Abstract
Three new depsipeptides, fijimycins A–C f(1–3), together with the known etamycin A (4), were isolated and identified from the fermentation broth of strain CNS-575, a Streptomyces sp. cultured from a marine sediment sample collected off Nasese, Fiji. The planar structures of the new fijimycins were assigned by combined interpretation of NMR and MS/MS spectroscopic data. These assignments were complicated by the fact that 1–3 occurred as complex amide conformational mixtures. The absolute configurations of the component amino acids were established using the Marfey’s method. Fijimycins A–C, and etamycin A, were shown to possess significant in vitro antibacterial activity against three methicillin-resistant Staphylococcus aureus (MRSA) strains with MIC100 values between 4–16 µg/mL.
Keywords: antibacterial depsipeptides, MRSA activity, etamycin
1. Introduction
Infectious disease caused by methicillin-resistant Staphylococcus aureus (MRSA) is a growing problem of potential urgency in both community and hospital settings.1 To seek new antibiotics to treat MRSA infection, we have initiated studies to examine the secondary metabolites of marine-derived and obligate marine actinomycetes. Recent studies have shown that these microorganisms are a rich source of novel secondary metabolites.2–6 The foundation of this research includes an international collaboration designed to explore the biological and natural product diversity of microorganisms cultured from marine samples collected around the islands of Fiji. As part of this program, we identified a crude fermentation extract of strain CNS-575, which showed appreciable antibiotic activity against methicillin-resistant Staphylococcus aureus. This strain was identified as a Streptomyces sp. based on 16S rDNA sequence analysis. Chemical investigations, reported here, led to the discovery of the fijimycins A–C (1–3), three new depsipeptides of the etamycin class, together with etamycin A (4) itself. Herein, we report the isolation, structure elucidation, and antibiotic activities of these three new compounds.
2. Results and Discussion
2.1. Structure elucidation of fijimycin A (1)
Fijimycin A was isolated as a white, amorphous solid. The molecular formula was assigned as C44H62N8O11 (corresponding to 18 degrees of unsaturation) on the basis of ESI high-resolution mass spectrometry data (obsd [M+Na]+ 901.4436, calcd 901.4430). The molecule was found to adopt two conformations as observed in the 1H NMR spectrum in CDCl3, acetonitrile-d3, methanol-d4, acetone-d6, and DMSO-d6. NMR data acquisition at elevated and depressed temperatures did not reduce the relative composition of the conformers. However, the two conformers were sufficiently well resolved in CDCl3 to permit assignment of the component amino acids and the peptide sequence. The 1H NMR spectrum displayed characteristics of a typical depsipeptide, illustrating three amide NH signals (δH 8.51, 8.22, 6.76), three N-methyl singlets (δH 3.23, 2.99, 2.96), eight α-amino protons (δH 6.65, 5.31, 5.21, 5.14, 4.87, 4.77, 3.68, 3.49), and one ester carbinol proton (δH 5.85). In the 13C NMR spectrum, eight amide or ester resonances (δC 173.6, 171.9, 170.4, 169.2, 168.6, 167.7, 167.5, 167.0) were observed. The IR spectrum of 1 showed intense absorption bands at 1745 and 1638 cm−1, confirming that fijimycin A was a depsipeptide.
Further analysis of 2D NMR spectroscopic data (gCOSY, gHSQC, gHMBC, and TOCSY) allowed eight subunits to be established: α-phenylsarcosine (PhSar), alanine (Ala), N, β-dimethylleucine (DiMeLeu), sarcosine (Sar), 4-hydroxyproline (Hyp), leucine (Leu), threonine (Thr), and 3-hydroxypicolinic acid (3HyPic). The amino acid sequence of 1 was assigned by HMBC analysis using correlations from the α-protons or the N-methyl protons to the carbonyl carbon resonances. Specifically, the linkage of PhSar and Ala was established by HMBC correlations from the N-methyl proton of PhSar (δH 2.99) and the α-proton of Ala (δH 5.14) to the Ala amide carbonyl (δc 171.9). Cross-peaks between the α-proton of Ala and carbonyl carbon (C-13) of DiMeLeu (δc 168.6) linked these two amino acids. This tripeptide fragment was linked to the Sar unit on the basis of HMBC correlations from the N-methyl proton of DiMeLeu (δH 2.96) to the Sar amide carbonyl (C-21, δc 167.7). The subsequent connection of Sar to Hyp was facilitated by HMBC correlations from the N-methyl singlet proton (H-23, δH 3.23) to the Hyp carbonyl carbon C-24 (δc 173.6). The connection of Leu and Hyp was achieved on the basis of an HMBC correlation from the δ-amino proton of Hyp (H-28, δH 3.74) to C-29 (δc 170.4). Further, an HMBC correlation from the α-proton of Leu (δH 4.77) and the α-proton of Thr (δH 4.87) to C-35 (δc 167.5) revealed the linkage of Leu to Thr. An HMBC correlation between the amide proton of Thr (δH 8.51) and C-39 (δc 167.0) linked the 3HyPic to Thr. Finally, the ring closure linkage was secured by a three-bond HMBC correlation from the H-37 proton of Thr (δH 5.85) to C-1 (δc 169.2), which allowed the planar structure of fijimycin A (1) to be assigned.
