Abstract
Single cell patterning holds important implications for biology, biochemistry, biotechnology, medicine, and bioinformatics. The challenge for single cell patterning is to produce small islands hosting only single cells and retaining their viability for a prolonged period of time. This study demonstrated a surface engineering approach that uses a covalently-bound short peptide as a mediator to pattern cells with improved single cell adhesion and prolonged cellular viability on gold patterned SiO2 substrates. The underlying hypothesis is that cell adhesion is regulated by the type, availability and stability of effective cell adhesion peptides, and thus covalently bound short peptides would promote cell spreading and thus, single cell adhesion and viability. The effectiveness of this approach and the underlying mechanism for the increased probability of single cell adhesion and prolonged cell viability by short peptides were studied by comparing cellular behavior of human umbilical cord vein endothelial cells on three model surfaces whose gold electrodes were immobilized with fibronectin, physically adsorbed Arg-Glu-Asp-Val-Tyr, and covalently-bound Lys-Arg-Glu-Asp-Val-Tyr, respectively. The surface chemistry and binding properties were characterized by reflectance Fourier transform infrared spectroscopy. Both short peptides were superior to fibronectin in producing adhesion of only single cells, while the covalently bound peptide also reduced apoptosis and necrosis of adhered cells. Controlling cell spreading by peptide binding domains to regulate apoptosis and viability represents a fundamental mechanism in cell-materials interaction and provides an effective strategy in engineering arrays of single cells.
Keywords: Single cell patterning, endothelial cells, peptides, cell adhesion, cellular viability
Introduction
The ability to position and probe single cells is of great interest in fundamental cell biology, cell-based biosensor technologies, medical diagnostics, and tissue engineering.1–6 The single-cell analysis is fundamental to comprehending many biological processes and mechanisms, as it reveals the response of each individual cell under stimulation.7 Critical cell-to-cell differences are lost in averaged bulk cell measurements.8 Patterning viable single cells on an addressable array of identical cell hosts (e.g., an array of microelectrodes with the same physical and chemical properties) would aid the statistical analysis of single cell behavior and cell-matrix interaction.9 In practical applications, particularly for screening, detection, or sensing systems, microarrays of single cells allow for rapid and inexpensive analyses, require minimal sample volumes, and provide high throughput data acquisition and portability.
Micropatterning of living cells on substrates has experienced a rapid growth in recent years. A number of techniques have been developed to produce micro-scale cell patterns. Typical examples include microcontact printing,10 microfluidic channels,11–13 elastomeric stencils,14 and elastomeric membranes.15 These approaches typically utilize mechanical devices to deliver proteins/peptides to guide cell adhesion or directly deposit cells on a substrate of single material.11, 12, 16–18 Despite the encouraging advances made with these techniques, patterning single cells on an microarray and retaining their viability for a prolonged period of time remain as a challenge. Single cell patterning requires an area for cell adhesion at a size comparable to an individual cell which is typically 10 to 20 μm, to minimize the probability of a second cell attachment. However, adhesion sites of such small areas tend to suppress cell spread and thus are prone to causing cell death. It was reported that cells could be geometrically switched between growth and apoptosis.19 Endothelial cells cultured on single islands coated with fibronectin spread and progressed through the cell cycle when the island area was larger than ~40 μm × 40 μm, but failed to extend and underwent apoptosis when cells were restricted to areas smaller than ~20 μm × 20 μm.
Here we report on creating microarrays of human umbilical cord vein endothelial cells (HU-VEC) on gold-patterned silicon oxide substrates using a covalently-bound short peptide, Lys-Arg-Glu-Asp-Val-Tyr (KREDVY), to mediate single cell adhesion and maintain cellular viability. To demonstrate the strength of this approach and elucidate the underlying mechanism, two control surfaces were prepared for comparison: one coated with fibronectin as an adhesive protein which is commonly used for receptor-mediated cell adhesion, and another physically bound with a short peptide, Arg-Glu-Asp-Val-Tyr (REDVY).
This study is based on the hypothesis that cell spreading is dictated by the type, conformation, availability and stability of effective cell adhesion peptides, and thus covalently bound short peptides would provide a extracellular matrix with a large number of uniform and effective binding sites to promote cell spreading and thus, single cell adhesion and viability on small islands (20 μm × 20 μm). The extracellular matrix (ECM) is known to play a regulatory role in cellular growth, differentiation, and apoptosis. A recent study showed that cell attachment and spreading are dictated by the balance of the availability of potential adhesion domains and cellular traction forces,20 suggesting the important role of cytoskeletal tension in regulating cell spreading and thus the cell fate (apoptosis, differentiation, and proliferation, etc). In this regard, the apoptosis of cells on 20 μm × 20 μm coated with fibronectin might be caused by both geometry effect19 and low binding efficiency of long-chain proteins, as long-chain proteins may not have an insufficient number and correct peptide orientation of binding sites to support cell spreading.
The endothelial cell was selected as model cell in this study because it plays an important role in angiogenesis and tissue repair, and has a broad range of applications in detection of bacteria, viruses, and toxins.21–23 Endothelial cells serve as major barriers separating the blood from tissue compartments whose interaction with bacteria defines the course of invasive infections and inflammatory responses.23
The surface immobilization of peptides/proteins was characterized by reflectance FTIR spectroscopy. Cell adhesion was studied by optical and confocal microscopes. Efficiency of signal cell patterning including single cell adhesion and coverage was quantified by examination of cells with DAPI stain. The cell viability over time (apoptosis and necrosis) was studied by apoptosis assay. Cells adhered on different substrates were immunochemically stained to identify cell-surface focal contacts for cell binding and cellular actin filaments for cell spreading.
