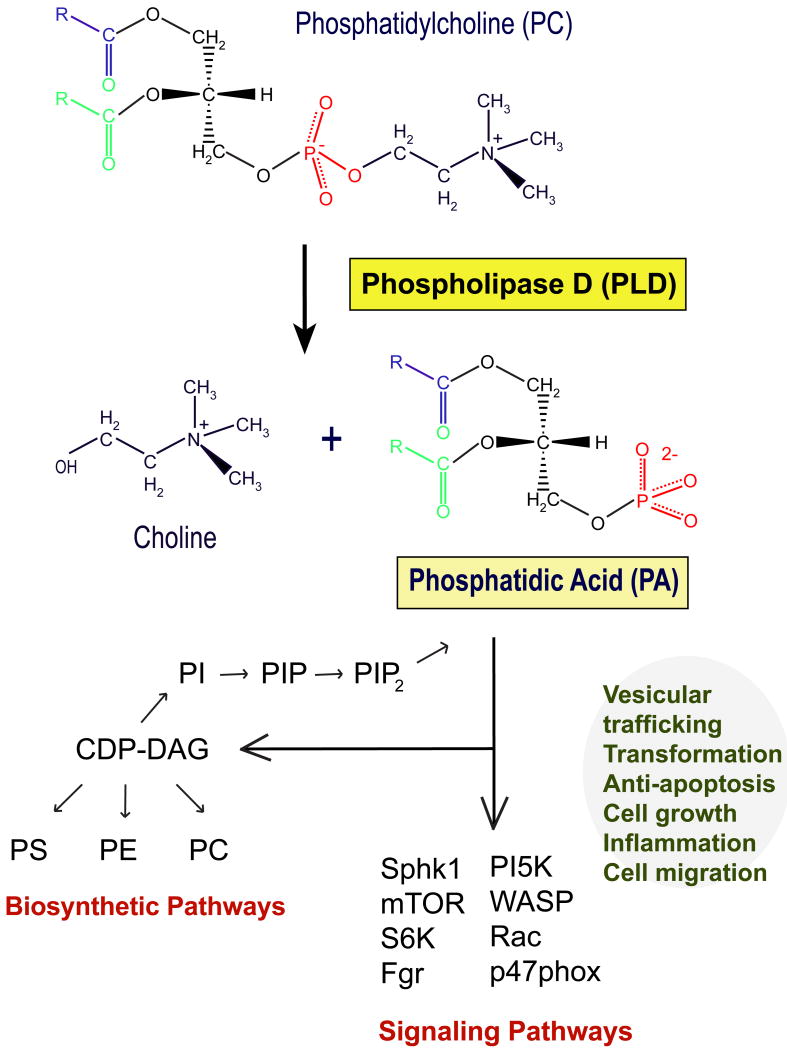
Figure 1. PLD chemical reaction.
A nucleophilic attack of water allows the breaking of the phosphodiester bond of the polar head of PC, which releases PA and choline in the transphosphatidylation reaction. PA is a metabolite that is at the center of the synthesis of other phospholipids. Apart from this, PA has a signaling mission in that it mediates several cellular functions, as indicated.
1.1. Cloning and genomic and enzymatic analyses
There are two genes in humans that encode for PLD, PLD1 and PLD2 (FIGURE 2). The PLD1 gene's location is 3q26 [20] and PLD2 gene's location is 17p13 [21]. Cloning and expression analysis of PLD1 was first reported in 1997 from mouse [22] and from rat brain [23], and it was recognized as part of a PLD superfamily [24]. Two alternately spliced forms of PLD1 were found and the purified proteins were activated by PKC and small GTPases [25]. PLD2 was also cloned from mouse [26] and from human B-cells [27]. PLD2 was expressed as a couple of splice variants that had equal enzymatic activity [28]. Genomic analysis of murine PLD1 and comparison to PLD2 revealed an unusual difference in gene size, with PLD1 being much larger than PLD2 [29]. PLD1 is a 1072 amino acid, 120 kDa protein, while PLD2 is a 933 amino acid, 106 kDa protein. Expression of the mammalian isoforms and detailed study of their regulation soon followed [2, 9, 30-36]. PLD1 and PLD2 generate structurally identical PA species in mammalian cells [37]. Their difference is on the regulation and the subcellular localization. Structurally, PLD1 bears a stretch of 120 amino acids (the “loop”) that is absent in PLD2 and confers a differential response to GTPases [38]. PLD1 was first found associated with caveolin-rich [39, 40] and Golgi-enriched membranes [41]. Specifically, PLD1 localizes to secretory granules and lysosomes and is translocated to the plasma membrane upon cellular stimulation [42]. On the other hand, PLD2 localizes in sarcolemmal membranes [43], plasma membranes [44] or the rims of the Golgi apparatus [45, 46].
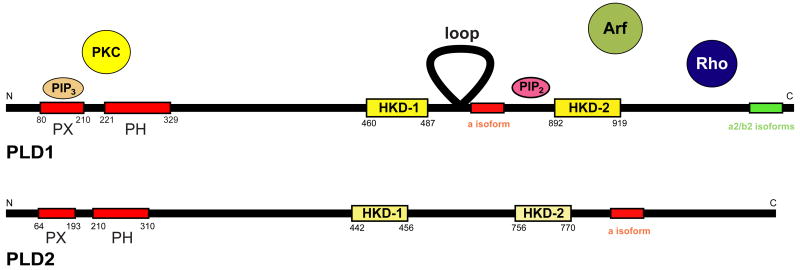
Figure 2. PLD main characteristics.
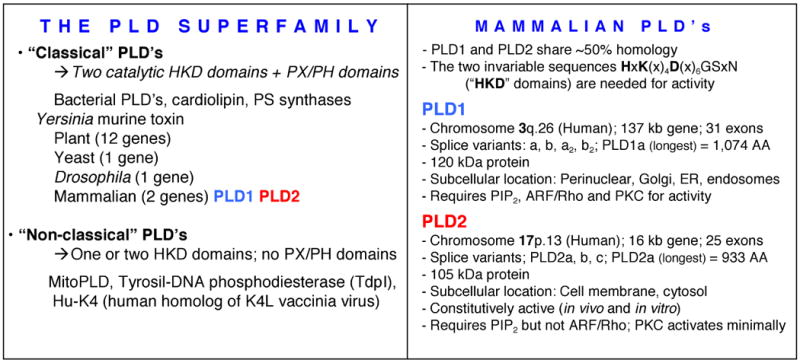
A HKD domain in the catalytic site defines the PLD superfamily. Mammalian PLD's are PLD1 and PLD2 and the chart compiles their main physical and cellular characteristics.
1.2. Catalytic and regulatory domains
The C terminus of mammalian PLD is required for catalytic activity [47] and it is there where the HKD domains reside. These are the “PLD signature” and both PLD1 and PLD2 isoforms have two of these HKD domains. An association of N- and C-terminal domains of PLD is required for catalytic activity [48]. Selective estrogen receptor (ER) modulators differentially regulate PLD catalytic activity in ER-negative breast cancer cells [49]. Multi-mono-ubiquitination has also been reported to be involved in enzymatic activity and palmitoylation [50]. Several regulatory domains have been recognized in PLD. A pleckstrin homology (PH) domain fold in the N-terminus was first indicated for mammalian PLD in [51]. The dependence on PIP2 was proven on [52] and this PH domain is essential for a PLD-aldolase interaction [53]. Two SH2-recognizable motifs are known to exist within the PH domain [54]. A phox domain is also at the N-terminus of PLD (219). PIP3 interacts with this domain and stimulates activity of PLD1 [55]. PLD binds to other proteins: PLCγ1 [56], Grb2 and Sos [57-59], cdc42 [60], Syk [61] and PIP-Kinase [62].
2. Functions of PLD
2.1. Vesicle trafficking
A clear role for PLD in membrane traffic was recognized early [63-65] and has been reported as essential for the oxidative burst [66] and transport proteins [67, 68]. The release of nascent secretory vesicles from the trans-Golgi network during exocytosis is dependent on PLD [69-73] in mast cells and regulatory T cells [74]. As for the mechanism, SCAMP2 interacts with Arf6 and PLD1 and links their function to exocytotic fusion pore formation [75]; both the RSK2 protein [76] and PA are essential for exocytosis [77, 78]. A role of PLD1 for receptor-mediated endocytosis has also been recognized [79, 80], and the PLD2 isoform also regulates endocytosis of angitensin, transferrin and G-protein coupled receptors [81-83]. The phox homology domain of PLD activates dynamin GTPase activity and accelerates EGF-R endocytosis [84], and similar to exocytosis, the presence of PA is needed for endocytosis [85]. Both these functions rely on actin remodeling and participation of the cytoskeleton for which PLD is also a key player [26, 64, 86, 87]. Other cellular functions that are mediated by the cytoskeleton are adhesion and ruffling. Both PLD1 [88, 89] and PLD2 [90] are involved in leukocyte adhesion, cell spreading [91] and cell membrane ruffling [45, 92].
2.2. Cell migration and phagocytosis
One of the major functions that PLD has been implicated with has been its role in inflammation, particularly migration and chemotaxis of leukocytes [93], but not confined to them as this seems to be a rather general biological phenomenon regulated by PLD. Thus, PLD-induced cell migration has been reported in Dictyostelium [94], epithelial cells (during wound healing) [95, 96], human cancer cells [97, 98], phagocytes [93, 99-101] and fibroblasts [102, 103]. Phosphocofilin [104], S6K [103], Rac2 [105] and the tyrosine kinase Fer [106, 107] are involved in PLD-mediated migration [107]. Also, a sequential regulation of DOCK2 dynamics by PA and PIP3 during neutrophil chemotaxis has been reported [107]. A major function of leukocytes, phagocytosis, is also regulated by PLD. This was demonstrated in response to complement-opsonized particles [108] and to M. tuberculosis or opsonized zymosan by human macrophages [109, 110]. PLD1 and PLD2 coordinately regulate macrophage phagocytosis [111] mediated by PA [112, 113] and RalA [114].
2.3. Apoptosis and proliferation
There is a large body of reports indicating an anti-apoptotic role of PLD [115-117], with an indication of signaling through increased expressions of Bcl-2 and Bcl-xL [118]. Apoptosis is enhanced [119] through farnesol inhibition of PLD signal transduction; PLD delays [120] or prevents apoptosis by inhibiting the expression of early growth response-1 and phosphatase and tensin homologue deleted on chromosome 10 [121]. As far as proliferation and mitogenesis, it was recognized early as PA had an effect on proliferation in osteoblastic [122] and pheochromocytoma PC12 cells [123]. PLD overcomes cell cycle arrest induced by high-intensity Raf signaling [124]. Transmodulation between PLD and c-Src enhances cell proliferation [125]. Survival signals generated by estrogen and PLD in MCF-7 breast cancer cells are dependent on c-myc [126] and c-myc stabilizes responses to estrogen and PLD in MCF-7 breast cancer cells [127]. Further, triptolide-induced suppression of PLD expression inhibits proliferation of MDA-MB-231 breast cancer cells [128]. A role for PLD2 phosphorylation has been proposed for the proliferation-inducing capabilities of PLD2 [129, 130].
