Abstract
Peptidyl-prolyl cis/trans isomerases (PPIases) are enzymes assisting protein folding and protein quality control in organisms of all kingdoms of life. In contrast to the other sub-classes of PPIases, the cyclophilins and the FK-506 binding proteins, little was formerly known about the parvulin type of PPIase in Archaea. Recently, the first solution structure of an archaeal parvulin, the PinA protein from Cenarchaeum symbiosum, was reported. Investigation of occurrence and frequency of PPIase sequences in numerous archaeal genomes now revealed a strong tendency for thermophilic microorganisms to reduce the number of PPIases. Single-domain parvulins were mostly found in the genomes of recently proposed deep-branching archaeal subgroups, the Thaumarchaeota and the ARMANs (archaeal Richmond Mine acidophilic nanoorganisms). Hence, we used the parvulin sequence to reclassify available archaeal metagenomic contigs, thereby, adding new members to these subgroups. A combination of genomic background analysis and phylogenetic approaches of parvulin sequences suggested that the assigned sequences belong to at least two distinct groups of Thaumarchaeota. Finally, machine learning approaches were applied to identify amino acid residues that separate archaeal and bacterial parvulin proteins from each other. When mapped onto the recent PinA solution structure, most of these positions form a cluster at one site of the protein possibly indicating a different functionality of the two groups of parvulin proteins.
Keywords: archaeal protein, Pin1, PPIase, single-domain parvulin, Thaumarchaeota
Introduction
Cis/trans isomerisation of peptidyl-prolyl moieties within proteins can be regarded to be a molecular switch and is widely accredited as a means by which cell cycle events, protein (de)activation, folding and quality control are triggered. As cis/trans isomerisation is a relatively slow process under moderate temperature, organisms of all three kingdoms of life have developed or maintained enzymes, the peptidyl-prolyl cis/trans isomerases (PPIases), that accelerate this protein folding step.1,2 To date, three non-homologous families of PPIases are known: cyclophilins, FK506-binding proteins (FKBPs) and parvulins. Although cyclophilins and FKBPs have been analysed in several archaeal genomes and assigned chaperone activity in vitro, the actual cellular functions of these PPIases are not fully understood.1
In contrast to cyclophilins and FKBPs, archaeal parvulin sequences only became available in the last few years by completion of the two genomes from Nitrosopumilus maritimus3 and Cenarchaeum symbiosum,4 and by the deposition of three near-complete genomes of ultra-small acidophilic micro-archaea from biofilms from the Berkeley pit.5,6 In addition to these genomes, parvulin sequences are available from metagenomic studies using shotgun sequencing approaches on different samples: these were samples from fresh- and surface water7,8 and samples collected from iron mines.9 This last study described the archaeal Richmond Mine acidophilic nanoorganisms (ARMANs), acidophilic ultra small nano-archaea, which are frequent hosts of replicating viruses.10 These microorganisms seem to build a clade at the bottom of the euryarchaeal branch6 and are therefore annotated as “undefined Euryarchaeota” in NCBI databases.
The group of mesophilic Crenarchaeota was recently recognised as a new deep-branching phylum, the Thaumarchaeota.11 Recently, Nitrosphaera gargensis has been described as the first moderately thermophilic thaumarchaeal species based on its 16S ribosomal DNA sequence.12 There have been several studies that added few fosmids or metagenomic contigs to the phylum,13–15 however, the resolution within the thaumarchaeal cluster remained poor.
The cellular function of archaeal parvulins has not yet been studied in detail. Recently, the first structure of an archaeal parvulin, PinA of Cenarcheaeum symbiosum, a psychrophilic organism living in symbiosis with the marine sponge Axiella mexicana has been solved.16 In the course of characterising structure and cellular function of archaeal single-domain parvulins (sdPars), we studied the occurrence and frequency of PinA proteins in different clades of the archaeal kingdom by a comparative genomics-based approach and defined by machine learning algorithms decisive structural features that separate bacterial and archaeal single-domain parvulins.
Results and Discussion
Distribution of prolyl isomerases in Archaea
We searched all available completely or nearly fully sequenced archaeal genomes for their content in prolyl isomerases (PPIases) to establish a basis for further phylogenetic analyses. A total of 98 genomes was investigated that can be grouped into 17 different groups including 13 established orders and the four recently proposed groups ARMANs,9 Thaumarchaeota,11 Korarchaeota17 and Nanoarchaeota.18 Their content in FK506 binding proteins (FKBPs), small and large cyclophilins, and parvulins is listed in Table 1; a detailed listing is given in Supplementary Table 1.
Table 1.
Occurrence of prolyl isomerases in archaeal genomes.
| Kingdom | Order | # | FKBPs |
Cyclophilins |
Single-domain parvulins | Temperature optimum | |
|---|---|---|---|---|---|---|---|
| Single-domain | Multi-domain | ||||||
| Crenarchaeota | Desulfurococcales | 8 | 1 per genome | None | None | None | (Hyper) thermophilic |
| Crenarchaeota | Thermoproteales | 8 | 1 per genome | None | None | None | (Hyper) thermophilic |
| Crenarchaeota | Sulfolobales | 12 | 1 per genome | None | None | None | (Hyper) thermophilic |
| Euryarchaeota | Archaeoglobales | 3 | 1 per genome | None | None | None | (Hyper) thermophilic |
| Euryarchaeota | Thermoplasmatales | 4 | 1 per genome | 1 in Picrophilus 1 in Ferroplasma none in Thermoplasma |
None | None | thermophilic |
| Euryarchaeota | Thermococcales | 9 | 1 in Pyrococcus 2 in Thermococcus |
None | None | None | Pyrococcus: hyperthermophilic; Thermococcus: thermophilic |
| Euryarchaeota | Halobacteriales | 13 | 1–3 per genome | 1 per genome | None | None | Mesophilic/moderatly thermophilic |
| Euryarchaeota | Methanococcales | 12 | 2 in Methanocaldococcus; 2–3 in Methanococcus |
None in Methanocaldococcus; 0–2 in Methanococcus |
None | None | Methanococcus: meophilic Methanocaldococcus: thermophilic |
| Euryarchaeota | Methanocellales | 1 | 4 | 1 | None | None | Mesophilic |
| Euryarchaeota | Methanopyrales | 1 | 1 | None | None | None | thermophilic |
| Euryarchaeota | Methanosarcinales | 7* | 4 in Methanosarcina; 4 in Methanohalophilus; 1 in Methanosaeta concilii; 2–3 in the other genomes |
1 per genome; 2 in Methanosaeta concilii | None | none; 1 in Methanosaeta concilii | Mesophilic |
| Euryarchaeota | Methanobacteriales | 8 | 1 per genome | 1 per genome | None | none | Mesophilic |
| Euryarchaeota | Methanomicrobiales | 6 | 1–4 per genome | 1–2 per genome | None | 1 in 4 genomes none in rest |
Mesophilic |
| Euryarchaeota | ARMAN | 3 | 2 in Micrarchaeum; 3 in Parvarchaeum |
None | None | 1 in two genomes none in 1 genome |
Mesophilic |
| Thaumarchaeota | – | 3* | 1 per genome | 1 per genome | 1 per genome; N. limnia seems to have none |
1 per genome | Mesophilic/psychrophilic |
| Korarchaeota | – | 1 | 1 | None | None | None | (Hyper) thermophilic |
| Nanoarchaeota | – | 1 | None | None | None | None | (Hyper) thermophilic |
| Caldiarchaeum | 1* | 1, 2 or 3 (ambiguously annotated) | None | None | None | (Hyper) thermophilic | |
Notes: Refer to Supplementary Table 1 for details.