Experimental
Materials
All the materials and chemicals were used as received. 11-mercaptoundecanoic acid 95% (11-MUA), 3-mercaptopropionic acid 99% (3-MPA), N-hydroxysuccinimide 97% (NHS), 1-ethyl-3-(3-dimethylamino-propyl) carbodiimide (EDAC), fibronectin protein, paraformaldehyde, glutaraldehyde, 4′, 6-Diamidino-2-phenyindole (DAPI) and anti vinculin-FITC were purchased from Sigma (St. Louis, MO). 2-[methoxy(polyethyleneoxy)propyl] trimethoxysilane (Mw = 460–590 Dalton) and Nanostrip 2X were purchased from Gelest (Morrisville, PA) and Cyantek (Fremont, CA.), respectively. Alexa Fluor® 594 phalloidin and Vybrant® Apoptosis assay Kit #2 were obtained from Molecular Probes (Eugene, OR). REDV (518.3 Dalton) and KREDVY (806.1 Dalton) were purchased from Synpep (Dublin, CA). Human umbilical cord vein endothelial cells (HUVECs) and cell culture supplies including EGM-2, HEPES-buffered saline, trypsin EDTA, and trypsin neutralizing solution were purchased from Clontics (Walkersville, MD).
Substrate preparation
Four-inch p-type silicon substrates with (100) orientation were cleaned with piranha (hydrogen peroxide/sulfuric acid 2:5 v/v) at 120°C for 10 minutes, dipped in HF, and rinsed with DI water thoroughly. A 1.1 μm layer of positive photoresist was then coated on the surface, and an array of 20 μm × 20 μm gold squares (electrodes) spaced 60 μm apart was patterned on silicon oxide substrates by conventional microfabrication as follows. A 10 nm layer of titanium (Ti) was then deposited onto the photoresist-developed substrates at a deposition rate of 0.3 Å/s. A gold film of 100 nm in thickness was subsequently deposited onto the Ti at a deposition rate of 5 Å/s. The photoresist was dissolved in acetone and the remaining metal film was lifted off. After lift-off, the surfaces were exposed to buffered oxide etch (HF/NH4F 5:1 v/v) for 60 sec. and rinsed with DI water to remove the native oxide on silicon regions, followed by oxidation under a dry oxygen flow for 6 hrs at 400°C, yielding a 60 Å oxide layer on the silicon regions.24 The gold-patterned silicon wafers were cut into 8 mm × 8 mm slides. To minimize surface contaminants and scratches, the silicon wafers were coated with a 2 μm layer of photoresist on their polished sides before cutting.
Surface modification
The surfaces were modified following a procedure established previously24 with modifications. The protective photoresist layer on gold-patterned silicon oxide substrates was removed by sonication in acetone, then in ethanol, and finally in DI water. The substrates were then placed in Nanostrip 2X solution (H2SO5) at room temperature for 20 min, and dried under nitrogen, which resulted in a hydroxyl layer on the silicon oxide surfaces. The gold patterned silicon oxide substrates were reacted with a 20 mM solution of alkane thiols of 11-mercaptoundecanoic acid (MUA) and 3-mercaptopropionic acid (MPA) (1:10 v/v) for 16 hrs to create a self-assembled monolayer (SAM) on gold squares. The use of the mixture of thiols is based on a previous study.25 The substrates were then exposed to PEG solution containing 3 m-PEG-silane and 1% triethylamine as a catalyst in deoxygenated toluene to passivate silicon oxide. The PEG reaction proceeded at 60°C for 18 hrs in nitrogen-filled flasks that were pre-treated with Sigmacote to minimize the side reaction of PEG with the flasks. The PEG-treated surfaces were cleaned by sonication in toluene and ethanol for 5 min each, followed by a rinse with DI water and drying under nitrogen. The substrates with a SAM of alkane thiol on gold and M-PEG-silane on silicon oxide were then immersed in an aqueous solution of 150 mM EDAC and 30 mM N-hydroxysuccinimide (NHS) for 30 min to attach the NHS group to the –COOH terminus of the SAMs. The substrates with NHS on gold and PEG on silicon oxide were sterilized with 70% ethanol for 15 min, and exposed to either fibronectin protein, REDV, or KREDVY peptide in a phosphate buffer solution (PBS, pH = 8.2) at a concentration of 0.1 mg/ml. The reaction continued at room temperature for 1 hr. To remove loosely bound moieties from the surface after each step of surface modification, the substrate was rinsed with its original solvent and DI water, respectively.
Surface characterization by FTIR
Surfaces coated with fibronectin protein or peptides were characterized using a Nicolet Magna 760 Fourier transform infrared (FTIR) spectroscope equipped with a FT-85 grazing angle sample compartment. FTIR absorption spectra of 750 scans were acquired at a resolution of 8 cm−1. The system was purged with dry air for 1 hr before each data collection to remove water vapor in the sample compartment. Spectra analysis was performed using standard Nicolet and Origin software.
Cell culture and adhesion
HUVEC cells were cultured in EGM-2 medium supplemented with bovine brain extract (BBE), hydrocortisone, hFGF-B, VEGF, R3-IGF-1, ascorbic acid, heparin, FBS, hEGF, and GA-1000. The final serum concentration was 2%. Cells were seeded on culture flasks at passage 1 and the medium was changed after 24 hrs. At 70% confluency, cells were subcultured as follows. After aspiration from culture flasks, cells were rinsed with 2–3 ml of HEPES-BSS buffer solution for 3 times, followed by incubation with 2 ml of trypsin/EDTA solution. The trypsinization process continued until ~90% of the cells were collected. After cells were released, the trypsin was neutralized in the flask with 4 ml of TNS, and the detached cells were transferred to a 15-ml sterile centrifuge tube. The harvested cells were centrifuged at 220 g for 5 min. Cells were diluted in growth medium. Cells at a concentration of 1.5 × 105 cells/ml were seeded on the substrates. The substrates were incubated for 18 hrs before fixation with a mixture of 2% glutaraldehyde and paraformaldehyde in phosphate buffer solution for optical microscopy and with 4% paraformaldehyde for fluorescence microscopy.