2.4. Differentiation and other functions
An increased mRNA expression of PLD isozymes was observed during granulocytic differentiation of HL60 cells [131, 132], astrocytes [133] or myocytes [134]. A specific participation of PLD2 has been reported [135]. The mechanism involves remodeling of actin cytoskeleton [134], the transcription factor CREB pathway [136] and mTOR-IGF2 [137]. Other key functions in which PLD has been associated are: regulation of transcription, as reported in T lymphoid Jurkat cells with activation of transcription factor AP-1 (29), STAT [138] and the transcription repression of the cyclin-dependent kinase inhibitor p21 gene [139], mitochondrial fusion [77, 140] and a role of PLD in development [141-143].
3. Regulation of PLD2
As indicated in the Introduction, the mechanisms of signal transduction involving PLD are based on a large body of work by many investigators, and two pathways have been delineated that would stimulate its enzymatic activity. One pathway relies on small GTPase proteins for signal transduction, as Arf, Rho and related proteins can regulate PLD activity [17, 84, 144, 145]. The other pathway relies on tyrosine kinases, as PLD can be activated by growth factors/mitogens like EGF, PDGF, insulin and serum [48, 125, 146-148]. PLD1 from fibroblasts or HL-60 granulocytes can be tyrosyl phosphorylated [149, 150]. As far as PLD2, it was initially believed that this isoform was constitutively active and in no particular need of regulation. Later reports documented that this was not the case, as Kim et al. [151] demonstrated that PIP2 did increase PLD2 activity considerably. Furthermore, given the biological relevance of this enzyme in key functions (membrane remodeling, cell growth and migration) it could not possibly always be turned “on”.
Both the catalytic (HKD) and the regulatory (PH and PX) domains were studied in detail in the PLD1 isoform (FIGURE 3), but PLD2 was traditionally studied in lesser detail and much less was known about its regulation. Our laboratory has been focusing on the study of PLD2 regulation in mammalian cells. Over the past few years, we have reported, in regards to the catalytic action of PLD that PA is a chemoattractant agent for leukcoytes that binds to and acts through the ribosomal S6 kinase (S6K). As for the regulatory sites, PX and PH, we have reported the discovery of the PLD2 interaction with Grb2 via the PX domain leading to de novo DNA synthesis, as well as actin polymerization, the regulation of tyrosine phosphorylation on the N-terminus of the protein and the interaction of PLD2 with Rac2 via the PH domain, which will now be reviewed.
Figure 3. PLD1 and PLD2 domains.
Extensive work from labs referenced in the text has mapped out the fine regulation of the PLD1 isoform (with small GTPases Arf and Rho; PKC and phospholipids), whereas the understanding of PLD2 regulation lagged behind until a few years ago.
3.1. PA is a chemoattractant agent for leukcoytes acting through S6K
Apart from being a pleiotropic lipidic second messenger in mammalian cells, we have reported that PA acts as a leukocyte chemoattractant, as it elicits actin polymerization and chemotaxis of human neutrophils and differentiated proleukemic HL-60 cells [152] (FIGURE 4). Based on experiments with silencing RNA and with PLD mutants, we have also shown that the mechanism for this involves the ribosomal S6 kinase (S6K) signaling enzyme. PA was found to specifically and saturably bind to and activate recombinant and immunoprecipitated endogenous S6K with a stoichiometry of 94:1 lipid/protein for activation [153]. PA also modulated S6K enzymatic activity and phosphorylation on residues T389 and T421/S424, as well as phosphorylation of p70S6K's natural substrate S6 protein on residues S235/S236. These results implicated PA as a nexus that brings together cell phospholipases and kinases. The binding of PA to S6K occurs in addition to the also known case of its specific binding to mTOR [154, 155].
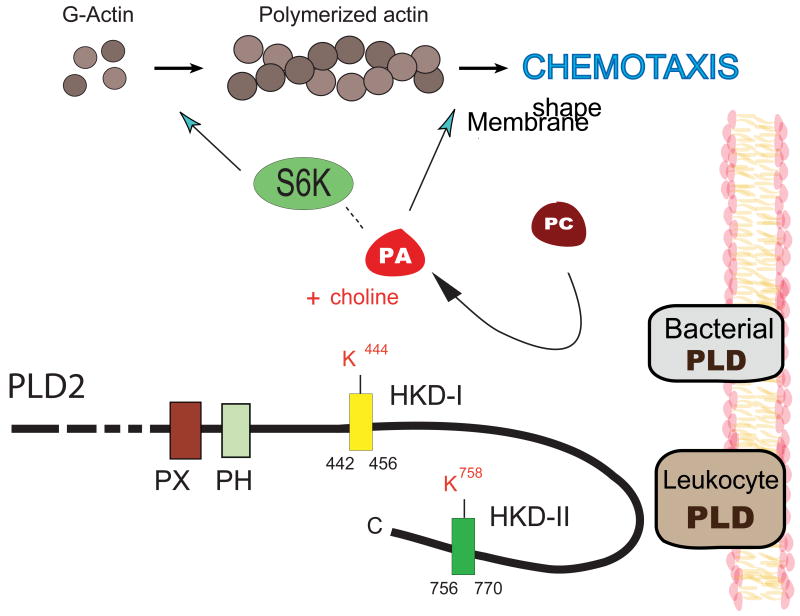
Figure 4. PA and S6K.
The formation of PA by PLD2 initiates a cascade of events that culminate in its binding to ribosomal S6 kinase (S6K) and subsequent actin polymerization and chemotaxis. Bacterial (extracellular) PLD can also produce PA that acts as a chemoattractant for leukocyte cells.
The results above were derived from exogenous PA added to a cell culture, but we also demonstrated that intracellular, PLD-derived PA, affected cell migration within the cell is being produced. In a sense, a gradient of chemoattractants (or PA itself) outside of the cell can be mimicked inside the cell by PA. We also demonstrated a connection between extracellular and intracellular PA. Using an EGFP-derived PA sensor (yeast soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) protein Spo20p PA-binding domain (PABD) cloned in a pEGFPC1 vector), we showed that exogenous PA, or PA generated in situ by bacterial (Streptomyces chromofuscus) PLD enters the cell and accumulates in vesicle-like cytoplasmic structures [152].
We surmised that the advantage for the cell to have PA as a chemoattractant is as follows: PA derived from PLD action, whether extracellularly (from microbial action as in S. chromofuscus's case) or intracellularly (from leukocytes), induces polymerization of actin, pseudopodia formation and chemotaxis in a process that is mediated by S6K activation. Phagocyte migration is paramount to the body's primary immune response. Neutrophils are attracted to the chemical markers released by damaged cells during the invasion by microorganisms. One of the unintended signals (at least from the point of view of the pathogen) could be the generation of PA in the cell membrane of the leukocyte. Some of this PA would further signal through S6K to help initiate membrane polarization and chemotaxis, events that are ultimately crucial to the neutrophil's mission of phagocytosis.
3.2. PLD2 and Grb2: Molecular Interactions
3.2.1. The discovery of Grb2 as a regulator of PLD2
In our early studies of regulation of PLD activation, we found the presence of phosphorylated PLD on tyrosine residues [156] that was later confirmed with the advent of better antibodies [57]. As we attempted to unravel the nature of the kinase(s) that phosphorylated PLD2, we noticed a new phenomenon in a COS-7 cell line that stably expressed myc-PLD2. After treating cells with PTP1b, we unexpectedly noted the presence of a low-molecular weight protein (∼29 kDa) in immunoblots that was increasingly tyrosyl-phosphorylated [57]. A literature search revealed that proteins of this size could play a role in tyrosine phosphorylation signaling, included, among others, the Src homology-2 (SH2)-containing growth factor receptor-bound protein 2 (Grb2).
Since Grb2 serves as an anchor between PTP1b and protein tyrosine kinases through the SH2 and SH3 domains [157], we investigated the possibility that Grb2 might play a role in regulating PLD2 phosphorylation. Indeed that was the case, as the addition of recombinant Grb2 to cell extracts elevated PY levels of myc-PLD2 (>10-fold). We also found that PLD2 exists in the living cell as a ternary complex with PTP1b and the docking protein Grb2. All these findings were reported in Horn et al. [57] and constituted the first implication ever of Grb2 in the PLD field.
3.2.2. The PLD2 residues Y169 and Y179 are regulated by Grb2 and Sos
We then established the mechanistic underpinnings of the PLD2/Grb2 association by identifying residues Y169 and Y179 in the PLD2 protein as being essential for the Grb2 interaction [58]. Y169 and Y179 are located within two consensus sites in the PX domain of PLD2 that mediate an SH2 interaction with Grb2. This was demonstrated with a SH2-deficient GST-Grb2-R86K mutant that failed to pull down PLD2 in vitro. We also created two point mutations in PLD2, PLD2-Y169F and PLD2-Y179F, that reduced Grb2 binding while simultaneous mutation of both residues completely abolished it. We also showed that Grb2 recruits to the PLD2-Grb2 dimer the GEF Sos, in effect implicating Sos in the PLD signaling pathway for the first time [58]. Mutation of either Y169 or Y179 to phenylalanine led to a decreased Sos interaction with PLD2. These results suggested that PLD2 residues Y169 and Y179, which bind Grb2, are responsible for Sos recruitment and that the complex PLD2/Grb2/Sos exists in vivo.
Zhao et al. [158] presented a different, but complementary perspective, one in which PA binds directly to Sos, thus opening the door for inclusion of EGF-R and PLD in the ERK pathway. In actuality, whether PLD2 binds to Grb2 and Grb2 serves as a link to Sos (16) or PA, synthesized by PLD binds directly to Sos [158] are two parts of the function of mitogen-stimulated cell activation that involves PLD, as highlighted in [159].
Following our original discovery of Grb2 and Sos regulating PLD2, we showed that the presence of Grb2 and its interaction with localized intracellular structures was essential for PLD2 activity and signaling in vivo [59]. In resting COS-7 cells, Grb2 serves to keep a fraction of PLD2 localized to the perinuclear Golgi region through its SH2 domain. Such co-localization results in changes in EGF-stimulated cells with more localization occurring in both the cytosol and the plasma membrane. We also demonstrated that a PLD2-Grb2 interaction affects DNA synthesis in a PI3K-dependent manner [129] and have continued our exploration of other biological consequences of a strong PLD2-Grb2 association, such as cell migration (FIGURE 5).