Please note that additional genome sequences became available very recently.
This comparison reveals that some PPIase subfamilies do not exist in Archaea: In contrast to bacterial and eukaryotic organisms, large multidomain FKBPs, multidomain cyclophilins (except Thaumarchaeota) and multidomain parvulins are absent in any archaeal genome sequenced so far. The only ubiquitous class of PPIases in Archaea are single-domain FKBPs except in Nanoarchaeum equitans which is special because of its dependency on Ignicoccus. While in all examined non-thaumarchaeal genomes, cyclophilins consist of a single PPIase domain of about 160 amino acids, the two known thaumarchaeal genomes of Nitrosopumilus maritimus and Cenarchaeum symbiosum additionally contain a protein with more than 500 amino acids carrying an N-terminal cyclophilin domain. No other conserved domains are found for the 545 aa C. symbiosum protein in a CDD search.19 In the 509 aa N. maritimus protein, there is a fragment of a putative Zn-dependent protease [CDD:COG4784] located in the middle of the protein. Although the function of this protein remains unclear, it separates the two thaumarchaeal species from the rest of the Archaea by its mere existence.
From Table 1, a certain tendency for the content of PPIases can be inferred: The higher the preferred growth temperature, the lower the content of PPIases in the genome. All species of the (hyper-)thermophilic Crenarchaeota contain only one FKBP-type PPIase per genome. The same applies to the thermophilic Korarchaeum cryptophilum and the hyperthermophilic euryarchaeal orders Archaeoglobales and Methanopyrales. Strikingly, even different species from the same orders differ in their PPIase content depending on their favoured temperature range. The Methanococcales include the hyperthermophilic Methanocaldococcus and the mesophilic Methanococcus species. Whereas the hyperthermophilic species contain only two FKBPs, mesophilic species additionally contain one or two cyclophilins or a third FKBP (eg, Methanococcus aeolicus Nankai-3). Cold-adapted microorganisms contain more than three prolyl isomerases eg, four PPIases are found in the psychrophilic archaeon Cenarchaeum symbiosum.16
Although there are exceptions from the ‘rule’ when going to lower temperatures, the correlation itself is not surprising and was previously suggested.1 At higher growth temperatures spontaneous Xaa-Pro bond isomerisation becomes faster and, hence, less enzymatic assistance in this process is needed. Although hyperthermophilic Archaea have reduced their PPIase repertoire to only one FKBP per genome, at least one isomerase seems to be absolutely crucial for them. Assuming nearly no difference between spontaneous and catalysed cis/trans isomerisation at elevated temperatures,1 this protein may serve a function other than being a PPIase.
In contrast to the total arsenal of PPIases, parvulin-type enzymes can only be found in three phyla including mesophilic microorganisms: the euryarchaeal Methanomicrobiales, the archaeal Richmond Mine acidophilic nanoorganisms (ARMAN) and the Thaumarchaeota. All Crenarchaeota, Korarchaeota and Nanoarchaeota species sequenced to date lack parvulin genes completely (Supplementary Table 1). Also very recent additions to the list of available archaeal genomes do not change this situation: The genomic sequence of another archaeon (Candidatus Caldiar-chaeum subterraneum) that was classified somewhere between Euryarchaeota and Crenarchaeota, does not contain any parvulins.20 On the other hand, a third thaumarchaeal genome (Candidatus Nitrosoarchaeum limnia SFB1) that was recently released,21 contains a single-domain parvulin highly similar to the parvulin from Nitrosopumilus maritimus (85% identity on the level of amino acids). Thirdly, the genome of a Methanosarcina species (Candidatus Methanosaeta concilii GP-6) containing a single-domain parvulin [NCBI RefSeq NC_015416.1] may indicate that the occurrence of parvulin coding sequences within the group of Euryarchaeota is not strictly confined to Methanomicrobiales.
Whereas only 9 percent of the 65 genomes of Euryarchaeota available at the time of analysis—including the above mentioned ARMAN and Methanomicrobiales—possess a parvulin gene, the two known thaumarchaeal genomes both contain exactly one parvulin gene. This parvulin comprises a single domain with a molecular weight of about 10 kDa. We refer to this class of parvulins as single-domain parvulins (sdPar). Single-domain parvulins are absent from eukaryotic genomes. It could be that the compartmentalised Eukarya need parvulin proteins with additional domains for cellular targeting, protein binding or anchoring like it is the case for the two human representatives Pin122,23 and Par14/17.24–27 In contrast to Eukarya, many bacterial genomes contain single-domain parvulins. With the exception of Lentisphaera araneosa and two Planctomycetes, all known parvulin-containing Bacteria belong to the subgroup of Proteobacteria. Most of these genomes possess a single sdPar; some species contain two, and only the extreme psychrophilic species Colwellia psychrerythraea (Alteromonadales) contains three sdPar-type parvulin genes, which again suggests a relationship between temperature and PPIase content.
Of note, we found bacterial multi-domain proteins of the PrsA type containing parvulin domains very similar to sdPars. However, these paralogous sequences were excluded from further analysis because no corresponding multi-domain parvulin protein sequences could be found in any archaeal genome.