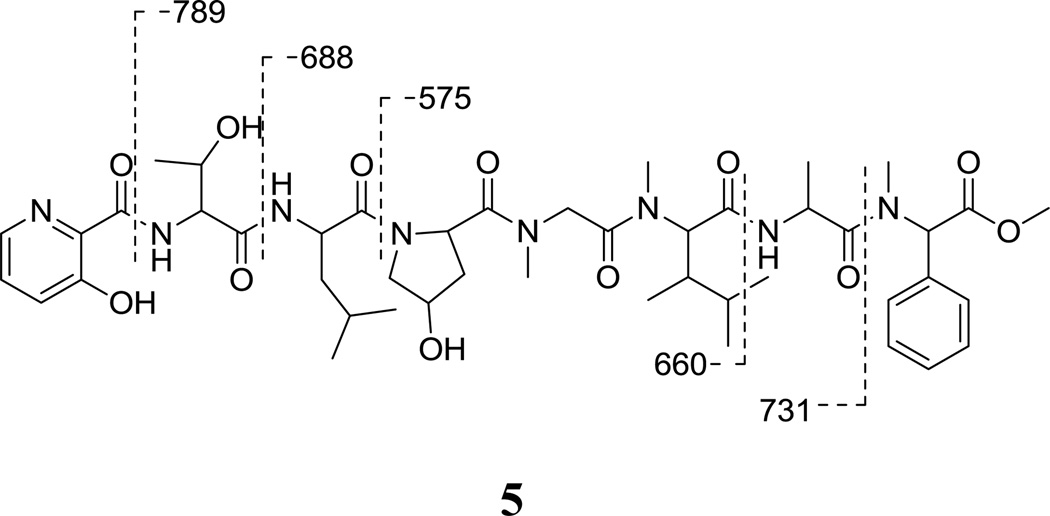
The ester linkage in fijimycin A was also confirmed by methanolysis to yield the methyl ester 5, (Figure 2; ESIMS [M+Na]+ m/z 933). Subsequent analysis on the ESIMS/MS of the methyl ester 5 indicated that the fragmentation patterns were consistent with the amino acid sequence as determined by NMR. Key fragments included those at m/z 789, 731, 688, 660 and 575, which indicated cleavages of the amide bonds between 3HyPic/Thr, PhSar/Ala, Thr/Leu, Ala/DiMeLeu, and Leu/Hyp, respectively.
Figure 2.
Structure of methanolysis product of fijimycin A (5) and mass spectrometric cleavage ions (m/z values) observed in the ESIMS/MS spectrum.
The absolute configurations of the amino acid units in 1 were determined by complete acid hydrolysis and HPLC-MSD analysis of the corresponding Marfey amino acid derivatives.7,8 Treatment of the hydrolysate of 1 (6 N HCl, 110 °C, 12 h) with N-(3-fluoro-2,4 -dinitrophenyl)-l-leucinamide (l-FDLA) yielded Marfey derivatives that were analyzed by LC-MS under positive-mode electrospray ionization (ESI). All the derivatives were identified by their retention times, molecular weights, UV spectra, and comparison of the appropriate FDLA derivatized d- and l-amino acid standards. In the hydrolysate, the retention times of the PhSar FDLA, Ala FDLA, DiMeLeu FDLA, Hyp FDLA, Leu FDLA and Thr FDLA, were 48.4, 37.9, 50.7, 31.5 52.5 and 32.3 min, respectively, which corresponded to the FDLA derivatives of d-α-phenylsarcosine, l-alanine, l-N, β-dimethylleucine, allo-hydroxy-d-proline, d-leucine, and l-threonine. Thus, the absolute configurations of the amino acid units in 1 were assigned. On the basis of these evaluations, fijimycin A (1) was defined as a stereoisomer of etamycin A containing a d-α-phenylsarcosine.