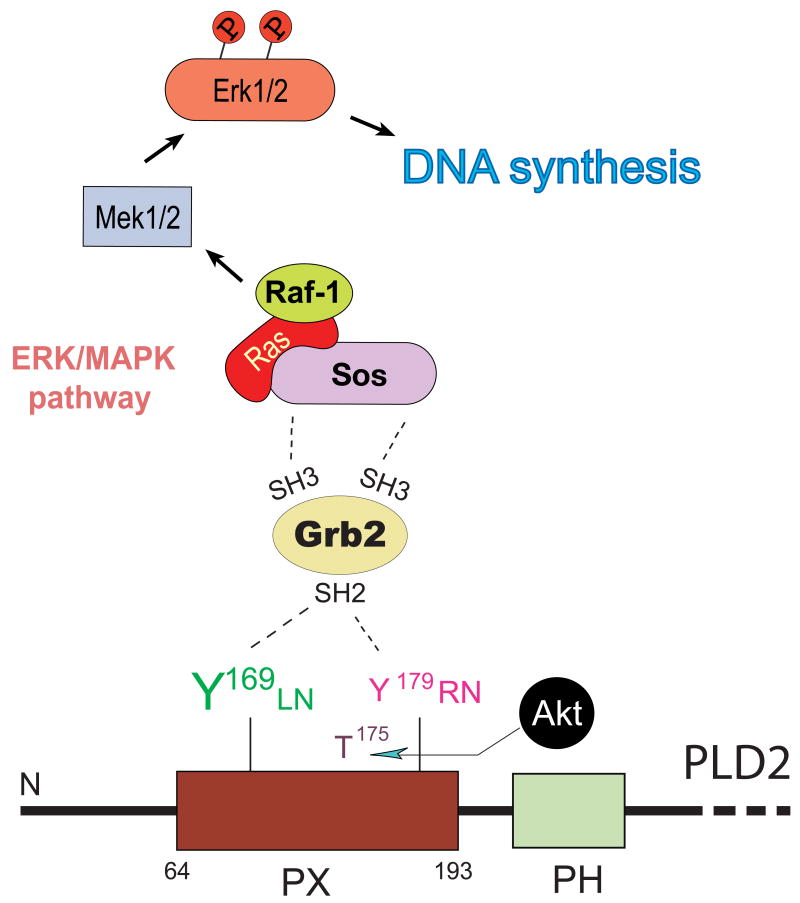
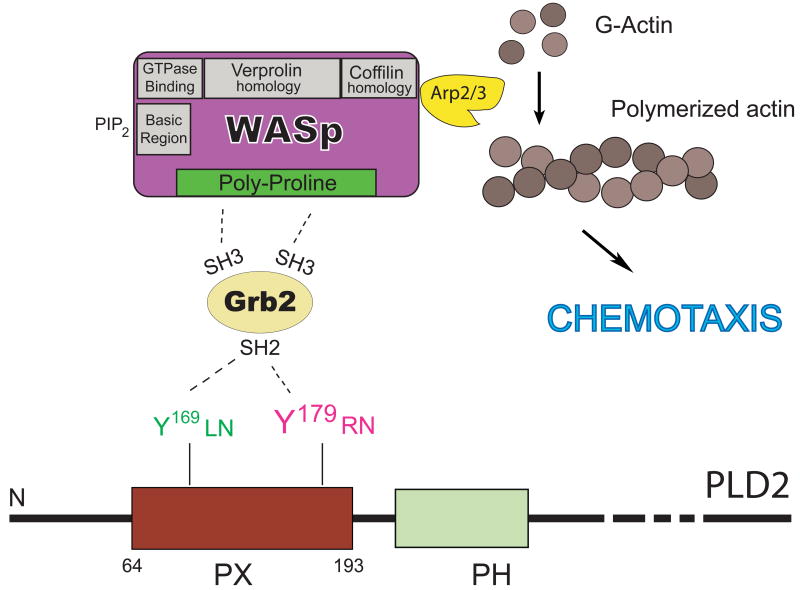
Figure 5. PLD2 binding to Grb2 at the PX domain: functional effect of DNA synthesis.
Identification of PLD2's Y169 and Y179, which bind to Grb2. Y169 mediates binding to Grb2, and its subsequent association to the GEF Sos mediates de novo DNA synthesis.
3.3. Functional Consequences of Grb2 biding to PLD2
The discovery of a PLD2-Grb2 protein-protein interaction results in three biological functions: actin-rich membrane ruffling, chemotaxis and phagocytosis, which we will describe in detail.
3.3.1. Membrane ruffling
The specific PLD2-Grb2 protein-protein interaction results in three biological functions of leukocytes (neutrophils and macrophages): cell membrane ruffling, chemotaxis and phagocytosis. Cell membrane ruffling or the formation of actin rich membrane protrusions occurs in cellular zones undergoing rapid reorganization of the plasma membrane and often precedes the formation of a lamellipodium and cell motility [160]. Stimulation by EGF results in the recruitment of Grb2 and Shc to cellular compartments that contained the receptor for EGF (EGF-R) [161].
We found that Grb2 cooperates with PLD2 in enhancing membrane ruffling, which is further increased when macrophages are stimulated with the growth factor M-CSF (23). PLD2 co-immunoprecipitates with Grb2 and does so to a greater extent in stimulated cells. M-CSF-stimulated cells exhibit dorsal ruffles that are normally associated with membrane endocytosis and indicated receptor-ligand internalization. The PLD2-Grb2 binding is at the level of the SH2 domain in Grb2, which results in enhanced PLD2 activity. A further cooperation with the Rho-GTPase Rac, stimulates actin polymerization and consequently membrane ruffle formation [162].
3.3.2. Chemotaxis
Since membrane ruffling precedes cell migration, the results of [162] lay the foundation for studying a PLD2-Grb2 association in chemotaxis. Immunofluorescence microscopy demonstrated that the PLD2-Grb2 protein complex localizes to lamellipodia during chemotaxis of murine macrophages via the Y169 residue of PLD2 [103]. As far as Grb2, the interaction was dependent on its SH2 domain since the SH2-domain deficient Grb2-R86K mutant impeded chemotaxis. Additionally, another protein-protein interaction that occurs in parallel to Grb2-PLD2 and positively affects PLD-mediated chemotaxis occurs via p70S6K. Immunofluorescent staining of S6K translocates from perinuclear regions and colocalizes with PLD2 in the cytosol following chemokine stimulation [153]. Interestingly, macrophages use both the Grb2/PLD2 and the PLD2/S6K pathways and at the full extent, whereas fibroblasts use only the former [153], giving an advantage to fast-moving macrophages vs. fibroblasts (cells that are only motile when required during wound healing).
3.3.3. Enter the world of WASP
The question that remains is that once PLD2 binds to Grb2 what happens next. The answer to this is that Grb2, as a bona fide docking protein, brings WASP to the PLD2 interaction through its two SH3 domains. The SH3 domains in Grb2 bind target proteins with proline-rich regions. One such candidate is the Wiskott-Aldrich syndrome protein (WASP), which is a major regulator of actin assembly and motility in leukocytes. Two important regions in WASP are its poly-proline rich domain and its carboxyl terminal verprolin homology/acidic domain (VCA). The latter enables the interaction of WASP with actin [163-167] and its role in podosome formation and chemotaxis [168, 169]. We hypothesized that once bound to PLD2 through SH2, the two SH3 domains of Grb2 could then interact with the poly-proline rich domain of WASP.
Our laboratory found that simultaneous cell transfection of PLD2, Grb2 and WASP had a robust enhancing effect on chemoattractant-mediated chemotaxis [103] or phagocytosis of antigen-coated erythrocytes [170]. An interaction of PLD2 with WASP through Grb2 was confirmed by immunoprecipitation and Western-blotting analysis and by immunofluorescence and co-localization of these three proteins in phagocytic cups and early endosomes. We have proposed that Grb2, a docking protein that is known to serve as a nexus for other signaling proteins (Sos in the ERK pathway, etc.), could bind to PLD2 (at the Y169 and Y179 sites) through its only SH2 domain and could also bind to WASP (at the proline-rich region) through its two SH3 domains and that this is a step required for chemotaxis and phagocytosis. FIGURE 6 shows a compilation of the participation of Grb2 in cell chemotaxis through WASP and actin polymerization.
Figure 6. A PLD2-Grb2-WASP trimeric complex and chemotaxis.
PLD2 binding to Grb2 initiates chemotaxis mediated by Y179. In this case Grb2 is a “docking” protein that is able to bring along WASP, through the SH3 domains. WASP, through the Arp2/3 complex activation, ensures actin polymerization and cell migration events.
3.4. Regulation of PLD2 by protein tyrosine kinases and phosphatases
3.4.1. Protein tyrosine kinases: a differential role for PLD1 and PLD2
The PLD1 isoform is phosphorylated on tyrosine residues, although some laboratories have indicated that this, in itself, does not lead to changes in lipase activity [150, 171-173]. The situation is different for PLD2, the isoform that this review addresses. It can be detected as a phosphotyrosine protein in vivo, and EGF, PDGF, insulin and other mitogens affect the level of tyrosyl phosphorylation of PLD2 [10]. The presence of total PLD activity in cell homogenates by tyrosyl phosphorylation has been previously documented [10, 156, 171]. PLD2 can form a complex with the EGF-R [48] or with Pyk2 and Src kinases [174]. Peroxide and EGF induce tyrosine phosphorylation of PLD and PKCα activation [172], while activation of PLD by 8-Br-cAMP is accomplished through Src, Ras and ERK [175]. Co-expression of PLD2 and the kinases Fyn or Fgr leads to an enhancement of PLD activation and degranulation of mast cells [176]. In addition to tyrosine kinases, PLD2 could be phosphorylated by the serine/threonine kinase AKT at residue T175 that serves to upregulate de novo DNA synthesis [129]. Phosphorylation can be seen in response to osmotic stress [177] and in cell proliferation [178].
3.4.2. Identification of new tyrosine target residues
Choi et al. [176] have found that PLD2 is specifically phosphorylated on residues Y11, Y14, Y165 and Y470. Mutation of Y470 resulted in a 50% decrease in PLD2 activation and suggests some partial loss of catalytic activity. Additionally, mutation of only Y14, but not the other three tyrosine residues, resulted in mislocalization of PLD2 as ascertained by immunofluorescence microscopy. Overexpression of PLD2-Y11F mutants lead to an enhancement in PLD2 activity in resting or EGF-activated cells [48]. Using LC-MS analyses of phosphopeptides, we found that Y296, Y511 and Y415 are vital to PLD2 regulation and can be phosphorylated by EGF-R, Src and JAK3, respectively, to yield either positive or negative effects on the lipase's activity [179].
3.4.3. Tyrosine activity levels in cancer cells
Elevation of either PLD1 or PLD2 has the potential to contribute to the progression of the malignant phenotype in cells that also have elevated levels of EGF-R or Src tyrosine kinases [180]. However, since overexpression of phosphorylation deficient mutants PLD2-Y11F [48]or PLD2-Y296F [179] increases PLD2 activity, we hypothesized that PLD2 activity in low-invasive breast cancer cell lines (such as MCF-7) is comparatively low compared to more invasive cells because it is downregulated by tyrosyl phosphorylation at Y296 via EGF-R. This low level of PLD activity can be increased by in vitro treatment with either JAK3 or Src that can target other tyrosine residues (for example Y415 or Y511) [179]. Additionally, JAK3 participates in the activation of PLD through the IL-8R and EGF-R pathways [181, 182], while Src participates in the activation of PLD through the Ras pathway [176]. Thus, a coordinated effort of at least three kinases (EGF-R, Src and JAK3) (along with phosphatases) plays a key role in regulating PLD2 in cancer cells and has been reviewed previously [19].
3.4.4. Phosphatases
As indicated earlier, PLD2 forms a ternary complex with both PTP1b and Grb2 [57]. Tyrosyl phosphorylation/dephosphorylation of PLD2 dictates whether PLD2 is activated or suppressed by certain signaling molecules and the participation of CD45 and PTP1b phosphatases [179]. Low concentrations of phosphatases that target Y296 (an inhibitory site) or Y511 (an ambivalent site) specifically result in positive regulation of PLD2 and high lipase activity. Contrarily, high concentrations of phosphatases or phosphatases that specifically target Y415 (an activator site) result in loss of lipase activity. A recent report from our lab [178] indicates that increased cell transformation in PLD2-overexpressing cells occurs as a result of increased de novo DNA synthesis induced by PLD2 with the specific tyrosine residues involved in these functions being Y179 and Y511 and also the participation of CD45 phosphatase. The balance between phosphorylation and dephosphorylation at these residues keep PLD2 highly regulated [19].