Genomic context of archaeal parvulins
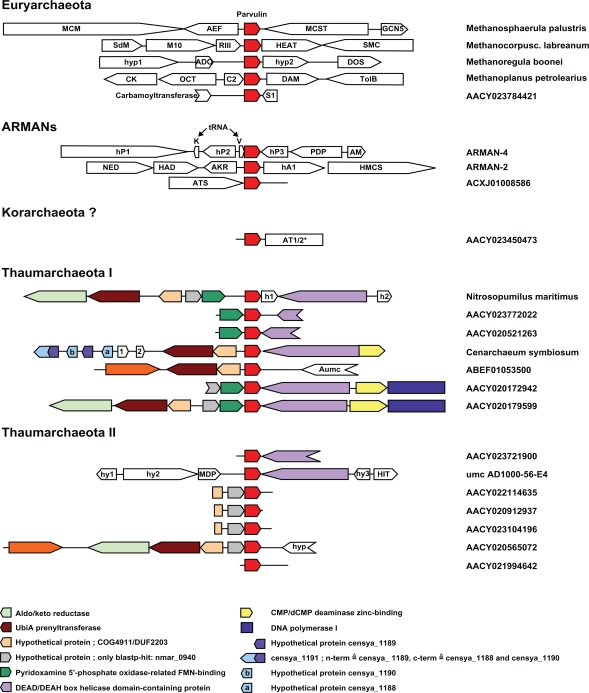
In order to characterise the relationship among archaeal parvulins, we examined the genomic context of the different parvulin loci in Archaea for conserved structures. In a first step, the genomic context of the parvulin locus was examined in the fully sequenced archaeal genomes. No conserved gene organisation was found within the six available genomes from the Methanomicrobiales and the ARMAN group (Fig. 1, upper half). In contrast, the two deposited thaumarchaeal genomes of Nitrosopumilus maritimus and Cenarchaeum symbiosum have an antisense DEAD/DEAH-box helicase containing protein (DHCP) downstream of the parvulin reading frame. The Nitrosopumilus genome carries an inserted hypothetical protein between parvulin and DHCP, which is missing in the Cenarchaeum genome. In 5′ direction from parvulin, both genomes contain two genes in opposite orientation, an UbiA prenyltransferase and a hypothetical protein with a conserved domain of unknown function DUF2203. In contrast to Cenarchaeum, the Nitrosopumilus genome has two more proteins inserted in between: the hypothetical protein nmar_0940 and a pyridoxamine 5′-phosphate oxidase-related FMN-binding protein (PPOX). Taken together, no conserved putative operons could be detected in the investigated genomes (Fig. 1, lower half).
Figure 1.
Genomic context analysis of the archaeal parvulin locus. The genetic background was analysed as described in the main text. White-backed arrows indicate genes occurring only once. Other colour codes are indicated within the figure. All abbreviations in this schematic are given below. 12 parvulin containing contigs starting with “AACY” from the Sorcerer II voyage,49 ACXJ01008586 from samples collected from thick floating biofilms in the Richmond Mine.6 ABEF01053500 from a subtropical gyre in 4000 m depth was deposited by Ed DeLong and colleagues from the Hawaiian research station ALOHA. The parvulin containing fosmid AD1000-56-E4 derives from plankton collected in 1000 m depth in the Adriatic Sea.8 The metagenomic contigs AACY023784421 and ACXJ01008586 were added to their groups on basis of their parvulin primary sequence, for ACXJ01008586 this stands in agreement with its origin. The marine metagenomic contig AACY023450473 contains 341 amino acids of the N-terminus of an aminotransferase class I/II. This sequence shows highest similarity to the protein YP_001737635 from Candidatus Korarchaeum cryptofilum OPF8 (39 percent amino acids identity over 331 amino acids). Hence, this contig may belong to the Korarchaeota. AACY023772022, AACY020521263, AACY020172942 and AACY020179599 were clustered with Nitrosopumilus maritimus, because they share the same PPOX gene preceding parvulin. Comparably, ABEF01053500 was grouped with Cenarchaeum symbiosum due to an antisense hypothetical protein DUF2203 preceding the parvulin gene. With 80% sequence identity of their parvulin proteins, Nitrosopumilus maritimus and Cenarchaeum symbiosum were grouped together in Thaumarchaeota I. AACY022114635, AACY020912937, AACY023104196 and AACY020565072 were also clustered due to the gene directly upstream of parvulin, the hypothetical protein homologous to nmar_0940. AACY021994642 was also grouped with these contigs because of its high parvulin primary sequence similarity. AACY023721900 and umc-AD1000-56-E4 have a totally different upstream region, but they can clearly been classified as Thaumarchaeota II due to the typical downstream DHCP reading frame and their parvulin primary sequence.
Abbreviations: 1, hypothetical protein censya_1187 (Cenarchaeum specific), [GeneID: 6371367], Cenarchaeum symbiosum A, [Ref.Seq.: NC_014820.1]; 2, hypothetical protein censya_1186 (Cenarchaeum specific), [GeneID: 6371366], Cenarchaeum symbiosum A, [Ref.Seq.: NC_014820.1]; ADC, acetolactate decarboxylase, [GeneID: 5411158], Methanoregula boonei 6A8, [Ref.Seq.: NC_009712.1]; AEF, auxin efflux carrier, [GeneID: 7271583], Methanosphaerula palustris E1-9c, [Ref.Seq.: NC_011832.1]; AKR, adenylate kinase related protein, [GenBank: EET90508.1], Candidatus Micrarchaeum acidiphilum ARMAN-2, [GenBank: GG697236.1]; AM, antibiotic biosynthesis monooxygenase, [GenBank: EEZ92921.1], Candidatus Parvarchaeum acidiphilum ARMAN-4, [GenBank: GG730045.1]; AT1/2, aminotransferase class I and II, n.a., n.a., [GenBank: AACY023450473.1]; ATS, asparagyl-tRNA-synthetase, n.a., n.a., [GenBank: ACXJ01008586.1]; Aumc, hypothetical protein (uncultured marine crenarchaeota-umc specific), n.a., n.a., [GenBank: ABEF01053500.1]; C2, cupin 2 conserved barrel, [GeneID: 9742629], Methanoplanus petrolearius DSM 11571, [Ref.Seq.: NC_014507.1]; CK, carbamate kinase, [GeneID: 9742627], Methanoplanus petrolearius DSM 11571, [Ref.Seq.: NC_014507.1]; DAM, DNA adenine methylase, [GeneID: 9742631], Methanoplanus petrolearius DSM 11571, [Ref.Seq.: NC_014507.1]; DOS, dihydropterate synt, [GeneID: 5411815], Methanoregula boonei 6A8, [Ref. Seq.: NC_009712.1]; GCN5, GCN5-related N-acetyltransferase, [GeneID: 7271586], Methanosphaerula palustris E1-9c, [Ref.Seq.: NC_011832.1]; h1, hypothetical protein nmar_0943 (Nitrosopumilus specific), [GeneID: 5773171], Nitrosopumilus maritimus SCM1, [Ref.Seq.: NC_010085.1]; h2, hypothetical protein nmar_0945 (Nitrosopumilus specific), [GeneID: 5774572], Nitrosopumilus maritimus SCM1, [Ref.Seq.: NC_010085.1]; hA1, hypothetical protein UNLARM2_0040 (ARMAN specific), [GenBank: EET90506.1], Candidatus Micrarchaeum acidiphilum ARMAN-2, [GenBank: GG697236.1]; HAD, HAD-superfamily hydrolase, [GenBank: EET90509.1], Candidatus Micrarchaeum acidiphilum ARMAN-2, [GenBank: GG697236.1]; HEAT, HEAT-repeat containing protein, [GeneID: 4795375], Methanocorpusculum labreanum Z, [Ref.Seq.: NC_008942.1]; HIT, histidin triade