Cell staining
Cells adhered on the substrates were fixed, permeabilized, and stained with Alexa Fluor® 594 phalloidin dye for F-actin staining (red) and immunostained with monoclonal anti-vinculin-FITC (green) followed by cellular staining with 6-Diamidino-2-phenyindole (DAPI, blue). Before fixation the substrates were washed with PBS to remove cell debris and loosely attached cells. Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature and permeabilized by treating cells with Triton X100 (0.1% in PBS) for 10 min. Following three washes with PBS the samples were incubated for 30 min with a 1X blocking buffer solution (5% (w/v) of nonfat dry milk in PBS containing 0.1% Tween-20) for background passivation. The actual staining was done in two steps. First, the primary antibody against vinculin (anti vinculin- FITC) was diluted in blocking buffer following the manufacture’s recommendations and treated with cells over night in dark at 4 °C. Next, the samples were washed three times with blocking buffer before cells were exposed to a phalloidin-AlexaFluor546 for 1 hr at RT. The samples were then washed with PBS three times and blown dry with air. A final treatment with gold anti-fade solution containing 6-Diamidino-2-phenyindole (DAPI) stained cell nuclei and preserved the fluorescence of the samples for confocal microscopy.
Cell viability assay: apoptosis and necrosis
Vybrant® apoptosis assay allows for the simultaneous visualization of viable, necrotic and apoptotic cells on substrates. Necrosis results from direct cell damage; apoptosis is genetically-programmed cell death in which cells effectively commit suicide. Green fluorescently labeled Annexin V protein (in the presence of calcium) specifically binds to the phosphatidylserine protein on membranes of apoptotic cells. Propidium iodide does not penetrate to either live or apoptotic cells, but rather, stains nuclei of necrotic cells in red. Cell-patterned substrates were washed twice with cold PBS and placed in 500 μl of Annexin V binding buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4). The substrates were incubated with 100 μl of Annexin V and 2 μl of PI solution for 15 min at room temperature, washed twice with binding buffer, and imaged with a fluorescence microscope.
Results and Discussion
Immobilization and characterization of adhesion protein and peptides
Silicon oxide surfaces patterned with gold squares of 20 μm × 20 μm were chemically modified to activate the gold squares (to host cells) with a self-assembled monolayer (SAM) of carboxyl-terminated alkane thiols as described in Experimental section. The silicon oxide background was passivated with a PEG coating resistant to protein adsorption and cell adhesion.24 The surface-modified substrates were then exposed to (a) fibronectin, (b) REDVY, or (c) KREDVY. This scenario is illustrated with one gold square in silicon oxide background shown in Figure 1 (left panel). Fibronectin and KREDVY with lysine residues (K) containing primary ∈-amino groups were covalently bound to the carboxylate-terminated SAM on the gold squares. This bonding was formed by activation of the terminal carboxylate group with an N-hydroxysuccinimide (NHS) ester intermediate, followed by the displacement of the NHS group by the lysine residues of proteins or peptides.24 REDVY, without lysine residues, was physically adsorbed on the gold squares. These processes produced three patterned surfaces of different surface chemistries and cell binding natures, each with an array of cell adhesive sites (gold squares) on a non-adhesive background (passivated silicon oxide).
Figure 1.
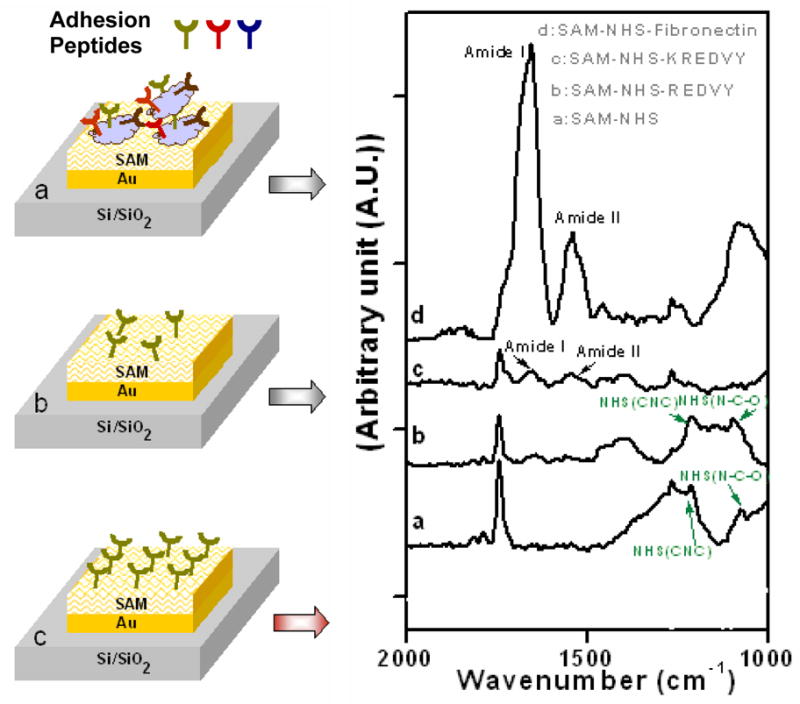
Left panel schematic representation of gold squares coated with: (a) fibronectin with multiple types of cell adhesion sequences, (b) physically adsorbed REDVY with REDV sequence, and (c) covalently bound KREDVY with REDV sequence. Right panel: Grazing angle FTIR absorption spectra of surfaces modified with various coatings.
The surface chemistries and binding properties of the three surfaces were characterized by reflectance FTIR spectroscopy. Figure 1 (right panel) shows the IR spectra acquired from the surfaces coated with (a) SAM-NHS, (b) SAM-NHS-REDVY, (c) SAM-NHS-KREDVY, and (d) SAM-NHS-fibronectin. Spectrum (a) has characteristic bands at 1078, 1222, 1370, and 1741 cm−1, obtained from the surface modified with SAM-NHS, a monolayer on which the protein or peptides were further immobilized. The high intensity band at 1741 cm−1 is attributed to asymmetric stretch of NHS carbonyls, and indicates successful covalent binding of NHS with the underlying alkane thiol SAM. The amide I and amide II peaks in spectrum (b) indicate the presence of associated peptide bonds on the surface. Physical adsorption of REDVY on the surface is confirmed by the presence of unreacted NHS groups with spectrum bands corresponding to νas (CNC) and ν (NCO) of NHS at 1222 cm−1 and 1078 cm−1 that would be otherwise absent due to the chemical bonding. Successful covalent binding between the lysine residue of KREDVY and NHS monolayer is characterized by the presence of the amide I and amide II bands at 1653 and 1543 cm−1 (spectrum (c)), the significant reduction of the intensity of carbonyl peak at 1741 cm−1 as well as the absence of bands at 1078, 1222, and 1370 cm−1 that correspond to NCO stretch, asymmetric CNC stretch, and symmetric CNC stretch of the NHS, respectively. However, the continued presence of a small peak at 1741 cm−1 suggests the incomplete conversion of SAM carboxylic groups to amides. A comparable result was reported by Frey et al. wherein an NHS layer had to be reacted with poly-lysine residue three times to convert most of the carboxylic groups to amide groups.26 Spectrum (d), acquired from the fibronectin-modified surface, confirms the covalent interactions of fibronectin with NHS-SAM. A full conversion of carboxylic groups to amides is characterized by the complete absence of peaks at 1741 cm−1 and 1222 cm−1 and the presence of strong amide I and amide II peaks.27 The high intensities of amide I and amide II bands indicate a high density of peptides on the surface, presumably due to the large amount of peptides in fibronectin.