2.2. Structure elucidation of fijimycin B (2)
Fijimycin B was obtained as a white, amorphous solid, which was determined to have the molecular formula C42H66N8O11 by analysis of HRESIMS data (obsd [M+Na]+ 881.4737, calcd 881.4743). The 1H NMR spectrum of fijimycin B was similar to that of 1. However, major differences were observed for both the aromatic and the aliphatic proton resonances. The disappearance of benzene signals suggested the lost of the α-phenylsarcosine (PhSar). Meanwhile, the 1H, COSY and TOCSY NMR spectra displayed a spin system consisting of two methyls at δ 0.83 and 0.93, a methine at δ 1.61, a methylene at δ 1.79, a methine at δ 5.44, corresponding to a leucine moiety. An HMBC correlation from the methine proton (1-H, δ 5.44) to the N-methyl carbonyl (C-7, δ 29.8) suggested the existence of an N-methylleucine (NMeLeu) residue. Using the same approach as in the assignment of 1, the overall structure was assigned by interpretation of 1D and 2D NMR and ESIMS/MS spectroscopic data. Excluding the NMeLeu unit, the identity and sequence of amino acids in fijimycin B were the same as in 1. Using Marfey derivitization and LC/MS analysis, the absolute configurations of amino acid units in 2 were determined as l-NMeLeu, l-Ala, l-DiMeLeu, d-HyP, d-Leu, and l-Thr.
2.3 Structure elucidation of fijimycin C (3)
Fijimycin C, a minor metabolite, was isolated as an amorphous white powder, which was assigned the molecular formula C44H62N8O12, by analysis of HRESIMS data (m/z [M+Na]+ 917.4365, calcd 917.4379). The molecular composition indicated one more oxygen atom compared to the formula of 1. Analysis of 1D and 2D NMR spectroscopic data allowed assignment of fijimycin C as 3. Analysis of the 1D proton and 2D COSY NMR spectra of 3 showed a spin system consisting of an amide proton doublet at δ 7.29, a methine at δ 5.27, a methylene at δ 3.82 and 3.93, an OH multiplet at δ 4.89, suggesting the alanine (Ala) moiety in 1 was replaced by a serine (Ser) unit. Marfey derivation and LC-MS analysis revealed the absolute configurations of amino acid units in 3 as l-PhSar, l-Ser, l-DiMeLeu, d-HyP, d-Leu, and l-Thr.
2.4. Antibiotic activities of fijimycins A–C
The antibiotic activities of fijimycins A–C (1–3) were examined in vitro against three MRSA strains, the hospital-associated strain (ATCC33591), the sequenced hospital-associated strain (Sanger 252), and the community-associated strain (UAMS1182). As shown in Table 4, fijimycins A, C and etamycin A exhibited strong antibiotic activities against the three MRSA strains in the concentration range of 4–32 µg/mL. However, fijimycin B showed weak inhibition against both ATCC33591 and UAMS1182, which indicated that the α-phenylsarcosine unit might be vital for significant antibacterial activity. The similar antimicrobial activities of the stereoisomers fijimycin A and etamycin A suggests that substituting d- for l-α-phenylsarcosine had little effect on the anti-MRSA activities.
Table 4.
Antibiotic activities (MIC100, µg/mL*) of fijimycins A–C and etamycin A against three MRSA strains
| Compound | ATCC33591 | Sanger 252 | UAMS1182 |
|---|---|---|---|
| Fijimycin A | 16 | 32 | 4 |
| Fijimycin B | >32 | n/a | >32 |
| Fijimycin C | 32 | n/a | 8 |
| Etamycin A | 16 | 16 | 4 |
MIC100 values represent the lowest drug concentration inhibiting 100% of bacterial growth.