protein, [GenBank: ACF09643.1], uncultured marine crenarchaeote AD1000-56-E4, [GenBank: EU686623.2]; HMCS, 3-hydroxy-3-methylglutaryl CoA synthase, [GenBank: EET90505.1], Candidatus Micrarchaeum acidiphilum ARMAN-2, [GenBank: GG697236.1]; hP1, hypothetical protein BJBARM4_0439 (parvarchaeum specific), [GenBank: EEZ92926.1], Candidatus Parvarchaeum acidiphilum ARMAN-4, [GenBank: GG730045.1]; hP2, hypothetical protein BJBARM4_0438 (parvarchaeum specific), [GenBank: EEZ92925.1], Candidatus Parvarchaeum acidiphilum ARMAN-4, [GenBank: GG730045.1]; hP3, hypothetical protein BJBARM4_0436 (parvarchaeum specific), [GenBank: EEZ92923.1], Candidatus Parvarchaeum acidiphilum ARMAN-4, [GenBank: GG730045.1]; hy1, hypothetical protein (specific for Thaumarchaeota and umcs), [GenBank: ACF09649.1], uncultured marine crenarchaeote AD1000-56-E4, [Gen-Bank: EU686623.2]; hy2, hypothetical protein (specific for Thaumarchaeota and umcs), [GenBank: ACF09648.1], uncultured marine crenarchaeote AD1000-56-E4, [GenBank: EU686623.2]; hy3, hypothetical protein (umc specific), [GenBank: ACF09644.1], uncultured marine crenarchaeote AD1000-56-E4, [GenBank: EU686623.2]; hyp, hypothetical protein with putative conserved domain DUF726, n.a., n.a., [GenBank: AACY020565072.1]; hyp1, hypothetical protein MBOO_0211 (no blastp-hit), [GeneID: 5411157], Methanoregula boonei 6A8, [Ref.Seq.: NC_009712.1]; hyp2, hypothetical protein MBOO_0214 (no blastp-hit), [GeneID: 5411814], Methanoregula boonei 6A8, [Ref.Seq.: NC_009712.1]; M10, methan mark 10, [GeneID: 4795594], Methanocorpusculum labreanum Z, [Ref.Seq.: NC_008942.1]; MCM, MCM family protein, [GeneID: 7271582], Methanosphaerula palustris E1-9c, [Ref.Seq.: NC_011832.1]; MCST, methyl-accepting chemotaxis sensory transducer with Pas/Pac sensor, [GeneID: 7271585], Methanosphaerula palustris E1-9c, [Ref.Seq.: NC_011832.1]; MDP, metal-dependent protease (COG 1310), [GenBank: ACF09647.1], uncultured marine crenarchaeote AD1000-56-E4, [GenBank: EU686623.2]; NED, NAD-dependent epimerase/dehydratase, [GenBank: EET90510.1], Candidatus Micrarchaeum acidiphilum ARMAN-2, [GenBank: GG697236.1]; OCT, ornithine carbamoyltransferase, [GeneID: 9742628], Methanoplanus petrolearius DSM 11571, [Ref.Seq.: NC_014507.1]; PDP, pirin-domain protein, [GenBank: EEZ92922.1], Candidatus Parvarchaeum acidiphilum ARMAN-4, [GenBank: GG730045.1]; RIII, ribonuclease III, [GeneID: 4795607], Methanocorpusculum labreanum Z, [Ref.Seq.: NC_008942.1]; S1, S1-tex like protein, n.a., n.a., [GenBank: AACY023784421.1]; SdM, SAM dependent methyltransferase, [GeneID: 4795613], Methanocorpusculum labreanum Z, [Ref.Seq.: NC_008942.1]; SMC, SMC-domain containing protein, [GeneID: 4795178], Methanocorpusculum labreanum Z, [Ref.Seq.: NC_008942.1]; TolB, TolB-like protein, [GeneID: 9742632], Methanoplanus petrolearius DSM 11571, [Ref.Seq.: NC_014507.1].
To extend this analysis to more archaeal sequences than the eight fully sequenced genomes mentioned above, the NCBI databases whole genome shotgun reads (wgs) and environmental samples (env_nt) were searched for contiguous sequences (contigs) containing single-domain parvulins using tBLASTn with the known archaeal parvulins as queries. This search yielded 14 additional sequences with sizes between 797 and 7533 bases (Fig. 1, lower half). Additionally, an annotated fosmid AD1000-56-E4 (35.5 kb) was found carrying an sdPar gene when searching the non-redundant protein sequences (nr) database by BLASTp using the N. maritimus sdPar as query.
In all fully sequenced archaeal organisms, only one parvulin gene was found per genome. Consequently, all newly found parvulin-containing contigs were treated as belonging to different (uncultivated) organisms and their genomic context was analysed as described. In eight of the metagenomic contigs the parvulin gene is found in close proximity to upstream genes (Fig. 1). In four contigs, a pyridoxamine 5′-phosphate oxidase-related FMN-binding protein (PPOX) is in direct neighbourhood to parvulin. Two of these four contigs also contain the same partial hypothetical protein 5′ to parvulin. This open reading frame is in direct vicinity to the parvulin locus in four other contigs, which lack the PPOX gene. Although the genome of Nitrosopumilus maritimus contains the reading frames for hypothetical protein nmar_0940, PPOX and sdPar (in this order), co-transcription is not likely there, because of an intergenic gap of more than 300 bases between parvulin and the PPOX gene. Hence, although the PPOX-gene could be co-transcribed with the parvulin in four contigs, this putative operon is most likely not conserved in all PPOX-containing organisms. Even though these findings give some hints for a polycistronic transcription including the parvulin message, it is not possible to make functional statements as one of the two found proteins is a hypothetical protein of unknown function.
Clustering of the highly conserved archaeal single-domain parvulins
Besides the search for putative operons, a combination of genomic context analysis and comparison of the parvulin primary sequence suggested a grouping of the metagenomic contigs into different clusters indicated in Figure 1 that we refer to as Thaumarchaeota I and II. Genomes and contigs were added to the group of putative Thaumarchaeota I when either sharing a PPOX gene immediately upstream of the parvulin sequence (similar to the N. maritimus sequence) or the hypothetical protein DUF2203 (related to the C. symbiosum sequence). All other contigs comprising parvulin primary sequences homologous to N. maritimus or C. symbiosum were added to the second group of presumed Thaumarchaeota II. Nevertheless, all these sequences may belong to the formerly proposed group I.1a of Thaumarchaeota11 due to their overall similarity and their common oceanic origin.