Cell adhesion on microarrays
To create microarrays of cell patterns, HUVEC cells were cultured on fibronectin or peptide modified substrates in standard culture media for 18 hrs. After cell culture, the cells were fixed and examined using differential interference contrast (DIC) reflectance optical microscopy. Both REDVY and KREDVY contain tetrapeptide REDV sequence specific to α4β1 receptors of HUVEC cells.28 Fibronectin has at least two types of cell binding sequences for HUVEC: the Arg-Gly-Asp (RGD) that would bind to the α5β1 and αvβ3 integrin receptors, and the tetrapeptide REDV that would bind to α4β1 receptors.28 When immobilized on the carboxylate-terminated gold surfaces, these cell adhesion protein and peptides would exhibit different binding domain configurations as shown conceptually for a gold square in Figure 1 (left panel). Because fibronectins are long-chain molecules, they would bind to the surface with random orientation of RGD and REDV domains (a), whereas short-chain REDVY and KREDVY peptides would exhibit much more ordered distribution and/or orientation (b and c). Additionally, the KREDVY peptide would exhibit a more uniform molecular orientation as a result of its covalent bonding with the underlying SAM layer and thus provide more uniform binding domains. Figure 2 shows the optical images of HUVEC cells adhered on the gold squares modified with fibronectin (a), REDVY (b) and KREDVY (c), respectively. The cells were seen to adhere mainly on the sites of gold squares, indicating a high degree of cell selectivity for all three surfaces. The fibronectin-modified surface exhibited a pattern of multiple-cell binding on gold squares and slight nonspecific cell adhesion onto the silicon oxide background around the gold squares. This is not surprising in light of the multiple types of cell binding sequences and randomly oriented binding domains of long-chain fibronectin protein. Cell adhesion, the morphology of adhered cells, and the extent of cell spreading are dictated by the availability, conformation, and distribution of cell binding domains. When a cell approaches a gold square covered with fibronectin protein molecules, it is confronted with binding domains of different types and orientations. It is conceivable that only a portion of the surface-bound protein molecules and a portion of peptide sequences in those molecules are involved in the cell binding process, presumably those cell binding sequences in the molecules that are oriented roughly at the same direction. Thus, the “effective” binding domains on a gold square for the adhesion of this particular cell can be only a small fraction of those physically presented on the gold square. A large number of cell binding sequences in fibronectin does not necessarily result in a large number of effective binding domains, due to the molecules bound in a random orientation to the surface.20 The spreading of the adhered cell over the gold square can also be hindered by the limited number of effective binding domains. Instead, the probability of binding a second cell to the same gold square is increased due to availability of free binding domains of different types and orientations.
Figure 2.
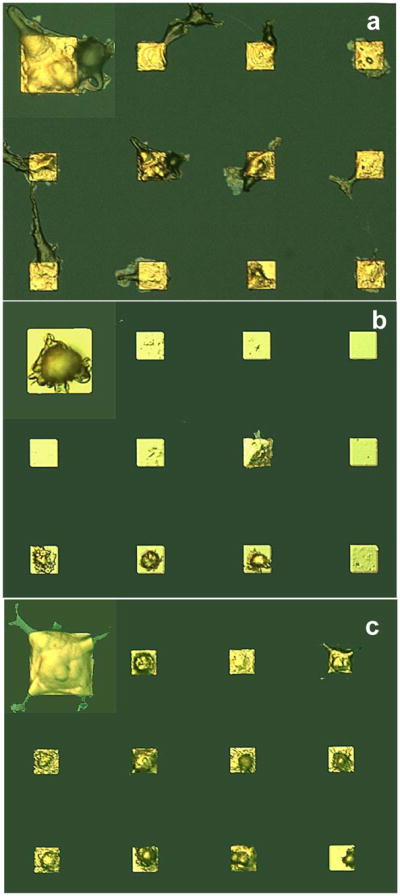
Optical micrographs of HUVEC cells patterned on gold electrodes of silicon oxide substrates with gold electrodes coated with (a) fibronectin, (b) physically adsorbed REDVY, and (c) covalently bound KREDVY. The insets show a magnified cell image for each case to reveal the cell morphology.
Cells attached on the surface coated with REDVY peptide exhibited a pattern with most of gold squares hosting one cell (Figure 2b), but with apparently low cell coverage (the number of gold squares occupied by cells versus the total number of gold squares). In addition, the cells barely spread across the gold squares. Two possibilities may account for this low cell coverage, both because of the weak physical binding between the REDVY peptide molecules and underlying SAM-NHS layer: (1) the detachment of REDVY peptide from the gold surface, resulting in an insufficient number of REDVY peptides available on the gold square to bind a cell; (2) the detachment of the cell-REDVY conjugates from the gold square after the cell bound to the REDVY peptides. Low cell spreading on the REDVY-modified surface can also be attributed to the physical binding of REDVY molecules. Cell spreading or migration requires the dynamic formation and dispersal of cell contacts with the extracellular matrix. For receptor-mediated cell adhesion, cell spreading on a surface is driven by the traction force set by adhesive molecules peripheral to the initial focal contacts between the cell and surface.20 As cell spreading proceeds, which continually stretches the cell and increases force, greater force is required to cause increased cell spreading. A straightforward explanation is that the physically-adsorbed REDVY molecules are unlikely to sustain such traction; instead, they are prone to be detached from the surface by the contractile force of the cell, resulting in the detachment of the REDVY molecules from the surface. Although the physically adsorbed REDVY peptides do not form robust binding with the underlying SAM, they do provide more effective and uniformly distributed binding sites than fibronectin due to their small molecular size. Thus, the majority of REDVY molecules on the gold square would participate in binding of the cell if they are not detached from the surface. Thus, after a cell has been bound, there would be too few binding domains left for binding of a second cell.