3.5. Regulation of PLD2 by Rac2: an unexpected negative effect
PLD1 enzymatic activity is regulated through phosphoinositides, PKC, ADP ribosylation factor (ARF), Rho, fatty acids, Ca2+ and protein phosphorylation. ARF was the first G-protein identified to activate PLD1 [144, 183]. Other small GTPases such as RhoA, Rac1 and Cdc42 in the presence of GTPγS directly associate with and stimulate PLD1 activity [184-187]. The Rho family of GTPases is also involved in indirect regulation of PLD1 enzymatic activity through stimulation of PI(4,5)P2 kinase, Rho kinase [42] or by intracellular translocation of the PLD isoforms [188-190]. It was shown that Rac1 does not regulate the activity of PLD2 [26, 191], however, the role of the other Rac isoform, Rac2, has not received full attention until recently. Our laboratory has concentrated on Rac2, as it is more directly implicated in chemotaxis, and has wondered if Rac2 would modulate the activity of PLD2, which is also implicated in cell migration.
3.5.1. A dual effect of Rac2 on PLD2
In [192] we have reported the surprising discovery of a dual effect of the GTPase on the lipase, whereby Rac2 affects PLD2 enzymatic activity both positively and negatively, which helps to explain the progression through chemotaxis in leukocytes. A positive feedback loop exists between Rac2 and PLD2 during early stages of stimulation (when phagocytes initiate cell migration). Conversely, Rac2 has a negative effect on PLD2 at late times of stimulation (when phagocytes are locked in position at the inflammation site) and when there is an overwhelming amount of Rac2 in the cell (FIGURE 7). Supporting the newly reported negative effect of Rac2 on PLD2, we found that endogenous PLD activity from bone marrow neutrophils and bone marrow-derived macrophages is high in Rac2-/- KO mice.
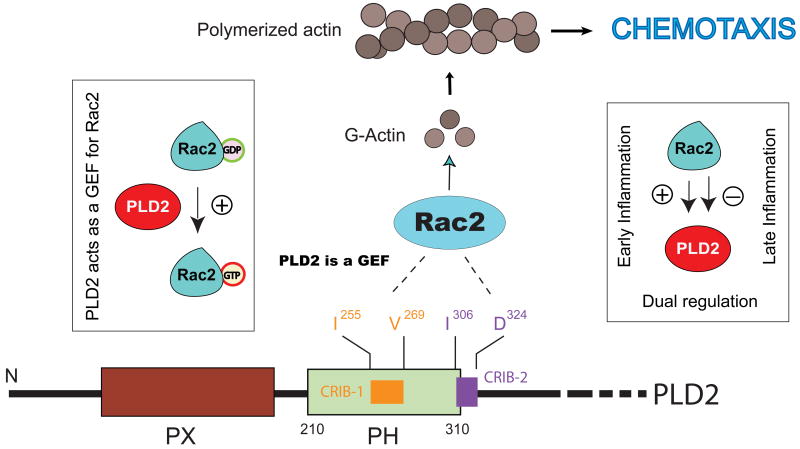
Figure 7. PLD2 and Rac: dual regulation.
PLD2, thru the PH domain, binds to Rac2 in two newly identified CRIB domains. The Rac2-PLD2 binding has opposite functional consequences depending on the temporal stage of cellular stimulation. Also, PLD2 can act as a GEF for Rac2, initiating a GDP-GTP exchange and thus activating Rac2.
3.5.2. How does Rac2 inhibit PLD2?
We have observed a fraction of fluorescence signals that localized in the plasma membrane for both Rac2-CFP and PLD2-YFP [192]. In some cases, these two signals do not overlap, but rather, they are extremely adjacent to each other, whereby Rac2 is closer to the plasma membrane than PLD2. This led us to believe that Rac2 exerts a negative effect on PLD2 because Rac2 occupies PLD2's PH domain, pulling PLD2 apart from the normal environment and out of the reach of PIP2 and other essential co-factors for enzymatic activity. This “terminates” PLD2 activity and, consequently, chemotaxis. Further proving this point, it was found that PLD2 activity could be rescued in cells in the presence of excess PIP2, which excludes Rac2 from the membrane.
As all this was observed in primary cells (neutrophils and macrophages), as well as in cell lines (macrophages) [192], we believe this dual mechanism could be extensive to a large array of migratory cells. Further, this represents an example of simplicity and economy of cell resources, as just two molecules can be sufficient for accomplishing a well-defined biological function.
3.5.3. Site of binding of Rac2 to PLD2
What we have just described indicates a profound relation between Rac2 and PLD2 and reinforces the idea that PLD and Rac small GTPases are integral to cell signaling [99, 193]. In a subsequent study [194], we demonstrated that Rac2 and PLD2 form a protein-protein complex in the cell, as they can both be immunoprecipitated with antibodies specific for the other protein and as a strong FRET signal in cells. The existence of FRET between the donor (CFP-Rac2) and the acceptor (YFP-PLD2) in vivo indicates that Rac2 and PLD2 are in close enough proximity (< 7 nm apart) so as to form stable complexes in the cell.
The PLD2-Rac2 interaction begged the question as to the specific site on PLD2 that binds to Rac2. To ascertain this, we generated several PLD2 mutants where we hypothesized that binding occurred that were based on other examples of Rac2 binding partners. A CRIB motif (for Cdc42/Rac Interactive Binding) has been shown to specifically bind the GTP-bound form of Cdc42 or Rac with a preference for Cdc42 [195-198]. The prototypes are the non-receptor tyrosine kinase, ACK, and the serine/threonine kinase, PAK [199]. The length of the consensus CRIB motif is approximately 16 amino acids containing a region of variable length between the two halves of the binding motif. The CRIB motif contains eight core amino acids with the sequence ISXPXXXXFXHXXHVG [200, 201]. However, proteins with one or two differences within the core sequence can still show binding to Cdc42/Rac [201].
3.5.4. Two new CRIB domains found in PLD2
A manual search of the PLD2 amino acid sequence yielded two putative CRIB-like candidate amino acid segments that contained the key amino acids of the consensus in the appropriate order. We named the first domain CRIB-1 (255ISFVQLFDPGFEVQGV270) and the second domain CRIB-2 (306ITELAQGPGRDFLQLHRHDSY326), which are located in and downstream from the PH domain of PLD2, respectively [194]. We demonstrated that CRIB deletion mutants PLD2-ΔCRIB1 and PLD2-ΔCRIB2 negated a PLD2-Rac2 association. We further determined that the apparent KD values for Rac2-WT binding to PLD2-WT are ∼10 nM and that the stoichiometry of the biding is ∼1:2 (PLD2:Rac2). We also found that in vitro, excess Rac2 had a negative effect on PLD2 activity as seen previously in cells [192]. FIGURE 7 shows a compilation of the participation of PLD2 in cell chemotaxis through the participation of all the players we have described so far: Grb2 (associated with the PX domain), Rac (associated with the PH domain), as well as WASP, S6K and the product of PLD reaction, PA.
3.6. PLD2 acting as a GEF: a novel and unexpected discovery
While we were studying the correlation between Rac2 and PLD2 and to our surprise, we found that Rac2 GTP-loading activity (as p21-binding-domain pulldown) was increased in the presence of PLD2 [194]. The positive effect of PLD2 on Rac2 GTP-loading activity led us to the novel discovery that for the first time documents that PLD2 can act as a GEF for the small GTPase Rac2 [202] (FIGURE 7). As currently known, there are three families of GEFs for monomeric GTPases: Cdc25, Dbl homology (DH) and Sec7 domains [203-205]. However, none of these proteins is a phospholipase. The new mechanism for GTPase activation by a phospholipase involves both PLD2 and the product of the lipase reaction, PA. As these findings were extended beyond leukocytes to other cells, we believe that this mechanism is universal in cell biology and defines a new class of GEF molecules never reported before.
3.6.1. Proving that PLD2 is a GEF
A series of GTP/GDP exchange experiments, which are the hallmark means to define a GEF similar to that described in reference [206], indicated that PLD2 elicited dissociation of [3H]-GDP from Rac2 and then binding of [35S]-GTPγS to Rac2 in the presence of PLD2. This effect is large enough to be meaningful (50% decrease for GDP dissociation and 300% increase for GTP association), had a reasonable kinetic behavior with a half-time of 10 min and saturation at 30 min and was augmented with increasing PLD2 concentrations. Of note, we found that PLD2 enhanced the GTP/GDP exchange of Rac2 in vitro in the absence of any other co-factors (i.e., the reaction mixtures had only recombinant proteins), indicating that PLD2 is a bona fide GEF for Rac2 [206]. Further, we reasoned that the result of PLD2 action on Rac2 has biological consequences. As Rac2 has been extensively implicated in cell migration and phagocytosis [207-210] in leukocytes, we found that PLD2 and Rac2 co-localize in the cell and lead to augmentation of normal physiological functions of leukocytes: adhesion, polarization, chemotaxis and phagocytosis.
3.6.2. The mechanism of PLD2 as a GEF
In [202], we reported that PA, the product of the PLD enzymatic reaction increases the capability of PLD being a GEF for Rac2. Thus, two things are needed to elicit full GEF action on Rac2: one was the protein-protein interaction between PLD2 and Rac2 (at the newly discovered CRIB domains) and the other was the presence of PA. The mechanism of enzymatic activation involves first the production of PA and then the activation of Rac as the order of reaction. Computer simulations have indicated an excellent level of alignment between a known GEF and PLD2's PH domain, especially three beta sheets around CRIB-1. We can speculate that PA could serve to displace the P-loop from binding to the phosphates of GDP by inserting itself in Rac2 (we have observed that PA binds and activates GTP loading) into the nucleotide-binding site as to sterically and electrostatically expel the GDP nucleotide by a push-pull mechanism.
Conclusions
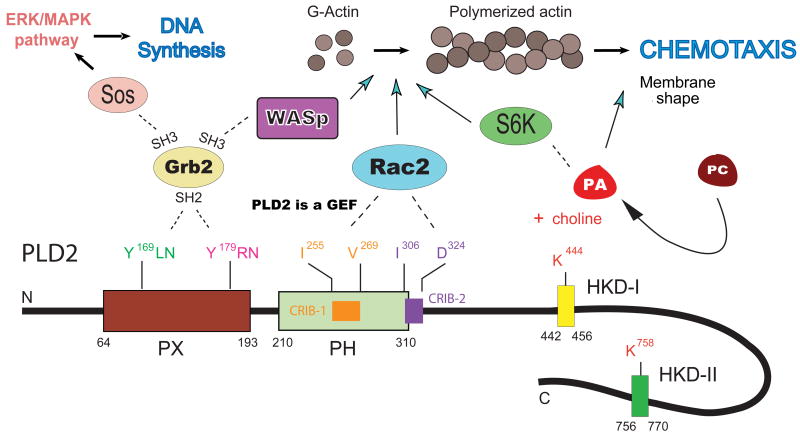
FIGURE 8 presents a summary of the results obtained in our laboratory over the course of the last few years, which have been discussed in detail in the previous sections. It shows an exquisite regulation of PLD2 by a wealth of interacting partners (S6K, Grb2, Sos, kinases, phosphatases, WASp and Rac2) that lead to regulated functionalities in disparate functions such as de novo DNA synthesis and modulation of inflammatory responses of leukocytes, such as adhesion, chemotaxis and phagocytosis. The model includes the surprise discovery of PLD as the first lipase to be considered as a GEF. A phospholipase such as PLD that exists already in the cell membrane that acts directly on Rac allows a quick response of the cell without intermediary signaling molecules and provides a great advantage in the transmission of information. This provides only the latest level of PLD2 regulation in a field that promises newer and exciting advances in the next few years.