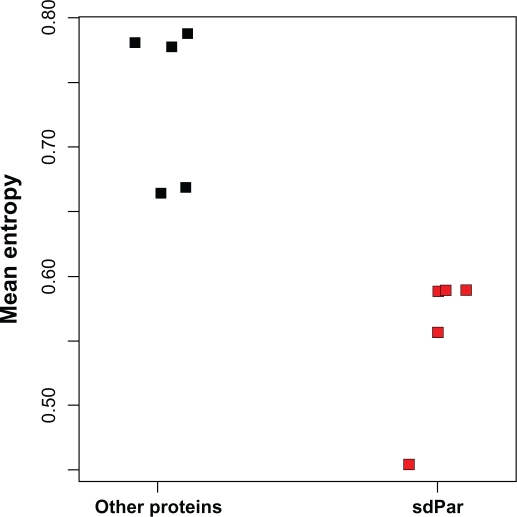
Analysis of genomic context from fully sequenced archaeal genomes and metagenomic data has now yielded archaeal parvulin sequences in 23 different genomic environments. This allowed us to compare the diversity of sdPar amino acid sequences with that of other proteins. Therefore, we computed the sequence entropy, averaged over all sequence positions, for multiple sequence alignments of sdPars and multiple sequence alignments of proteins UbiA, DUF, hyp, PPOX, and DHCP from the corresponding organisms. Figure 2 shows that the mean sequence entropy of sdPars is significantly lower than that of other proteins from corresponding organisms (P = 0.01 in Mann-Whitney test). Thus, single-domain parvulins were found to be significantly more conserved than their genomic neighbours.
Figure 2.
Mean Shannon entropy of the archaeal parvulin and its genomic neighbours. At the parvulin locus of N. maritimus, we found the following neighbouring proteins to be present in at least 5 different contigs: UbiA, DUF, hyp, PPOX, and DHCP. The mean Shannon entropy (unit: bit) of these sequences was calculated as a measure of sequence diversity and compared with the same measure for sdPar from the corresponding organisms.
Abbreviations: UbiA, UbiA prenyltransferase; DUF, hypothetical protein of unknown function (COG4911/DUF2203); hyp, hypothetical protein nmar_0940; PPOX, pyridoxamine 5′-phosphate oxidase-related FMN-binding protein; Par, parvulin; DHCP, DEAD/DEAH box containing protein.
Relationship among archaeal parvulins
The high sequence conservation of sdPar proteins in Archaea tempted us to find out whether this short protein sequence (around 100 amino acids) allows the calculation of reasonable maximum likelihood phylogeny (MLP). Although parvulin cannot substitute for established phylogenetic markers like small subunit RNA, it can probably deliver valuable insights into the relationship within recently proposed archaeal subgroups.
All available single-domain parvulins of Bacteria and Archaea were collected to generate an initial dataset of 326 sdPar sequences. Next, 59 bacterial paralogous sequences were removed, identified as such by calculating trees with the unfiltered dataset and comparing the results with a tree based on small subunit ribosomal RNA.28 Additionally, 26 parvulin sequences were filtered out from bacterial orders where less than 50 percent of fully or near fully sequenced genomes contained a single-domain parvulin that may have emerged from horizontal gene transfer (HGT). Trimming resulted in a final data-set of 241 sequences (218 bacterial genomes, eight archaeal genomes, one fosmid and 14 metagenomic contigs), which were aligned with T-Coffee29 and used to calculate maximum likelihood phylogeny with PhyML.30 Supplementary Figure 1 shows the resulting tree that was evaluated by 1000 bootstraps and rooted by Escherichia coli SlyD, an FKBP with structural analogy to the parvulin fold but without any sequential similarities.2,31,32
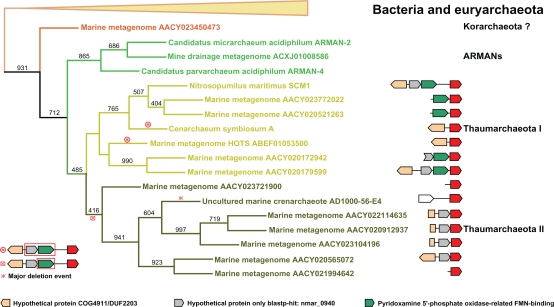
Please note that the procedure described so far has assumed all single-domain parvulins to be monophyletic and under-estimates horizontal gene transfer (HGT) events. However, a recent paper33 has revealed that HGTs have been frequent events within marine uncultured planktonic archaea that Thaumarchaeota are part of. This paper also assigns a bacterial origin to the sdPar sequence of the archaeal fosmid AD1000-56-E4. Hence, we further concentrated on the relationships within the smaller archaeal sub-tree excluding all euryarchaeal sequences (Fig. 3). For this sub-tree to be monophyletic we have three main indications: (1) the respective sequences form a cluster set apart from other sdPar sequences by high bootstrap values (931/1000); (2) when using NmPinA or CsPinA sequences as seed for BLASTp searches within the nr database, the thaumarchaeal sdPars always constitute the very first hits with very small e values and (3) their genomic context is highly conserved.
Figure 3.
Archaeal branch of an MLP tree combined with genomic context. The figure displays an expanded section of the maximum likelihood phylogeny tree from Figure 3. The outgroup has been omitted and the whole bacterial clade has been collapsed for clarity. Red signs indicate three deletion events suggested by the genomic context: The deletion of the hyp0940 and PPOX genes is described in the main text. The putative PPOX deletion seems to be a basal event for the Thaumarchaeota II subgroup. The large genetic rearrangement concerning the uncultured marine Crenarchaeota fosmid AD1000-56-E4 makes this sequence unique in the group of Thaumarchaeota. Next to the Thaumarchaeota, the corresponding parvulin loci with the available genomic contexts are displayed. The groups predicted from the genomic context are also well defined in the MLP tree.
Hence, it seems feasible to use single-domain parvulins as marker to detect and reclassify novel members of the recently proposed archaeal phyla of ARMANs and Thaumarchaeota. As one of the above mentioned metagenomic contigs (AACY023784421) does not show similarity to any of the ARMAN or thaumarchaeal sequences in its genomic context and its sdPar sequence clusters more with the four annotated Methanomicrobiales, this sequence may be of euryarchaeal origin.
The parvulin sequence from the marine metagenomic contig AACY023450473 is different from all other sdPar sequences in the MLP tree. This contig contains a putative korarchaeal aminotransferase downstream of the sdPar reading frame (Fig. 1) and hence may be classified as a korarchaeal sequence. The respective microorganism may be the first mesophilic to moderately thermophilic korarchaeon as all archaeal parvulins found until now are from mesophilic species. Alternatively, it could be the first parvulin of a thermophilic species, as all Korarchaeota known are (hyper-)thermophilic. As this contig was from marine surface water samples, the first interpretation seems to be more likely. In either case, the existence of a parvulin within a korarchaeal genome supports that Korarchaeota may be genetic hybrids between Euryarchaeota and Crenarchaeota.17
The sdPar sequences from all available ARMAN species are as a group highly similar to the respective proteins from Thaumarchaeota. This may indicate a closer relationship of ARMANs to Thaumarchaeota than to other Archaea. This would be in agreement with a recently reported 16s ribosomal RNA tree including the ARMANs where they branch very early from the euryarchaeal clade;6 but thaumarchaeal sequences were not enclosed in that tree. The ARMAN parvulins are similarly conserved as the thaumarchaeal within their group (score 69.7 and score 70.2, respectively), but unlike the Thaumarchaeota, they have no similarities to each other in their genomic context at all. Whether the ARMANs form another clade like Thaumarchaeota, Nanoarchaeota and Korarchaeota, or whether they belong to one of these clades remains to be elucidated. However, the comparison of the contained parvulin sequences makes a euryarchaeal annotation (“undefined Euryarchaeota”) of the group of extremely acidophilic organisms (ARMANs) rather unlikely.