On the surface modified with short KREDVY peptide, highly specific single-cell adhesion and higher cell coverage were observed (Figure 2c). This surface has the most uniform and robust cell binding sites (Figure 1c) as a result of the covalent binding of KREDVY molecules on the surface. In addition to having all the advantageous properties of the REDVY-modified surface for cell binding, the covalently bound KREDVY is less susceptible to detachment. The fully spread cell morphology (Figure 2c) also suggests that most of the available binding domains on the gold square have participated in the cell binding, and that the covalently bound KREDVY peptide provides sufficient traction force for cell spreading. Thus, once a cell is bound to the gold square, there would remain insufficient free space or binding domains for adhesion of a second cell. Small peptides have additional advantages over proteins in that they are less susceptible to cellular proteolysis and thermal degradation, and thus most of their active domains are available for cell adhesion.29
The cell adhesion models shown in Figure 1 and the discussion above were further validated by cell stain assays. Cells adhered on the substrates were fixed, permeabilized, and stained with DAPI (blue), immunostain containing anti vinculin-FITC (green), Alexa Fluor® 594 phalloidin dye (red) to reveal the distributions of nuclei, cell-surface focal contacts, and cellular actin filaments, respectively. Figure 3 shows the fluorescence images of cells adhered on the gold squares coated with fibronectin (a), REDVY (b), and KREDVY (c). Panel 1 (top) is the overlay of images of Panel 2 (cell nuclei), Panel 3 (focal contacts) and Panel 4 (actin filaments). Cells on the three surfaces generated different morphological, cytoskeletal and adhesion signals. The nucleus images in Panel 2 show that two cells were attached on the gold square coated with fibronectin (a) and single cells on both REDVY and KREDVY modified surfaces (b and c). A close look at Panel 3 reveals that two cells on the fibronectin modified surface (a) formed fewer focal contacts (per cell) with the substrate than did cells on the KREDVY modified surfaces (c), even though many cell binding sequences (hence cell adhesion domains) are available in fibronectin molecules. This supports the viewpoint noted above that only a portion of the available binding sequences in fibronectin participated in binding of a specific cell. The availability of cell binding domains, plus partial coverage of the gold square by a first-arrival cell, allows for the adherence of a second cell via a different type and/or orientation of binding domains. Spreading of the cells on the fibronectin-modified surface, as shown by the actin filaments (Panel 4a), extended beyond the gold square boundary.
Figure 3.
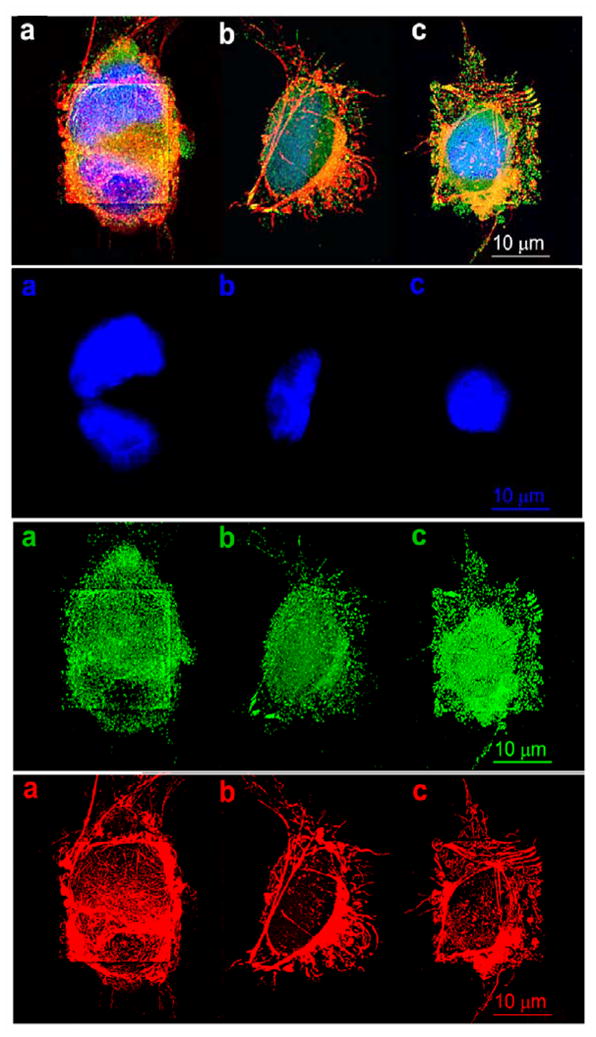
Fluorescent confocal microscopic images of HUVEC cells adhered on gold patterns coated with (a) fibronectin, (b) REDVY peptide, and (c) KREDVY peptide. Top panel is the trichromatic fluorescence images of the cells stained with DAPI, immunostain (monoclonal anti-vinculin-FITC conjugate), Alexa Fluor® 594 phalloidin dye for nuclei (blue), F-actin (red), and vinculin (green), respectively. The lower three panels are the images of separate channels: #2 = nuclei, #3 = vinculin, and #4 = actin.