Figure 8. A summary of PLD2 regulation.
This model is a summary of all data in previous Figures 4-7, indicating our laboratory's inroads into the understanding of PLD2 regulation at the molecular level and the discovery of a plethora of associated partners: Grb2, Sos, WASP, Rac2 and their sites of binding (kinases and phosphatases are not shown in this diagram, but they were reviewed earlier [19]).
Acknowledgments
Dr. Cambronero's research is supported by grants HL056653 from the National Institutes of Health and from the Boonshoft School of Medicine (BSOM) Grant 229102-20053. The author wishes to thank Madhu Mahankali and Karen Henkels for their help with the references and proofreading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown HA, Henage LG, Preininger AM, Xiang Y, Exton JH. Methods Enzymol. 2007;434:49–87. doi: 10.1016/S0076-6879(07)34004-4. [DOI] [PubMed] [Google Scholar]
- 2.Frohman MA, Sung TC, Morris AJ. Biochim Biophys Acta. 1999;1439(2):175–186. doi: 10.1016/s1388-1981(99)00093-1. [DOI] [PubMed] [Google Scholar]
- 3.Exton JH. Biochim Biophys Acta. 1994;1212(1):26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- 4.English D. Cell Signal. 1996;8(5):341–347. doi: 10.1016/0898-6568(95)00076-3. [DOI] [PubMed] [Google Scholar]
- 5.Frohman MA, Morris AJ. Curr Biol. 1996;6(8):945–947. doi: 10.1016/s0960-9822(02)00634-6. [DOI] [PubMed] [Google Scholar]
- 6.Exton JH. J Biol Chem. 1997;272(25):15579–15582. doi: 10.1074/jbc.272.25.15579. [DOI] [PubMed] [Google Scholar]
- 7.Morris AJ, Frohman MA, Engebrecht J. Anal Biochem. 1997;252(1):1–9. doi: 10.1006/abio.1997.2299. [DOI] [PubMed] [Google Scholar]
- 8.Morris AJ, Hammond SM, Colley C, Sung TC, Jenco JM, Sciorra VA, Rudge SA, Frohman MA. Biochem Soc Trans. 1997;25(4):1151–1157. doi: 10.1042/bst0251151. [DOI] [PubMed] [Google Scholar]
- 9.Frohman MA, Morris AJ. Chem Phys Lipids. 1999;98(1-2):127–140. doi: 10.1016/s0009-3084(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 10.Houle MG, Bourgoin S. Biochim Biophys Acta. 1999;1439(2):135–149. doi: 10.1016/s1388-1981(99)00090-6. [DOI] [PubMed] [Google Scholar]
- 11.Cockcroft S. Cell Mol Life Sci. 2001;58(11):1674–1687. doi: 10.1007/PL00000805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDermott M, Wakelam MJ, Morris AJ. Biochem Cell Biol. 2004;82(1):225–253. doi: 10.1139/o03-079. [DOI] [PubMed] [Google Scholar]
- 13.Jenkins GM, Frohman MA. Cell Mol Life Sci. 2005;62(19-20):2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geny B, Cockcroft S. Biochem J. 1992;284(Pt 2):531–538. doi: 10.1042/bj2840531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Billah MM. Curr Opin Immunol. 1993;5(1):114–123. doi: 10.1016/0952-7915(93)90090-f. [DOI] [PubMed] [Google Scholar]
- 16.Conricode KM, Smith JL, Burns DJ, Exton JH. FEBS Lett. 1994;342(2):149–153. doi: 10.1016/0014-5793(94)80490-7. [DOI] [PubMed] [Google Scholar]
- 17.Bae CD, Min DS, Fleming IN, Exton JH. J Biol Chem. 1998;273(19):11596–11604. doi: 10.1074/jbc.273.19.11596. [DOI] [PubMed] [Google Scholar]
- 18.Kook S, Exton JH. Cell Signal. 2005;17(11):1423–1432. doi: 10.1016/j.cellsig.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Cambronero J. Scientific World Journal. 2010;10:1356–1369. doi: 10.1100/tsw.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SH, Chun YH, Ryu SH, Suh PG, Kim H. Cytogenet Cell Genet. 1998;82(3-4):224. doi: 10.1159/000015105. [DOI] [PubMed] [Google Scholar]
- 21.Park SH, Ryu SH, Suh PG, Kim H. Cytogenet Cell Genet. 1998;82(3-4):225. doi: 10.1159/000015106. [DOI] [PubMed] [Google Scholar]
- 22.Colley WC, Altshuller YM, Sue-Ling CK, Copeland NG, Gilbert DJ, Jenkins NA, Branch KD, Tsirka SE, Bollag RJ, Bollag WB, Frohman MA. Biochem J. 1997;326(Pt 3):745–753. doi: 10.1042/bj3260745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SK, Provost JJ, Bae CD, Ho WT, Exton JH. J Biol Chem. 1997;272(46):29263–29271. doi: 10.1074/jbc.272.46.29263. [DOI] [PubMed] [Google Scholar]
- 24.Sung TC, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht J, Frohman MA. Embo J. 1997;16(15):4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond SM, Jenco JM, Nakashima S, Cadwallader K, Gu Q, Cook S, Nozawa Y, Prestwich GD, Frohman MA, Morris AJ. J Biol Chem. 1997;272(6):3860–3868. doi: 10.1074/jbc.272.6.3860. [DOI] [PubMed] [Google Scholar]
- 26.Colley WC, Sung TC, Roll R, Jenco J, Hammond SM, Altshuller Y, Bar-Sagi D, Morris AJ, Frohman MA. Curr Biol. 1997;7(3):191–201. doi: 10.1016/s0960-9822(97)70090-3. [DOI] [PubMed] [Google Scholar]
- 27.Lopez I, Arnold RS, Lambeth JD. J Biol Chem. 1998;273(21):12846–12852. doi: 10.1074/jbc.273.21.12846. [DOI] [PubMed] [Google Scholar]
- 28.Redina OE, Frohman MA. Biochem J. 1998;331(Pt 3):845–851. doi: 10.1042/bj3310845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redina OE, Frohman MA. Gene. 1998;222(1):53–60. doi: 10.1016/s0378-1119(98)00465-x. [DOI] [PubMed] [Google Scholar]
- 30.Min DS, Park SK, Exton JH. J Biol Chem. 1998;273(12):7044–7051. doi: 10.1074/jbc.273.12.7044. [DOI] [PubMed] [Google Scholar]
- 31.Sung TC, Altshuller YM, Morris AJ, Frohman MA. J Biol Chem. 1999;274(1):494–502. doi: 10.1074/jbc.274.1.494. [DOI] [PubMed] [Google Scholar]
- 32.Meier KE, Gibbs TC, Knoepp SM, Ella KM. Biochim Biophys Acta. 1999;1439(2):199–213. doi: 10.1016/s1388-1981(99)00095-5. [DOI] [PubMed] [Google Scholar]
- 33.Gibbs TC, Meier KE. J Cell Physiol. 2000;182(1):77–87. doi: 10.1002/(SICI)1097-4652(200001)182:1<77::AID-JCP9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Redina O, Altshuller YM, Yamazaki M, Ramos J, Chneiweiss H, Kanaho Y, Frohman MA. J Biol Chem. 2000;275(45):35224–35232. doi: 10.1074/jbc.M003329200. [DOI] [PubMed] [Google Scholar]
- 35.Horn JM, Lehman JA, Alter G, Horwitz J, Gomez-Cambronero J. Biochim Biophys Acta. 2001;1530(1):97–110. doi: 10.1016/s1388-1981(00)00172-4. [DOI] [PubMed] [Google Scholar]
- 36.Xie Z, Ho WT, Exton JH. Biochim Biophys Acta. 2002;1580(1):9–21. doi: 10.1016/s1388-1981(01)00168-8. [DOI] [PubMed] [Google Scholar]
- 37.Pettitt TR, McDermott M, Saqib KM, Shimwell N, Wakelam MJ. Biochem J. 2001;360(Pt 3):707–715. doi: 10.1042/0264-6021:3600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Riebeling C, Bourgoin S, Shields D. Biochim Biophys Acta. 2008;1781(8):376–382. doi: 10.1016/j.bbalip.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Czarny M, Lavie Y, Fiucci G, Liscovitch M. J Biol Chem. 1999;274(5):2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- 40.Xu L, Shen Y, Joseph T, Bryant A, Luo JQ, Frankel P, Rotunda T, Foster DA. Biochem Biophys Res Commun. 2000;273(1):77–83. doi: 10.1006/bbrc.2000.2907. [DOI] [PubMed] [Google Scholar]
- 41.Ktistakis NT, Brown HA, Sternweis PC, Roth MG. Proc Natl Acad Sci U S A. 1995;92(11):4952–4956. doi: 10.1073/pnas.92.11.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown FD, Thompson N, Saqib KM, Clark JM, Powner D, Thompson NT, Solari R, Wakelam MJ. Curr Biol. 1998;8(14):835–838. doi: 10.1016/s0960-9822(98)70326-4. [DOI] [PubMed] [Google Scholar]
- 43.Park JB, Kim JH, Kim Y, Ha SH, Yoo JS, Du G, Frohman MA, Suh PG, Ryu SH. J Biol Chem. 2000;275(28):21295–21301. doi: 10.1074/jbc.M002463200. [DOI] [PubMed] [Google Scholar]
- 44.Sarri E, Pardo R, Fensome-Green A, Cockcroft S. Biochem J. 2003;369(Pt 2):319–329. doi: 10.1042/BJ20021347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freyberg Z, Bourgoin S, Shields D. Mol Biol Cell. 2002;13(11):3930–3942. doi: 10.1091/mbc.02-04-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonoda H, Okada T, Jahangeer S, Nakamura S. J Biol Chem. 2007;282(47):34085–34092. doi: 10.1074/jbc.M705593200. [DOI] [PubMed] [Google Scholar]
- 47.Liu MY, Gutowski S, Sternweis PC. J Biol Chem. 2001;276(8):5556–5562. doi: 10.1074/jbc.M006404200. [DOI] [PubMed] [Google Scholar]
- 48.Slaaby R, Jensen T, Hansen HS, Frohman MA, Seedorf K. J Biol Chem. 1998;273(50):33722–33727. doi: 10.1074/jbc.273.50.33722. [DOI] [PubMed] [Google Scholar]
- 49.Eisen SF, Brown HA. Mol Pharmacol. 2002;62(4):911–920. doi: 10.1124/mol.62.4.911. [DOI] [PubMed] [Google Scholar]
- 50.Yin H, Gui Y, Du G, Frohman MA, Zheng XL. J Biol Chem. 2010;285(18):13580–13588. doi: 10.1074/jbc.M109.046359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holbrook PG, Geetha V, Beaven MA, Munson PJ. FEBS Lett. 1999;448(2-3):269–272. doi: 10.1016/s0014-5793(99)00366-x. [DOI] [PubMed] [Google Scholar]
- 52.Hodgkin MN, Masson MR, Powner D, Saqib KM, Ponting CP, Wakelam MJ. Curr Biol. 2000;10(1):43–46. doi: 10.1016/s0960-9822(99)00264-x. [DOI] [PubMed] [Google Scholar]
- 53.Kim JH, Lee S, Lee TG, Hirata M, Suh PG, Ryu SH. Biochemistry. 2002;41(10):3414–3421. doi: 10.1021/bi015700a. [DOI] [PubMed] [Google Scholar]