The thaumarchaeal parvulin sequences are highly interrelated. This is in agreement with the recent acceptance of this group as a secluded deep branching phylum.11 Analysis of the genomic context suggests some major genetic rearrangements within this group. A comparison of the fully sequenced genomes of this group reveals a deletion of the hyp0940 and PPOX genes, present in N. maritimus, from the genome of the psychrophilic C. symbiosum.
Based solely on parvulin’s primary protein sequence, we were able to properly group the metagenomic contig AACY021994642 with only 804 bases, which is little more than half as long as the small ribosomal subunit RNA of N. maritimus (1409 bases). The other contigs of similar size, AACY023104196 (942 bases), AACY020912937 (797 bases) and AACY022114635 (989 bases), also contained information about the genomic context and could therefore be determined more reliably.
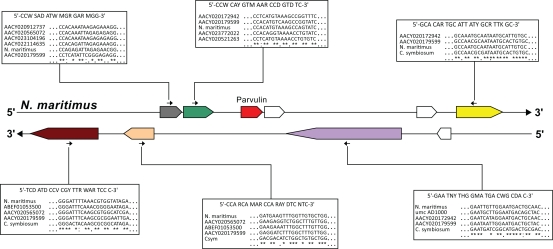
Our parvulin-based assignment of metagenomic contigs to distinct archaeal subgroups is of particular interest as it adds putative new members to the recently proposed phylum of Thaumarchaeota. Little is known today about the dimension, the diversity and the evolutionary relationships within this phylum, in spite of its important role in geochemical cycles in all marine surface waters on this planet. Using not only a single protein as a phylogenetic marker, but also its whole genomic locus including a variety of different and alternating genes gives additional opportunities of deepening the understanding of the phylum of Thaumarchaeota. One example how the toolset we deliver could be used in further studies is shown in Figure 4. Based on our analysis of the genomic context of parvulins, we propose sensible primers, which could be helpful for further studies. All proposed primers in neighbouring coding regions are within a 3 kb distance to the parvulin gene.
Figure 4.
Proposed primers for further metagenomic analyses. To get these primers the nucleotide sequences of the parvulin surrounding genes have been aligned with ClustalW. Several requirements (length between 18 and 24 bases, average GC content over 40%, average salt adjusted melting temperature between 50 °C and 65 °C) has been applied. The resulting primers for genes surrounding the thaumarchaeal parvulin are shown in this figure. For positions that were ambiguous, the respective IUPAC code for degenerate bases have been used: A or C, M; A or G, R; A or T, W; G or C, S; C or T, Y; G or T, K; A, G or C, V; A, C or T, H, A, G or T, D; G, C or T, B; A, G, C or T, N.
Comparing bacterial and archaeal parvulin proteins
Orthogonal to analyses of the PPIase repertoire within archaeal genomes, the genomic context and the degree of sequence conservation of archaeal parvulin proteins, we wanted to rationalise differences between these two groups of proteins on the level of amino acids. Qualitatively, protein parameters were compared for the parvulin proteins from Escherichia coli [PDB:1JNS] and Cenarchaeum symbiosum [PDB:2QRS]. The two proteins have similar isoelectric points: 9.23 and 9.59, respectively. However, the archaeal protein has more charged residues than the bacterial one (Asp+Glu/Arg+Lys: 12/18 relative to 9/13).
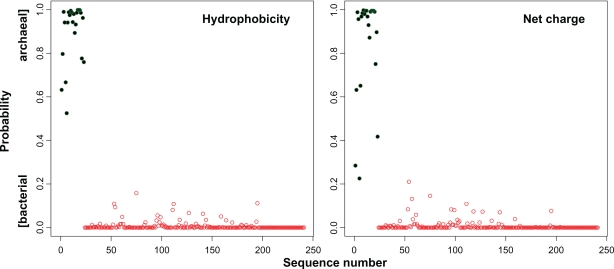
A more efficient and unbiased way to analyse differences between whole groups of protein sequences is to apply machine learning techniques.34 We used a random forest (RF)35 to differentiate our dataset of 241 archaeal and bacterial parvulin sequences. First, the sequences were encoded using the hydrophobicity descriptor and the net charge descriptor, respectively (Fig. 5).36 The encoded sequences were used as input for the RF classifiers. The classifiers trained with the hydrophobicity and the net charge descriptors were perfectly able to distinguish the two classes. Remarkably, very good classification accuracy can already be achieved with a linear separator, such as a linear support vector machine (data not shown). For further analysis, we used the so-called importance values generated in the RF that project the complex classifier onto more easily intelligible contributions by single sequence positions.
Figure 5.
A random forest (RF) can be trained to discriminate between bacterial and archaeal single-domain parvulins. Two descriptors were used (hydrophobicity and net charge) to describe the protein sequences. The very same dataset used for the MLP tree was used here. Prediction scale represents “0.0”, bacterial, and “1.0”, archaeal. The separation according to the MLP tree is represented in green (archaeal) and red (bacterial). For the hydrophobicity descriptor, the RF perfectly separate the two classes (F1 score = 1.0). For the net charge descriptor, the RF reaches an F1 score of 0.979 (cut off = 0.2).
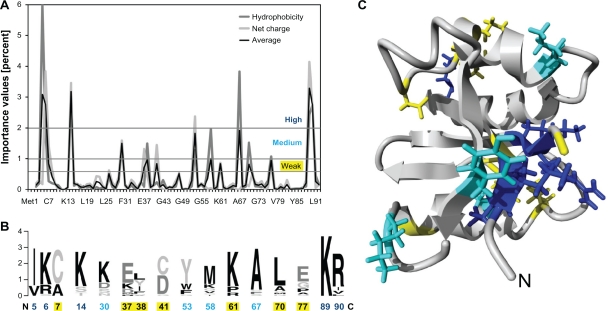
The positions were grouped into highly, medium or weakly important positions (Fig. 6A). A sequence logo was created from the 23 archaeal proteins for all these positions (Fig. 6B). It shows that the positions identified in the RF are largely conserved within the archaeal subgroup. A notable exception is Cys/Asp41 in the catalytic centre.2 Nine of the 15 identified positions include charged amino acids in the archaeal proteins. This is in agreement with the notion above that an archaeal parvulin contains many more charged residues than its bacterial counterpart. Notably, the identified positions are unevenly distributed. After mapping of the corresponding residues onto the recently published structure of the PinA protein from C. symbiosum, these residues form a charged patch on one side of the protein (Fig. 6C). This may represent a special feature of archaeal parvulins pointing towards a negatively charged binding partner. As C. symbiosum is a psychrophilic archaeon, this feature could also be a further hallmark of cold adaption of psychrophilic parvulins.16
Figure 6.