The cell on the REDVY modified surface has fewer focal contacts with the substrate than the cell on the surface with KREDVY, because the physically adsorbed peptide is prone to detachment from the surface. The cell is not fully spread to cover the entire gold square for the same reason as identified by its actin filament image shown in Panel 4b. The cell on the KREDVY modified surface formed the most dense and uniform focal contacts with the substrate, and the short peptide molecules confined the cell spreading to the gold square, as shown in the cell filament image (Panel 4c), leaving no additional cell binding domains or space for adhesion of a second cell. The dense KREDVY peptide molecules also facilitate preferential attachment of actin filaments terminated at the square edges and vinculin proteins concentrated on the domains around cell nucleus and the edges of the square, resulting in a fully spread cellular morphology, a phenomenon also observed in other studies.30, 31
Quantification of cell coverage and single cell adhesion
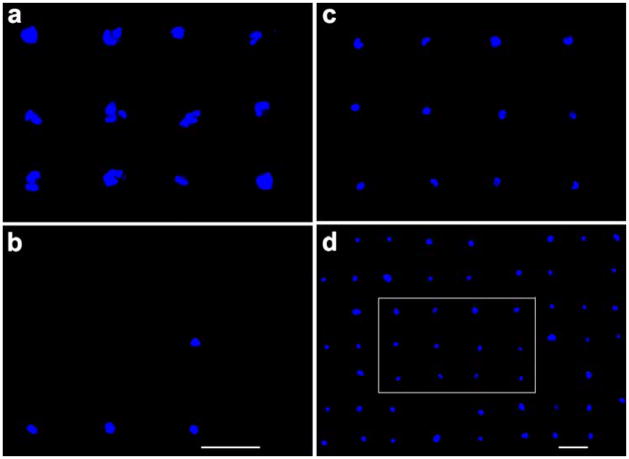
Although covalently bound KREDVY peptides considerably increased the probability of single cell adhesion as compared to protein-mediated cell adhesion, multiple cells were still seen to adhere to individual gold squares on a small number of them. Thus, quantification of single cell coverage is necessary for understanding the capability of this peptide-mediated cell adhesion process. HUVEC cells were cultured on fibronectin and peptide modified substrates in standard culture media for 18 hrs and stained with DAPI for cell nuclei to emit blue fluorescence. Fluorescence microscopy was used to identify single or multiple cells on gold squares. Cell coverage and single cell population for each type of surface were quantified from 378 gold squares (3 samples × 2 regions of interest with 63 squares per region). Cell coverage was calculated from the ratio of gold squares covered with cells to the total number of the squares in the region of interest, and single cell ratio was obtained by dividing the number of gold squares covered with single cells to the total number of gold squares covered with cells (single or multiple). Exemplary images of adhered cells on three model surfaces are shown in Figure 4, and quantification results are shown in Table 1. As expected, the fibronectin and KREDVY modified surfaces have higher cell coverage than the REDVY modified surface due to larger number binding domains available on the former two surfaces than on the latter. Both peptide modified surfaces have much higher single cell ratios than the fibronectin modified surface. Clearly, the KREDVY modified surfaces is the best candidate for single cell patterning in light of both cell coverage and single cell ratio. As a side note, for electrical recording, it is easy to distinguish between single and multiple cells, allowing either type of sites to be observed selectively.
Figure 4.
Fluorescent images of HUVEC cells cultured on gold-patterned silicon oxide substrates with gold squares coated with fibronectin (a: 10× objectives), physically adsorbed REDVY (b: 10 × objectives), and covalently bound KREDVY (c: 10× objective, d: 5× objective). Image (c) was captured from image (d) in the area surrounded by the white rectangle. Scale bars are 60 μm in all images.
Table 1.
Quantification of cell coverage for HUVEC cells adhered on gold-patterned silicon dioxide substrates with gold squares modified with fibronectin, REDV, and KREDV, respectively.
| Surface coating | % Cell coverage | % ratio of single-cell sites/total cell sites |
|---|---|---|
| Fibronectin | 83.47 ± 3.20 | 27.41 ± 3.64 |
| REDV | 56.28 ± 3.03 | 62.25 ± 3.23 |
| KREDV | 78.40 ± 3.96 | 72.17 ± 2.49 |
Cellular viability on patterned surfaces: apoptosis and necrosis
Retaining cell viability after cell adhesion is essential for cell biology studies and biomedical applications such as cell-based sensors and drug screening microarrays. The viability of cells patterned on the three model surfaces were studied with an apoptosis assay noted in Experimental section. Figure 5 shows fluorescence images of HUVEC cells on gold patterns immobilized with (a) Fibronectin, (b) REDVY peptide, and (c) KREDVY peptide after 7 days of cell adhesion. Cells on the gold pattern coated with fibronectin appeared apoptotic and necrotic (top panel). This might be caused by the competitive adhesion of multiple cells on a limited surface area of the gold squares, hindering individual cell growth and survival. This result suggests that large molecule proteins such as fibronectin can result in unpredictable behavior of adhered cells. Since the conformation of fibronectin protein on the gold surface varies from site to site, the degree of interactions of HUVEC cells with the binding peptides of the protein and the number of cells attached on each site can differ from pattern to pattern.
Figure 5.
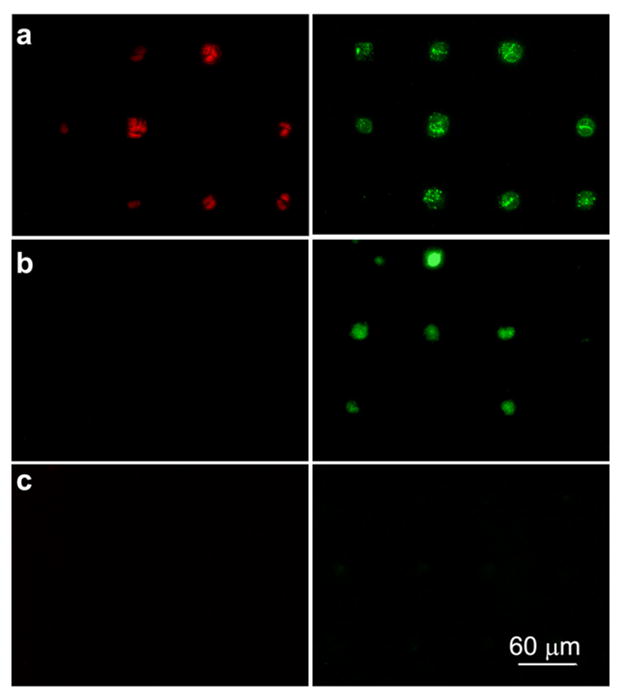
Fluorescence images of HUVEC cells on the gold patterns immobilized with (a) Fibronectin, (b) REDVY peptide, and (c) KREDVY peptide after 7 days of cell adhesion. Apoptotic cells fluoresce green, necrotic cells fluoresce red, and live cells show little or no fluorescence. Images were taken from triplicate substrates for each type of surfaces.