- 54.Ohguchi K, Banno Y, Nakagawa Y, Akao Y, Nozawa Y. J Cell Physiol. 2005;205(3):444–451. doi: 10.1002/jcp.20421. [DOI] [PubMed] [Google Scholar]
- 55.Lee JS, Kim JH, Jang IH, Kim HS, Han JM, Kazlauskas A, Yagisawa H, Suh PG, Ryu SH. J Cell Sci. 2005;118(Pt 19):4405–4413. doi: 10.1242/jcs.02564. [DOI] [PubMed] [Google Scholar]
- 56.Jang IH, Lee S, Park JB, Kim JH, Lee CS, Hur EM, Kim IS, Kim KT, Yagisawa H, Suh PG, Ryu SH. J Biol Chem. 2003;278(20):18184–18190. doi: 10.1074/jbc.M208438200. [DOI] [PubMed] [Google Scholar]
- 57.Horn J, Lopez I, Miller MW, Gomez-Cambronero J. Biochem Biophys Res Commun. 2005;332(1):58–67. doi: 10.1016/j.bbrc.2005.04.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Di Fulvio M, Lehman N, Lin X, Lopez I, Gomez-Cambronero J. Oncogene. 2006;25(21):3032–3040. doi: 10.1038/sj.onc.1209340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Fulvio M, Frondorf K, Henkels KM, Lehman N, Gomez-Cambronero J. J Mol Biol. 2007;367(3):814–824. doi: 10.1016/j.jmb.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoon MS, Cho CH, Lee KS, Han JS. Biochem Biophys Res Commun. 2006;347(3):594–600. doi: 10.1016/j.bbrc.2006.06.111. [DOI] [PubMed] [Google Scholar]
- 61.Lee JH, Kim YM, Kim NW, Kim JW, Her E, Kim BK, Kim JH, Ryu SH, Park JW, Seo DW, Han JW, Beaven MA, Choi WS. Blood. 2006;108(3):956–964. doi: 10.1182/blood-2005-10-009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Divecha N, Roefs M, Halstead JR, D'Andrea S, Fernandez-Borga M, Oomen L, Saqib KM, Wakelam MJ, D'Santos C. Embo J. 2000;19(20):5440–5449. doi: 10.1093/emboj/19.20.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cockcroft S. Chem Phys Lipids. 1996;80(1-2):59–80. doi: 10.1016/0009-3084(96)02546-7. [DOI] [PubMed] [Google Scholar]
- 64.Cross MJ, Roberts S, Ridley AJ, Hodgkin MN, Stewart A, Claesson-Welsh L, Wakelam MJ. Curr Biol. 1996;6(5):588–597. doi: 10.1016/s0960-9822(02)00545-6. [DOI] [PubMed] [Google Scholar]
- 65.Roth MG. Traffic. 2008;9(8):1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x. [DOI] [PubMed] [Google Scholar]
- 66.Melendez AJ, Bruetschy L, Floto RA, Harnett MM, Allen JM. Blood. 2001;98(12):3421–3428. doi: 10.1182/blood.v98.12.3421. [DOI] [PubMed] [Google Scholar]
- 67.Huang P, Altshuller YM, Hou JC, Pessin JE, Frohman MA. Mol Biol Cell. 2005;16(6):2614–2623. doi: 10.1091/mbc.E04-12-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faugaret D, Chouinard FC, Harbour D, El azreq MA, Bourgoin SG. Biochem Pharmacol. 2010;81(1):144–156. doi: 10.1016/j.bcp.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 69.Chen YG, Siddhanta A, Austin CD, Hammond SM, Sung TC, Frohman MA, Morris AJ, Shields D. J Cell Biol. 1997;138(3):495–504. doi: 10.1083/jcb.138.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stutchfield J, Cockcroft S. Biochem J. 1993;293(Pt 3):649–655. doi: 10.1042/bj2930649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morgan CP, Sengelov H, Whatmore J, Borregaard N, Cockcroft S. Biochem J. 1997;325(Pt 3):581–585. doi: 10.1042/bj3250581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Choi WS, Kim YM, Combs C, Frohman MA, Beaven MA. J Immunol. 2002;168(11):5682–5689. doi: 10.4049/jimmunol.168.11.5682. [DOI] [PubMed] [Google Scholar]
- 73.Hitomi T, Zhang J, Nicoletti LM, Grodzki AC, Jamur MC, Oliver C, Siraganian RP. Blood. 2004;104(13):4122–4128. doi: 10.1182/blood-2004-06-2091. [DOI] [PubMed] [Google Scholar]
- 74.Mead KI, Zheng Y, Manzotti CN, Perry LC, Liu MK, Burke F, Powner DJ, Wakelam MJ, Sansom DM. J Immunol. 2005;174(8):4803–4811. doi: 10.4049/jimmunol.174.8.4803. [DOI] [PubMed] [Google Scholar]
- 75.Liu L, Liao H, Castle A, Zhang J, Casanova J, Szabo G, Castle D. Mol Biol Cell. 2005;16(10):4463–4472. doi: 10.1091/mbc.E05-03-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeniou-Meyer M, Liu Y, Begle A, Olanich ME, Hanauer A, Becherer U, Rettig J, Bader MF, Vitale N. Proc Natl Acad Sci U S A. 2008;105(24):8434–8439. doi: 10.1073/pnas.0710676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Choi SY, Huang P, Jenkins GM, Chan DC, Schiller J, Frohman MA. Nat Cell Biol. 2006;8(11):1255–1262. doi: 10.1038/ncb1487. [DOI] [PubMed] [Google Scholar]
- 78.Zeniou-Meyer M, Zabari N, Ashery U, Chasserot-Golaz S, Haeberle AM, Demais V, Bailly Y, Gottfried I, Nakanishi H, Neiman AM, Du G, Frohman MA, Bader MF, Vitale N. J Biol Chem. 2007;282(30):21746–21757. doi: 10.1074/jbc.M702968200. [DOI] [PubMed] [Google Scholar]
- 79.Shen Y, Xu L, Foster DA. Mol Cell Biol. 2001;21(2):595–602. doi: 10.1128/MCB.21.2.595-602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koch T, Brandenburg LO, Schulz S, Liang Y, Klein J, Hollt V. J Biol Chem. 2003;278(11):9979–9985. doi: 10.1074/jbc.M206709200. [DOI] [PubMed] [Google Scholar]
- 81.Du G, Huang P, Liang BT, Frohman MA. Mol Biol Cell. 2004;15(3):1024–1030. doi: 10.1091/mbc.E03-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Padron D, Tall RD, Roth MG. Mol Biol Cell. 2006;17(2):598–606. doi: 10.1091/mbc.E05-05-0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koch T, Wu DF, Yang LQ, Brandenburg LO, Hollt V. J Neurochem. 2006;97(2):365–372. doi: 10.1111/j.1471-4159.2006.03736.x. [DOI] [PubMed] [Google Scholar]
- 84.Preininger AM, Henage LG, Oldham WM, Yoon EJ, Hamm HE, Brown HA. Mol Pharmacol. 2006;70(1):311–318. doi: 10.1124/mol.105.021451. [DOI] [PubMed] [Google Scholar]
- 85.Antonescu CN, Danuser G, Schmid SL. Mol Biol Cell. 2010;21(16):2944–2952. doi: 10.1091/mbc.E10-05-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ha KS, Exton JH. J Cell Biol. 1993;123(6 Pt 2):1789–1796. doi: 10.1083/jcb.123.6.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lukowski S, Lecomte MC, Mira JP, Marin P, Gautero H, Russo-Marie F, Geny B. J Biol Chem. 1996;271(39):24164–24171. doi: 10.1074/jbc.271.39.24164. [DOI] [PubMed] [Google Scholar]
- 88.Mor A, Wynne JP, Ahearn IM, Dustin ML, Du G, Philips MR. Mol Cell Biol. 2009;29(12):3297–3306. doi: 10.1128/MCB.00366-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Powner DJ, Payne RM, Pettitt TR, Giudici ML, Irvine RF, Wakelam MJ. J Cell Sci. 2005;118(Pt 13):2975–2986. doi: 10.1242/jcs.02432. [DOI] [PubMed] [Google Scholar]
- 90.Iyer SS, Agrawal RS, Thompson CR, Thompson S, Barton JA, Kusner DJ. J Immunol. 2006;176(6):3686–3696. doi: 10.4049/jimmunol.176.6.3686. [DOI] [PubMed] [Google Scholar]
- 91.Chae YC, Kim KL, Ha SH, Kim J, Suh PG, Ryu SH. Mol Cell Biol. 2010;30(21):5086–5098. doi: 10.1128/MCB.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shen Y, Zheng Y, Foster DA. Biochem Biophys Res Commun. 2002;293(1):201–206. doi: 10.1016/S0006-291X(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 93.Gomez-Cambronero J, Di Fulvio M, Knapek K. J Leukoc Biol. 2007;82(2):272–281. doi: 10.1189/jlb.0107033. [DOI] [PubMed] [Google Scholar]
- 94.Zouwail S, Pettitt TR, Dove SK, Chibalina MV, Powner DJ, Haynes L, Wakelam MJ, Insall RH. Biochem J. 2005;389(Pt 1):207–214. doi: 10.1042/BJ20050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mazie AR, Spix JK, Block ER, Achebe HB, Klarlund JK. J Cell Sci. 2006;119(Pt 8):1645–1654. doi: 10.1242/jcs.02858. [DOI] [PubMed] [Google Scholar]
- 96.Block ER, Klarlund JK. Mol Biol Cell. 2008;19(11):4909–4917. doi: 10.1091/mbc.E08-01-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Y, Rodrik V, Toschi A, Shi M, Hui L, Shen Y, Foster DA. J Biol Chem. 2006;281(23):15862–15868. doi: 10.1074/jbc.M600660200. [DOI] [PubMed] [Google Scholar]
- 98.Kim JH, Kim HW, Jeon H, Suh PG, Ryu SH. J Biol Chem. 2006;281(23):15747–15756. doi: 10.1074/jbc.M509844200. [DOI] [PubMed] [Google Scholar]
- 99.Lehman N, Di Fulvio M, McCray N, Campos I, Tabatabaian F, Gomez-Cambronero J. Blood. 2006;108(10):3564–3572. doi: 10.1182/blood-2006-02-005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagasaki A, Inotsume K, Kanada M, Uyeda TQ. Cell Struct Funct. 2008;33(1):27–33. doi: 10.1247/csf.07042. [DOI] [PubMed] [Google Scholar]
- 101.Carrigan SO, Pink DB, Stadnyk AW. J Leukoc Biol. 2007;82(6):1575–1584. doi: 10.1189/jlb.0806528. [DOI] [PubMed] [Google Scholar]
- 102.Pilquil C, Dewald J, Cherney A, Gorshkova I, Tigyi G, English D, Natarajan V, Brindley DN. J Biol Chem. 2006;281(50):38418–38429. doi: 10.1074/jbc.M601670200. [DOI] [PubMed] [Google Scholar]