Random forest approach to identify special features of archaeal parvulins. A random forest was used to distinguish archaeal and bacterial parvulins. (A) importance values were derived from the random forest analysis and plotted on the primary sequence of the parvulin protein from Cenarchaeum symbiosum. (B) the identified positions of all 23 archaeal proteins were used to create a protein logo. (C) mapping of important residues on the structure [PDB:2QRS] of the C. symbiosum parvulin.
Conclusion
Thaumarchaeota are known to play a crucial role in geochemical cycles in surface regions of non-coastal marine freshwater; their total number was estimated to be 1028 cells worldwide.37 These organisms do not belong to the phylum of Crenarchaeota, but form another deep-branching clade.11 Similar to Korarchaeota17 they possess genes related to Crenarchaeota as well as euryarchaeal genes. At the time of analysis, only two fully sequenced genomes of Thaumarchaeota are available3,4 and the dimension of this phylum is unknown. Recent studies have assigned some metagenomic contigs or fosmids to the thaumarchaeal phylum,13–15 but it remained impossible to group virtual organisms within the phylum.
Searching all available archaeal genomes for their PPIase content led to a correlation between the growth temperature of an organism and its PPIase content. There is a strong tendency for thermophilic microorganisms to reduce the total number of PPIases.
By examining the genomic context of sdPars, we could classify groups within the underrepresented and largely uncultivated archaeal subgroup of Thaumarchaeota and reveal that parvulin is significantly higher conserved than its genomic neighbours. Our work related the novel group of acidophilic Richmond Mine archaeal nanoorganism (ARMAN) much closer to the Thaumarchaeota than previously anticipated. Taken together, our study significantly expands the phylum of Thaumarchaeota by metagenomic sequences, allows a first grouping of these organisms and reveals important amino acid residues, or a molecular phenotype, that separate archaeal and bacterial parvulins from each other.
Methods
Searching genomes for their PPIase repertoire
First all fully and near fully genomes of Archaea were listed using the NCBI genomes database.38 Next, the following E. coli proteins were used as queries: SlyD-type-FKBP: [GeneID: 947859]; FkpA-type-FKBP: [GeneID: 947870]; trigger factor: [GeneID: 945081]; Cyclophilins: [GeneID: 949038]; sdPar: [GeneID:948285]; SurA: [GeneID: 94481]. Queries for PrsA and NifM were taken from the organisms Staphylococcus aureus (PrsA:[GeneID: 5560626]) and Azotobacter vinelandii (NifM: [GeneID: 7759132]). The genomes were searched by examining their annotated proteins or by searching the whole genomes with the different queries using BLASTp.39 Positive, but not annotated hits were verified using the Conserved Domain Database (CDD).19
Retrieving sdPar-containing metagenomic data and measurement of diversity
Metagenomic contigs were found by using the known and annotated archaeal parvulins as queries for tBLASTn searches39 in the whole genome shotgun (wgs) and the environmental samples (env) databases.38 The length of the sdPar reading frame was determined using ORF finder.40 Contigs were examined for the parvulin-surrounding genomic context using the same toolset.
For the evaluation of mean sequence entropies we computed with T-Coffee29 pairs of multiple sequence alignments for a certain protein sequence (UbiA, DUF, hyp, PPOX, DHCP) taken from a set of organisms and of sdPar taken from the same set. For each multiple sequence alignment we computed the Shannon entropy using the R-package bio3d41 and averaged this entropy over all alignment positions. The R-script for the computation of entropy and all multiple sequence alignments used as input for the script are provided as supplementary material (file Supp_entropy.zip).
Phylogenetic calculations
An initial dataset of 326 parvulin sequences was used for phylogenetic calculations. This dataset contained 16 sequences that were N-terminally truncated and one metagenomic sequence that has been C-terminally truncated to the sequence matching the sdPar sequences of Nitrosopumilus maritimus and Cenarchaeum symbiosum. The FKBP SlyD from E. coli was used as an outgroup representing an unrelated prolyl isomerase with conserved fold. These sequences were aligned using T-Coffee.29 Phylogeny was estimated by maximum-likelihood using PhyML 3.030 with 1000 bootstraps. A consensus tree was derived from this dataset using the program consense of the Phylip suite42 defining SlyD manually as outgroup. This tree was plotted using FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree). After removal of paralogues and sequences from clades where parvulins are not well represented, the 241 remaining sequences were used for another MLP calculation as described above.
Machine learning approach
Parvulins were first compared with respect to protein parameters43 and surface exposure44 as has been described.45 The filtered dataset of 241 single-domain parvulins was then used to train a linear support vector machine and a random forest. Therefore, N- and C-termini were trimmed on the basis of a multiple-sequence alignment and a loop of 15 amino acids was removed from sequences of Photobacterium species as it occurred only there. This S(E/Q)ALK(K/L) KNNNLRGLI loop might functionally correspond to the phosphate-binding loop KHSQSRRPSS-WRQEKITRTK of the Pin1 structure [PDB:1NMV];23 however, it is more similar to the KVKSKKSD-KEGLD extension seen in the Staphylococcus aureus PrsA parvulin that does not bind phosphorylated substrates [PDB:2JZV].46 The remaining sequences were projected to a length of 92 amino acids.
The 241 protein sequences were encoded using the hydrophobicity descriptor and the net charge descriptor, respectively.36 The encoded sequences were used as input for the linear support vector machine and the random forest35 as implemented in the R packages kern-lab and randomForest.47 The classifier models were evaluated by ten-fold leave-one-out cross-validation. As performance measurement we used the area under the receiver operating curve (AUC)48 and the F1 score, the harmonic mean of precision and recall:
with TP: true positives, FP: false positives, FN: false negatives.
Random forests estimate the importance of each sequence position for the classification process.35 Importance values from random forests using hydrophobicity and net charge descriptors were averaged and classified into highly (>2%), medium (1%–2%) and weakly (0.6%–1%) important residues. These were mapped on the NMR structure [PDB:2QRS] of the sdPar of C. Symbiosum.16
Acknowledgments
We thank Bettina Siebers for constructive and critical reading of the manuscript. Die Brücke is acknowledged for fruitful discussions. This study was funded in part by DFG grant BA1624/7-1 to PB. Publication costs for this article were covered by the open access publishing program from Deutsche Forschungsgemeinschaft.