No cell necrosis was observed on REDVY and KREDVY modified surfaces after 7 days of cell adhesion. However, cells adhered on the physically-adsorbed peptide (REDVY) became apoptotic at day 7. This might be attributable to the detachment of REDVY peptides from the gold surface over time, particularly when the cell culture medium was replenished, which started apoptosis. A similar observation has been reported for anchorage-dependent endothelial cells that undergo apoptosis when detached from extracellular matrix substrates.32 Cells adhered on the gold pattern modified with KREDVY remained viable throughout 7 days (i.e., no fluorescence signals), indicating that the covalently bound KREDVY peptide not only facilitates single-cell adhesion through peptide mediated adhesion, but also supports prolonged cell attachment and viability.
In a study to identify the critical parameter for endothelial cells to switch between growth and apoptosis, it was found that that a decrease in cell adhesion area would deleteriously restrict cell spreading and that a 20 μm square island coated with fibronectin would lead to cell death.19 In the present study, a similar conclusion was reached in that endothelial cells on 20 μm squares coated with fibronectin underwent apoptosis (Figure 5a). However, this study further revealed that the surfaces covalently bound with short peptide molecules on 20 μm squares can promote cell spreading and suppress the apoptosis. This observation suggests that not only the cell adhesion area but also the surface chemistry plays an important role in cellular viability, and a surface bound with adhesive short peptides may help retain cellular viability for a prolonged time. Indeed, the type of the ECM has been found to strongly influence the apoptosis,33 and a recent study showed that cell spreading and attachment are dictated by the balance of the availability of potential adhesion domains and cellular traction forces.20 The cell biology involved in the enhancement of the cellular viability by short peptides needs further investigation to reveal more in-depth underlying mechanisms, which is beyond the scope of the current study.
Conclusions
We have developed a simple surface modification strategy to fabricate microarrays of single endothelial cells on gold patterned silicon oxide substrates using a short peptide as a mediator. By comparing cellular adhesion behavior and viability on three model surfaces of different chemistries and cellular binding characteristics, we found that covalently attached KREDVY peptides help cell spreading, increase cell viability, and help localize single cells to 20 μm × 20 μum islands. The performance of covalently bound KREDVY is superior to that of physisorbed REDVY peptide or fibronectin. This study further showed that cell adhesion, spreading, apoptosis and necrosis are influenced not only by the size of surface area that confines the cell but also by the surface chemistry. This finding is particularly interesting in fundamental biology and significant in engineering arrays of single cells.
This cell patterning method is cost effective, requires no device components for depositing proteins or cells, and thus reduces the possibility of protein denaturation and potential damage to cells. Importantly, the method is able to pattern a cell in an area as small as comparable to the cell size while retaining cell viability for prolonged time thus considerably increasing the probability of single cell adhesion, an attribute that has not been demonstrated in previous studies. Study of single cell behavior offers insights to the cell biology, and microarrays of single cells integrated with real-time data acquisition technology may potentially provide a platform for development of cell-based sensors and for fundamental studies of cell processes and mechanisms such as signaling pathways, mutation, gene transfection, and defense mechanism, on either an isolated single cell or statistical basis.
Acknowledgments
This work is supported by a NIH/NIGMS grant (R01 GM075095), the University of Washington Engineered Biomaterials Research Center (NSF-EEC 9529161), and a National Lawrence Berkeley Laboratory ALS Doctoral Fellowship. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098 at Lawrence Berkeley National Laboratory (LBNL). The facilities at Fred Hutchinson Cancer Research Center and UWEB Optical Microscopy and Image Analysis shared resources funded by NSF (EEC-9872882) are acknowledged. We thank Joyce Tseng, Lisamarie Ramos, Yumiko Kusuma, and Ryan Kosai for laboratory assistance.
References
- 1.Chen CS, Alonso JL, Ostuni E, Whitesides GM, Ingber DE. Cell shape provides global control of focal adhesion assembly. Biochemical and Biophysical Research Communications. 2003;307(2):355–361. doi: 10.1016/s0006-291x(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 2.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 3.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411(6833):107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 4.Pancrazio JJ, Gray SA, Shubin YS, Kulagina N, Cuttino DS, Shaffer KM, Eisemann K, Curran A, Zim B, Gross GW, O’Shaughnessy TJ. A portable microelectrode array recording system incorporating cultured neuronal networks for neurotoxin detection. Biosensors & Bioelectronics. 2003;18(11):1339–1347. doi: 10.1016/s0956-5663(03)00092-7. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Sekhon NS, Borninski J, Chen N, Rubinsky B. Instantaneous, quantitative single-cell viability assessment by electrical evaluation of cell membrane integrity with microfabricated devices. Sensors and Actuators a-Physical. 2003;105(1):31–39. [Google Scholar]
- 6.Svedhem S, Dahlborg D, Ekeroth J, Kelly J, Hook F, Gold J. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir. 2003;19(17):6730–6736. [Google Scholar]
- 7.Chiou PY, Ohta AT, Wu MC. Massively parallel manipulation of single cells and microparticles using optical images. Nature. 2005;436(7049):370–372. doi: 10.1038/nature03831. [DOI] [PubMed] [Google Scholar]