- 103.Knapek K, Frondorf K, Post J, Short S, Cox D, Gomez-Cambronero J. Mol Cell Biol. 2010;30(18):4492–4506. doi: 10.1128/MCB.00229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Han L, Stope MB, de Jesus ML, Oude Weernink PA, Urban M, Wieland T, Rosskopf D, Mizuno K, Jakobs KH, Schmidt M. Embo J. 2007;26(19):4189–4202. doi: 10.1038/sj.emboj.7601852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chae YC, Kim JH, Kim KL, Kim HW, Lee HY, Heo WD, Meyer T, Suh PG, Ryu SH. Mol Biol Cell. 2008;19(7):3111–3123. doi: 10.1091/mbc.E07-04-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Itoh T, Hasegawa J, Tsujita K, Kanaho Y, Takenawa T. Sci Signal. 2009;2(87):ra52. doi: 10.1126/scisignal.2000393. [DOI] [PubMed] [Google Scholar]
- 107.Nishikimi A, Fukuhara H, Su W, Hongu T, Takasuga S, Mihara H, Cao Q, Sanematsu F, Kanai M, Hasegawa H, Tanaka Y, Shibasaki M, Kanaho Y, Sasaki T, Frohman MA, Fukui Y. Science. 2009;324(5925):384–387. doi: 10.1126/science.1170179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fallman M, Andersson R, Andersson T. J Immunol. 1993;151(1):330–338. [PubMed] [Google Scholar]
- 109.Kusner DJ, Hall CF, Schlesinger LS. J Exp Med. 1996;184(2):585–595. doi: 10.1084/jem.184.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Serrander L, Fallman M, Stendahl O. Inflammation. 1996;20(4):439–450. doi: 10.1007/BF01486745. [DOI] [PubMed] [Google Scholar]
- 111.Iyer SS, Barton JA, Bourgoin S, Kusner DJ. J Immunol. 2004;173(4):2615–2623. doi: 10.4049/jimmunol.173.4.2615. [DOI] [PubMed] [Google Scholar]
- 112.Corrotte M, Chasserot-Golaz S, Huang P, Du G, Ktistakis NT, Frohman MA, Vitale N, Bader MF, Grant NJ. Traffic. 2006;7(3):365–377. doi: 10.1111/j.1600-0854.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 113.Greco E, De Spirito M, Papi M, Fossati M, Auricchio G, Fraziano M. Biochem Biophys Res Commun. 2006;347(4):963–969. doi: 10.1016/j.bbrc.2006.06.186. [DOI] [PubMed] [Google Scholar]
- 114.Corrotte M, Nyguyen AP, Harlay ML, Vitale N, Bader MF, Grant NJ. J Immunol. 2010;185(5):2942–2950. doi: 10.4049/jimmunol.0903138. [DOI] [PubMed] [Google Scholar]
- 115.Kang JH, Shin I, Han JS. Exp Mol Med. 1998;30(1):21–27. doi: 10.1038/emm.1998.3. [DOI] [PubMed] [Google Scholar]
- 116.Lee SD, Lee BD, Han JM, Kim JH, Kim Y, Suh PG, Ryu SH. J Neurochem. 2000;75(3):1053–1059. doi: 10.1046/j.1471-4159.2000.0751053.x. [DOI] [PubMed] [Google Scholar]
- 117.Yamada M, Banno Y, Takuwa Y, Koda M, Hara A, Nozawa Y. Biochem J. 2004;378(Pt 2):649–656. doi: 10.1042/BJ20031398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Oh KJ, Lee SC, Choi HJ, Oh DY, Kim SC, Min do S, Kim JM, Lee KS, Han JS. J Cell Biochem. 2007;101(6):1409–1422. doi: 10.1002/jcb.21260. [DOI] [PubMed] [Google Scholar]
- 119.Taylor MM, Macdonald K, Morris AJ, McMaster CR. Febs J. 2005;272(19):5056–5063. doi: 10.1111/j.1742-4658.2005.04914.x. [DOI] [PubMed] [Google Scholar]
- 120.Lee SY, Kim JW, Jin JO, Song MG, Park JI, Min do S, Kwak JY. Biochem Biophys Res Commun. 2006;347(4):1039–1047. doi: 10.1016/j.bbrc.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 121.Kim J, Lee YH, Kwon TK, Chang JS, Chung KC, Min DS. Cancer Res. 2006;66(2):784–793. doi: 10.1158/0008-5472.CAN-05-1316. [DOI] [PubMed] [Google Scholar]
- 122.Carpio LC, Dziak R. Prostaglandins Leukot Essent Fatty Acids. 1998;59(2):101–109. doi: 10.1016/s0952-3278(98)90088-6. [DOI] [PubMed] [Google Scholar]
- 123.Lee CS, Bae YS, Lee SD, Suh PG, Ryu SH. Neurosci Lett. 2001;313(3):117–120. doi: 10.1016/s0304-3940(01)02233-9. [DOI] [PubMed] [Google Scholar]
- 124.Joseph T, Bryant A, Frankel P, Wooden R, Kerkhoff E, Rapp UR, Foster DA. Oncogene. 2002;21(22):3651–3658. doi: 10.1038/sj.onc.1205380. [DOI] [PubMed] [Google Scholar]
- 125.Ahn BH, Kim SY, Kim EH, Choi KS, Kwon TK, Lee YH, Chang JS, Kim MS, Jo YH, Min DS. Mol Cell Biol. 2003;23(9):3103–3115. doi: 10.1128/MCB.23.9.3103-3115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rodrik V, Zheng Y, Harrow F, Chen Y, Foster DA. Mol Cell Biol. 2005;25(17):7917–7925. doi: 10.1128/MCB.25.17.7917-7925.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rodrik V, Gomes E, Hui L, Rockwell P, Foster DA. FEBS Lett. 2006;580(24):5647–5652. doi: 10.1016/j.febslet.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kang DW, Lee JY, Oh DH, Park SY, Woo TM, Kim MK, Park MH, Jang YH, Min do S. Exp Mol Med. 2009;41(9):678–685. doi: 10.3858/emm.2009.41.9.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Di Fulvio M, Frondorf K, Gomez-Cambronero J. Cell Signal. 2008;20(1):176–185. doi: 10.1016/j.cellsig.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee JS, Kim IS, Kim JH, Cho W, Suh PG, Ryu SH. PLoS One. 2009;4(9):e7090. doi: 10.1371/journal.pone.0007090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nakashima S, Ohguchi K, Frohman MA, Nozawa Y. Biochim Biophys Acta. 1998;1389(3):173–177. doi: 10.1016/s0005-2760(97)00153-7. [DOI] [PubMed] [Google Scholar]
- 132.Di Fulvio M, Gomez-Cambronero J. J Leukoc Biol. 2005;77(6):999–1007. doi: 10.1189/jlb.1104684. [DOI] [PubMed] [Google Scholar]
- 133.Rujano MA, Pina P, Servitja JM, Ahumada AM, Picatoste F, Farres J, Sabria J. Biochem Biophys Res Commun. 2004;316(2):387–392. doi: 10.1016/j.bbrc.2004.02.057. [DOI] [PubMed] [Google Scholar]
- 134.Komati H, Naro F, Mebarek S, De Arcangelis V, Adamo S, Lagarde M, Prigent AF, Nemoz G. Mol Biol Cell. 2005;16(3):1232–1244. doi: 10.1091/mbc.E04-06-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sylvia VL, Schwartz Z, Del Toro F, DeVeau P, Whetstone R, Hardin RR, Dean DD, Boyan BD. Biochim Biophys Acta. 2001;1499(3):209–221. doi: 10.1016/s0167-4889(00)00120-8. [DOI] [PubMed] [Google Scholar]
- 136.Banno Y, Nemoto S, Murakami M, Kimura M, Ueno Y, Ohguchi K, Hara A, Okano Y, Kitade Y, Onozuka M, Murate T, Nozawa Y. J Neurochem. 2008;104(5):1372–1386. doi: 10.1111/j.1471-4159.2007.05085.x. [DOI] [PubMed] [Google Scholar]
- 137.Yoon MS, Chen J. J Cell Sci. 2008;121(Pt 3):282–289. doi: 10.1242/jcs.022566. [DOI] [PubMed] [Google Scholar]
- 138.Kim YR, Byun HS, Won M, Park KA, Kim JM, Choi BL, Lee H, Hong JH, Park J, Seok JH, Kim DW, Shong M, Park SK, Hur GM. BMC Cancer. 2008;8:144. doi: 10.1186/1471-2407-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kwun HJ, Lee JH, Min DS, Jang KL. FEBS Lett. 2003;544(1-3):38–44. doi: 10.1016/s0014-5793(03)00446-0. [DOI] [PubMed] [Google Scholar]
- 140.Jensen RE, Sesaki H. Nat Cell Biol. 2006;8(11):1215–1217. doi: 10.1038/ncb1106-1215. [DOI] [PubMed] [Google Scholar]
- 141.LaLonde M, Janssens H, Yun S, Crosby J, Redina O, Olive V, Altshuller YM, Choi SY, Du G, Gergen JP, Frohman MA. BMC Dev Biol. 2006;6:60. doi: 10.1186/1471-213X-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gregory P, Kraemer E, Zurcher G, Gentinetta R, Rohrbach V, Brodbeck U, Andres AC, Ziemiecki A, Butikofer P. Bone. 2005;37(2):139–147. doi: 10.1016/j.bone.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 143.Peng JF, Rhodes PG. Int J Dev Neurosci. 2000;18(6):585–589. doi: 10.1016/s0736-5748(99)00112-4. [DOI] [PubMed] [Google Scholar]
- 144.Brown HA, Gutowski S, Moomaw CR, Slaughter C, Sternweis PC. Cell. 1993;75(6):1137–1144. doi: 10.1016/0092-8674(93)90323-i. [DOI] [PubMed] [Google Scholar]
- 145.Cockcroft S, Thomas GM, Fensome A, Geny B, Cunningham E, Gout I, Hiles I, Totty NF, Truong O, Hsuan JJ. Science. 1994;263(5146):523–526. doi: 10.1126/science.8290961. [DOI] [PubMed] [Google Scholar]
- 146.Lu Z, Hornia A, Joseph T, Sukezane T, Frankel P, Zhong M, Bychenok S, Xu L, Feig LA, Foster DA. Mol Cell Biol. 2000;20(2):462–467. doi: 10.1128/mcb.20.2.462-467.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Han JM, Kim Y, Lee JS, Lee CS, Lee BD, Ohba M, Kuroki T, Suh PG, Ryu SH. Mol Biol Cell. 2002;13(11):3976–3988. doi: 10.1091/mbc.E02-02-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hess JA, Ross AH, Qiu RG, Symons M, Exton JH. J Biol Chem. 1997;272(3):1615–1620. doi: 10.1074/jbc.272.3.1615. [DOI] [PubMed] [Google Scholar]
- 149.Marcil J, Harbour D, Naccache PH, Bourgoin S. J Biol Chem. 1997;272(33):20660–20664. doi: 10.1074/jbc.272.33.20660. [DOI] [PubMed] [Google Scholar]
- 150.Min DS, Kim EG, Exton JH. J Biol Chem. 1998;273(45):29986–29994. doi: 10.1074/jbc.273.45.29986. [DOI] [PubMed] [Google Scholar]
- 151.Kim JH, Kim Y, Lee SD, Lopez I, Arnold RS, Lambeth JD, Suh PG, Ryu SH. FEBS Lett. 1999;454(1-2):42–46. doi: 10.1016/s0014-5793(99)00745-0. [DOI] [PubMed] [Google Scholar]
- 152.Frondorf K, Henkels KM, Frohman MA, Gomez-Cambronero J. J Biol Chem. 2010;285:15837–15847. doi: 10.1074/jbc.M109.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lehman N, Ledford B, Di Fulvio M, Frondorf K, McPhail LC, Gomez-Cambronero J. Faseb J. 2007;21(4):1075–1087. doi: 10.1096/fj.06-6652com. [DOI] [PubMed] [Google Scholar]
- 154.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Science. 2001;294(5548):1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- 155.Chen J, Fang Y. Biochem Pharmacol. 2002;64(7):1071–1077. doi: 10.1016/s0006-2952(02)01263-7. [DOI] [PubMed] [Google Scholar]