Abbreviations
- aa
amino acid
- ARMAN(s)
archaeal Richmond Mine acidophilic nanoorganism(s)
- CDD
conserved domain database
- contig
a contiguous stretch of aligned sequence
- FKBP
FK506 binding protein, one class of PPIases
- fosmid
DNA fragments in a vector based on the bacterial F-plasmid
- HGT
horizontal gene transfer
- MLP
maximum likelihood phylogeny
- PPIase
(peptidyl-) prolyl isomerase
- RF
random forest
- sdPar
single-domain parvulin
Footnotes
Authors’ Contributions
CL and JWM conceived this study. CL performed extensive database research. JvdB and DH performed MLP calculations. DH applied machine learning approaches. CL, JWM and PB analysed the data and wrote the paper. All authors read and approved the final manuscript.
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Maruyama T, Suzuki R, Furutani M. Archaeal peptidyl prolyl cis-trans isomerases (PPIases) update 2004. Front Biosci. 2004;9:1680–720. doi: 10.2741/1361. [DOI] [PubMed] [Google Scholar]
- 2.Mueller JW, Bayer P. Small family with key contacts: par14 and par17 parvulin proteins, relatives of pin1, now emerge in biomedical research. Perspect Medicin Chem. 2008;2:11–20. doi: 10.4137/pmc.s496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker CB, de lT, Jr, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci U S A. 2010;107:8818–23. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hallam SJ, Konstantinidis KT, Putnam N, et al. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc Natl Acad Sci U S A. 2006;103:18296–301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker BJ, Comolli LR, Dick GJ, et al. Enigmatic, ultrasmall, uncultivated Archaea. Proc Natl Acad Sci U S A. 2010;107:8806–11. doi: 10.1073/pnas.0914470107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dick GJ, Andersson AF, Baker BJ, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10:R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen JA. Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol. 2007;5:e82. doi: 10.1371/journal.pbio.0050082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, et al. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–86. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- 9.Baker BJ, Tyson GW, Webb RI, et al. Lineages of acidophilic archaea revealed by community genomic analysis. Science. 2006;314:1933–5. doi: 10.1126/science.1132690. [DOI] [PubMed] [Google Scholar]
- 10.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 2009;3:159–67. doi: 10.1038/ismej.2008.99. [DOI] [PubMed] [Google Scholar]
- 11.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–52. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 12.Hatzenpichler R, Lebedeva EV, Spieck E, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci U S A. 2008;105:2134–9. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brochier-Armanet C, Gribaldo S, Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol Direct. 2008;3:54. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der OJ. Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct. 2009;4:39. doi: 10.1186/1745-6150-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spang A, Hatzenpichler R, Brochier-Armanet C, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–40. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Jaremko Ł, Jaremko M, Elfaki I, Mueller JW, Ejchart A, Bayer P, Zhukov I. Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases. J Biol Chem. 2011 Feb 25;286(8):6554–65. doi: 10.1074/jbc.M110.160713. Epub 2010 Dec 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkins JG, Podar M, Graham DE, et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc Natl Acad Sci U S A. 2008;105:8102–7. doi: 10.1073/pnas.0801980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–7. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Anderson JB, Chitsaz F, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–10. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunoura T, Takaki Y, Kakuta J, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–7. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 23.Bayer E, Goettsch S, Mueller JW, et al. Structural analysis of the mitotic regulator hPin1 in solution: insights into domain architecture and substrate binding. J Biol Chem. 2003;278:26183–93. doi: 10.1074/jbc.M300721200. [DOI] [PubMed] [Google Scholar]
- 24.Rulten S, Thorpe J, Kay J. Identification of eukaryotic parvulin homologues: a new subfamily of peptidylprolyl cis-trans isomerases. Biochem Biophys Res Commun. 1999;259:557–62. doi: 10.1006/bbrc.1999.0828. [DOI] [PubMed] [Google Scholar]
- 25.Uchida T, Fujimori F, Tradler T, Fischer G, Rahfeld JU. Identification and characterization of a 14 kDa human protein as a novel parvulin-like peptidyl prolyl cis/trans isomerase. FEBS Lett. 1999;446:278–82. doi: 10.1016/s0014-5793(99)00239-2. [DOI] [PubMed] [Google Scholar]
- 26.Mueller JW, Kessler D, Neumann D, et al. Characterization of novel elongated Parvulin isoforms that are ubiquitously expressed in human tissues and originate from alternative transcription initiation. BMC Mol Biol. 2006;7:9. doi: 10.1186/1471-2199-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler D, Papatheodorou P, Stratmann T, et al. The DNA binding parvulin Par17 is targeted to the mitochondrial matrix by a recently evolved prepeptide uniquely present in Hominidae. BMC Biol. 2007;5:37. doi: 10.1186/1741-7007-5-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schleper C, Jurgens G, Jonuscheit M. Genomic studies of uncultivated archaea. Nat Rev Microbiol. 2005;3:479–88. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- 29.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–17. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 31.Sekerina E, Rahfeld JU, Muller J, et al. NMR solution structure of hPar14 reveals similarity to the peptidyl prolyl cis/trans isomerase domain of the mitotic regulator hPin1 but indicates a different functionality of the protein. J Mol Biol. 2000;301:1003–17. doi: 10.1006/jmbi.2000.4013. [DOI] [PubMed] [Google Scholar]
- 32.Weininger U, Haupt C, Schweimer K, et al. NMR solution structure of SlyD from Escherichia coli: spatial separation of prolyl isomerase and chaperone function. J Mol Biol. 2009;387:295–305. doi: 10.1016/j.jmb.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 33.Brochier-Armanet C, Deschamps P, Lopez-Garcia P, Zivanovic Y, Rodriguez-Valera F, Moreira D. Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J. 2011 doi: 10.1038/ismej.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heider D, Hauke S, Pyka M, Kessler D. Insights into the classification of small GTPases. Adv Appl Bioinf Chem. 2010;3:15–24. doi: 10.2147/aabc.s8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 36.Kawashima S, Pokarowski P, Pokarowska M, Kolinski A, Katayama T, Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36:D202–5. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–10. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 38.Sayers EW, Barrett T, Benson DA, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques. 2000;28:1102–4. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 41.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–6. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 42.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. 2005.
- 43.Gasteiger E, Hoogland C, Gattiker A, et al. Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM, editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [Google Scholar]
- 44.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 45.Grum D, van den Boom J, Neumann D, Matena A, Link NM, Mueller JW. A heterodimer of human 3′-phospho-adenosine-5′-phosphosulphate (PAPS) synthases is a new sulphate activating complex. Biochem Biophys Res Commun. 2010;395:420–5. doi: 10.1016/j.bbrc.2010.04.039. [DOI] [PubMed] [Google Scholar]
- 46.Heikkinen O, Seppala R, Tossavainen H, et al. Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA—implications for the catalytic mechanism of parvulins. BMC Struct Biol. 2009;9:17. doi: 10.1186/1472-6807-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2010. [Google Scholar]
- 48.Fawcett T. An introduction to ROC analysis. Pattern Recognition Letters. 2006;27:861–74. [Google Scholar]
- 49.Yooseph S, Sutton G, Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.