- 8.Teruel MN, Meyer T. Parallel single-cell monitoring of receptor-triggered membrane translocation of a calcium-sensing protein module. Science. 2002;295(5561):1910–1912. doi: 10.1126/science.1065028. [DOI] [PubMed] [Google Scholar]
- 9.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297(5584):1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 10.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Micropatterned surfaces for control of cell shape, position, and function. Biotechnology Progress. 1998;14(3):353–363. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 11.Chiu DT, Jeon N, Huang S, Kane RS, Wargo CJ, Choi IS, Ingber DE, White-sides GM. Patterned deposition of cells and proteins onto surfaces by using three-dimensional microfluidic systems. PNAS. 2000;97(6) doi: 10.1073/pnas.040562297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Folch A, Toner M. Microengineering of cellular interactions. Annual review of biomedical engineering. 2000;2:227–256. doi: 10.1146/annurev.bioeng.2.1.227. [DOI] [PubMed] [Google Scholar]
- 13.Whitesides GM, Ostuni E, Takayama S, Jiang XY, Ingber DE. Soft lithography in biology and biochemistry. Annual Review of Biomedical Engineering. 2001;3:335–373. doi: 10.1146/annurev.bioeng.3.1.335. [DOI] [PubMed] [Google Scholar]
- 14.Folch A, Jo BH, Hurtado O, Beebe DJ, Toner M. Microfabricated elastomeric stencils for micropatterning cell cultures. J Biomed Mater Res. 2000;52(2):346–53. doi: 10.1002/1097-4636(200011)52:2<346::aid-jbm14>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 15.Ostuni E, Kane R, Chen CS, Ingber DE, Whitesides GM. Patterning Mammalian Cells Using Elastomeric Membranes. Langmuir. 2000;16:7811–7819. [Google Scholar]
- 16.Folch A, Toner M. Cellular Micropatterns on Biocompatible Materials. Biotechnology Progress. 1998;14:388–392. doi: 10.1021/bp980037b. [DOI] [PubMed] [Google Scholar]
- 17.Ostuni E, Chen SC, Ingber ED, Whitesides MG. Selective Deposition of Proteins and Cells in Arrays of Microwells. Langmuir. 2001;17:2828–2834. [Google Scholar]
- 18.Jimbo Y, Robinson HPC, Kawana A. Simultaneous Measurement of Intracellular Calcium and Electrical-Activity from Patterned Neural Networks in Culture. IEEE Transactions on Biomedical Engineering. 1993;40(8):804–810. doi: 10.1109/10.238465. [DOI] [PubMed] [Google Scholar]
- 19.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276(5317):1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 20.Lehnert D, Wehrle-Haller B, David C, Weiland U, Ballestrem C, Imhof BA, Bastmeyer M. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. Journal of Cell Science. 2004;117(1):41–52. doi: 10.1242/jcs.00836. [DOI] [PubMed] [Google Scholar]
- 21.Kirkpatrick CJ, Otto M, Van Kooten T, Krump V, Kriegsmann J, Bittinger F. Endothelial cell cultures as a tool in biomaterial research. J Mater Sci Mater Med. 1999;10(10/11):589–94. doi: 10.1023/a:1008946615663. [DOI] [PubMed] [Google Scholar]
- 22.Kirkpatrick CJ, Wagner M, Hermanns I, Klein CL, Kohler H, Otto M, van Kooten TG, Bittinger F. Physiology and cell biology of the endothelium: a dynamic interface for cell communication. Int J Microcirc Clin Exp. 1997;17(5):231–40. doi: 10.1159/000179235. [DOI] [PubMed] [Google Scholar]
- 23.Bermpohl D, Halle A, Freyer D, Dagand E, Braun JS, Bechmann I, Schroder NWJ, Weber JR. Bacterial programmed cell death of cerebral endothelial cells involves dual death pathways. Journal Of Clinical Investigation. 2005;115(6):1607–1615. doi: 10.1172/JCI23223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veiseh M, Zhang M. Effect of silicon oxidation on long-term cell selectivity of cell-patterned Au/SiO2 platforms. J Am Chem Soc. 2006;128(4):1197–203. doi: 10.1021/ja055473q. [DOI] [PubMed] [Google Scholar]
- 25.Patel N, Davies CM, Hartshorne M, Heaton JR, Roberts JC, Tendler JBS, Williams MP. Immobilization of Protein Molecules onto Homogeneous and Mixed Carboxylate-Terminated Self-Assembled Monolayers. Langmuir. 1997;13:6485–6490. [Google Scholar]
- 26.Frey BL, Corn RM. Covalent attachment and derivatization of poly(L-lysine) monolayers on gold surfaces as characterized by polarization-modulation FT-IR spectroscopy. Analytical Chemistry. 1996;68(18):3187–3193. [Google Scholar]
- 27.Infrared analysis of peptides and proteins: principles and applications. American Chemical Society; Massachusetts, Boston: 2000. p. ix.p. 190. [Google Scholar]
- 28.Massia SP, Hubbell JA. Vascular Endothelial-Cell Adhesion and Spreading Promoted by the Peptide Redv of the Iiics Region of Plasma Fibronectin Is Mediated by Integrin Alpha-4-Beta-1. Journal of Biological Chemistry. 1992;267(20):14019–14026. [PubMed] [Google Scholar]
- 29.Massia SP, Hubbell JA. Covalent Surface Immobilization of Arg-Gly-Asp-Containing and Tyr-Ile-Gly-Ser-Arg-Containing Peptides to Obtain Well-Defined Cell-Adhesive Substrates. Analytical Biochemistry. 1990;187(2):292–301. doi: 10.1016/0003-2697(90)90459-m. [DOI] [PubMed] [Google Scholar]
- 30.Parker KK, Brock AL, Brangwynne C, Mannix RJ, Wang N, Ostuni E, Geisse NA, Adams JC, Whitesides GM, Ingber DE. Directional control of Lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB J. 2002;16:1195–1204. doi: 10.1096/fj.02-0038com. [DOI] [PubMed] [Google Scholar]
- 31.Wang N, Ostuni E, Whitesides GM, Ingber DE. Micropatterning tractional forces in living cells. Cell Motility and the Cytoskeleton. 2002;52(2):97–106. doi: 10.1002/cm.10037. [DOI] [PubMed] [Google Scholar]
- 32.Fukai F, Mashimo M, Akiyama K, Goto T, Tanuma S, Katayama T. Modulation of apoptotic cell death by extracellular matrix proteins and a fibronectin-derived antiadhesive peptide. Exp Cell Res. 1998;242(1):92–9. doi: 10.1006/excr.1998.4076. [DOI] [PubMed] [Google Scholar]
- 33.Boudreau N, Sympson CJ, Werb Z, Bissell MJ. Suppression of Ice and Apoptosis in Mammary Epithelial-Cells by Extracellular-Matrix. Science. 1995;267(5199):891–893. doi: 10.1126/science.7531366. [DOI] [PMC free article] [PubMed] [Google Scholar]