- 156.Gomez-Cambronero J. J Interferon Cytokine Res. 1995;15(10):877–885. doi: 10.1089/jir.1995.15.877. [DOI] [PubMed] [Google Scholar]
- 157.Goldstein BJ, Bittner-Kowalczyk A, White MF, Harbeck M. J Biol Chem. 2000;275(6):4283–4289. doi: 10.1074/jbc.275.6.4283. [DOI] [PubMed] [Google Scholar]
- 158.Zhao C, Du G, Skowronek K, Frohman MA, Bar-Sagi D. Nat Cell Biol. 2007;9(6):706–712. doi: 10.1038/ncb1594. [DOI] [PubMed] [Google Scholar]
- 159.Hancock JF. Nat Cell Biol. 2007;9(6):615–617. doi: 10.1038/ncb0607-615. [DOI] [PubMed] [Google Scholar]
- 160.Small JV, Stradal T, Vignal E, Rottner K. Trends Cell Biol. 2002;12(3):112–120. doi: 10.1016/s0962-8924(01)02237-1. [DOI] [PubMed] [Google Scholar]
- 161.Sorkin A. Biochem Soc Trans. 2001;29(Pt 4):480–484. doi: 10.1042/bst0290480. [DOI] [PubMed] [Google Scholar]
- 162.Mahankali M, Peng HJ, Cox D, Gomez-Cambronero J. Cell Signal. 2011;23(8):1291–1298. doi: 10.1016/j.cellsig.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang H, Schaff UY, Green CE, Chen H, Sarantos MR, Hu Y, Wara D, Simon SI, Lowell CA. Immunity. 2006;25(2):285–295. doi: 10.1016/j.immuni.2006.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Snapper SB, Meelu P, Nguyen D, Stockton BM, Bozza P, Alt FW, Rosen FS, von Andrian UH, Klein C. J Leukoc Biol. 2005;77(6):993–998. doi: 10.1189/jlb.0804444. [DOI] [PubMed] [Google Scholar]
- 165.Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW. PLoS Biol. 2006;4(2):e38. doi: 10.1371/journal.pbio.0040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Takenawa T, Suetsugu S. Nat Rev Mol Cell Biol. 2007;8(1):37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 167.Benesch S, Lommel S, Steffen A, Stradal TE, Scaplehorn N, Way M, Wehland J, Rottner K. J Biol Chem. 2002;277(40):37771–37776. doi: 10.1074/jbc.M204145200. [DOI] [PubMed] [Google Scholar]
- 168.Dovas A, Gevrey JC, Grossi A, Park H, Abou-Kheir W, Cox D. J Cell Sci. 2009;122(Pt 21):3873–3882. doi: 10.1242/jcs.051755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jones GE, Zicha D, Dunn GA, Blundell M, Thrasher A. Int J Biochem Cell Biol. 2002;34(7):806–815. doi: 10.1016/s1357-2725(01)00162-5. [DOI] [PubMed] [Google Scholar]
- 170.Kantonen S, Mahankali M, Hatton N, Park H, Cox D, Gomez-Cambronero J. Mol Cell Bio. 2011 doi: 10.1128/MCB.05684-11. Sep 19. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pochet S, Metioui M, Grosfils K, Gomez-Munoz A, Marino A, Dehaye JP. Cell Signal. 2003;15(1):103–113. doi: 10.1016/s0898-6568(02)00059-1. [DOI] [PubMed] [Google Scholar]
- 172.Min DS, Ahn BH, Jo YH. Mol Cells. 2001;11(3):369–378. [PubMed] [Google Scholar]
- 173.Ho WT, Xie Z, Zhao ZJ, Exton JH. Cell Signal. 2005;17(6):691–699. doi: 10.1016/j.cellsig.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 174.Banno Y, Ohguchi K, Matsumoto N, Koda M, Ueda M, Hara A, Dikic I, Nozawa Y. J Biol Chem. 2005;280(16):16319–16324. doi: 10.1074/jbc.M410903200. [DOI] [PubMed] [Google Scholar]
- 175.Yoon MS, Koo JB, Hwang JH, Lee KS, Han JS. FEBS Lett. 2005;579(25):5635–5642. doi: 10.1016/j.febslet.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 176.Choi WS, Hiragun T, Lee JH, Kim YM, Kim HP, Chahdi A, Her E, Han JW, Beaven MA. Mol Cell Biol. 2004;24(16):6980–6992. doi: 10.1128/MCB.24.16.6980-6992.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Blum JJ, Lehman JA, Horn JM, Gomez-Cambronero J. J Eukaryot Microbiol. 2001;48(1):102–110. doi: 10.1111/j.1550-7408.2001.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 178.Henkels KM, Short S, Peng HJ, Di Fulvio M, Gomez-Cambronero J. Biochem Biophys Res Commun. 2009;389(2):224–228. doi: 10.1016/j.bbrc.2009.08.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Henkels KM, Peng HJ, Frondorf K, Gomez-Cambronero J. Mol Cell Biol. 2010;30(9):2251–2263. doi: 10.1128/MCB.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Joseph T, Wooden R, Bryant A, Zhong M, Lu Z, Foster DA. Biochem Biophys Res Commun. 2001;289(5):1019–1024. doi: 10.1006/bbrc.2001.6118. [DOI] [PubMed] [Google Scholar]
- 181.Henkels KM, Frondorf K, Gonzalez-Mejia ME, Doseff AL, Gomez-Cambronero J. FEBS Lett. 2011;585(1):159–166. doi: 10.1016/j.febslet.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Henkels KM, Farkaly T, Mahankali M, Segall JE, Gomez-Cambronero J. J Mol Biol. 2011;408(5):850–862. doi: 10.1016/j.jmb.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Brown HA, Gutowski S, Kahn RA, Sternweis PC. J Biol Chem. 1995;270(25):14935–14943. doi: 10.1074/jbc.270.25.14935. [DOI] [PubMed] [Google Scholar]
- 184.Manser E, Chong C, Zhao ZS, Leung T, Michael G, Hall C, Lim L. J Biol Chem. 1995;270(42):25070–25078. doi: 10.1074/jbc.270.42.25070. [DOI] [PubMed] [Google Scholar]
- 185.Singer WD, Brown HA, Bokoch GM, Sternweis PC. J Biol Chem. 1995;270(25):14944–14950. doi: 10.1074/jbc.270.25.14944. [DOI] [PubMed] [Google Scholar]
- 186.Yamazaki M, Zhang Y, Watanabe H, Yokozeki T, Ohno S, Kaibuchi K, Shibata H, Mukai H, Ono Y, Frohman MA, Kanaho Y. J Biol Chem. 1999;274(10):6035–6038. doi: 10.1074/jbc.274.10.6035. [DOI] [PubMed] [Google Scholar]
- 187.Du G, Altshuller YM, Kim Y, Han JM, Ryu SH, Morris AJ, Frohman MA. Mol Biol Cell. 2000;11(12):4359–4368. doi: 10.1091/mbc.11.12.4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Powner DJ, Wakelam MJ. FEBS Lett. 2002;531(1):62–64. doi: 10.1016/s0014-5793(02)03410-5. [DOI] [PubMed] [Google Scholar]
- 189.Powner DJ, Hodgkin MN, Wakelam MJ. Mol Biol Cell. 2002;13(4):1252–1262. doi: 10.1091/mbc.01-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Schmidt M, Voss M, Weernink PA, Wetzel J, Amano M, Kaibuchi K, Jakobs KH. J Biol Chem. 1999;274(21):14648–14654. doi: 10.1074/jbc.274.21.14648. [DOI] [PubMed] [Google Scholar]
- 191.Lopez I, Arnold RS, Lambeth JD. J Biol Chem. 1998;273(21):12846–12852. doi: 10.1074/jbc.273.21.12846. [DOI] [PubMed] [Google Scholar]
- 192.Peng HJ, Henkels KM, Mahankali M, Marchal C, Bubulya P, Dinauer MC, Gomez-Cambronero J. Mol Cell Biol. 2011;31(11):2227–2240. doi: 10.1128/MCB.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dinauer MC. Curr Opin Hematol. 2003;10(1):8–15. doi: 10.1097/00062752-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 194.Peng HJ, Henkels KM, Mahankali M, Dinauer MC, Gomez-Cambronero J. J Biol Chem. 2011 doi: 10.1074/jbc.M110.206672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang B, Zheng Y. J Biol Chem. 1998;273(40):25728–25733. doi: 10.1074/jbc.273.40.25728. [DOI] [PubMed] [Google Scholar]
- 196.Takai Y, Sasaki T, Matozaki T. Physiol Rev. 2001;81(1):153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- 197.Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. Nature. 1995;376(6542):702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- 198.Aspenstrom P, Lindberg U, Hall A. Curr Biol. 1996;6(1):70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- 199.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. Nature. 1994;367(6458):40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 200.Burbelo PD, Drechsel D, Hall A. J Biol Chem. 1995;270(49):29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- 201.Martin GA, Bollag G, McCormick F, Abo A. Embo J. 1995;14(17):4385. doi: 10.1002/j.1460-2075.1995.tb00113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mahankali M, Peng HJ, Henkels KM, Dinauer MC, Gomez-Cambronero J. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1114692108. accepted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bui QT, Golinelli-Cohen MP, Jackson CL. Mol Genet Genomics. 2009;282(4):329–350. doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mitin N, Rossman KL, Der CJ. Curr Biol. 2005;15(14):R563–574. doi: 10.1016/j.cub.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 205.Rossman KL, Der CJ, Sondek J. Nat Rev Mol Cell Biol. 2005;6(2):167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 206.Hart MJ, Eva A, Evans T, Aaronson SA, Cerione RA. Nature. 1991;354(6351):311–314. doi: 10.1038/354311a0. [DOI] [PubMed] [Google Scholar]
- 207.Bokoch GM. Eur J Haematol. 1993;51(5):313–317. doi: 10.1111/j.1600-0609.1993.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 208.Bokoch GM, Bohl BP, Chuang TH. J Biol Chem. 1994;269(50):31674–31679. [PubMed] [Google Scholar]
- 209.Bokoch GM, Zhao T. Antioxid Redox Signal. 2006;8(9-10):1533–1548. doi: 10.1089/ars.2006.8.1533. [DOI] [PubMed] [Google Scholar]
- 210.Gomez-Cambronero J, Horn J, Paul CC, Baumann MA. J Immunol. 2003;171(12):6846–6855. doi: 10.4049/jimmunol.171.12.6846. [DOI] [PubMed] [Google Scholar]