3. Discussion
Etamycin A, also called virifogrisein I, was first isolated from cultures of a terrestrial Streptomyces species by Heinermann and Bartz in 1954. Subsequently, etamycin A was more completely characterized in 1958 by Sheehan.9 As one of the important members of group B streptogramins, etamycin A exhibited considerable activity against gram-positive bacteria, as well as Mycobacterium tuberculosis. Etamycin found marginal use in Europe as a clinical antibacterial agent and as a feed additive in agriculture.10 In a recent paper, the MRSA activities of etamycin were described in detail.11 Fijimycins A–C are new members of the etamycin class of antibacterial cyclic depsipeptides. 1H NMR studies of fijimycin A using different solvents revealed that fijimycin A exists as an equilibrium mixture of at least two or more stable conformers at room temperature. Similarly, etamycin A appeared as three distinct conformers in the NMR spectral data and the presence of conformers was not affected by changes of NMR solvent or acquisition temperature.11 The presence of conformers might be attributed to the cis and trans isomerization of the Hyp peptide bonds.12–14
4. Experimental Section
4.1. General experimental procedures
Optical rotations were measured using a JASCO P-2000 polarimeter with a 1 cm cell. IR spectra were acquired on a Perkin-Elmer 1600 series FTIR spectrometer. 1H, 13C, and 2D NMR spectroscopic data were obtained on Varian Inova 500 MHz and Varian Inova 300 MHz NMR spectrometers. HRESIMS data were recorded on a Thermo LTQ Orbitrap XL mass spectrometer, and ESIMS/MS data were recorded on a Thermo LCQ deca mass spectrometer. Low resolution LC-MS spectra were obtained on a Hewlett-Packard HP1100 integrated LC-MS system with a reversed-phase C18 column (Agilent, 4.6 mm × 100 mm, 5 µm) at a flow rate of 0.7 mL/min. Reversed phase HPLC separations were performed using a semipreparative C18 Phenomenex Luna 5 µm (10 mm × 250 mm) column with a CH3CN/H2O gradient solvent system. Preparative HPLC separations were performed using a Waters model 4000 system with a UV variable-wavelength detector, monitoring at 254 nm, and employing a C18 Nova-Pak 6 µm, 60 Å (40 mm × 300 mm) column. Amino acid standards of known configurations used to determine the absolute stereochemistry of fijimycins were purchased from Sigma-Aldrich, INC.
4.2. Bacterial isolation and identification
Strain CNS-575 was obtained from a marine sediment sample collected at a depth of ca. 0.5 m from the Nasese shoreline, Viti Levu, Fiji. The sediment was first heat-shocked at 55 °C for 6 minutes and then directly plated into medium M1 (10 g starch, 4 g yeast extract, 2 g peptone, 1 L filtered seawater) supplemented with 0.5 µg/ml rifampicin and 100 µg/L cycloheximide as previously described.15 The strain was identified as a Streptomyces sp. based on 16S rRNA gene sequence analysis (accession number GQ325662) and shares greatest similarity (99.2%) with the marine sponge-derived species S. haliclonae (accession number AB473556).
4.3. Fermentation and Extraction
Streptomyces sp., strain CNS-575, was cultured at 27 °C for 6 days with shaking at 215 rpm in 40 × 1 L volumes of the liquid medium (A1Bfe+C) [composed of 10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3·4H2O, 100 mg KBr, per 1 L seawater]. Pre-sterilized Amberlite XAD-7 resin (20 g/L) was added at the end of the fermentation period to adsorb extracellular secondary metabolites. The culture and resin were shaken at low speed for two additional hours. The resin and most of the cell mass were collected by filtration through cheesecloth and washed with DI water to remove salts. The resin, cell mass, and cheesecloth were then soaked for 2 h in acetone, after which the acetone extract was filtered and the solvent removed under vacuum to give 8.6 g of solid material from a 40 L culture.
4.4. Isolation of fijimycins A–C (1–3)
The crude acetone extract (8.6 g) was fractionated by Si gel open column chromatography eluting with a step gradient from 10% ethyl acetate in isooctane to 15% methanol in ethyl acetate to give nine fractions. Fraction six (3.9 g), which contained the depsipeptides, was subjected to reversed-phase HPLC (Phenomenex Luna C18, 10 mm × 250 mm, 5 µm) with 65% aqueous CH3CN at 2.5 mL/min flow rate and UV detection at 254 nm. Fijimycins A (1, 65 mg), B (2, 9.6 mg), and C (3, 3.2 mg) were eluted at 27.7, 40.5, and 17.5 min, respectively.
4.5. Fijimycin A
Fijimycin A was isolated as an amorphous white powder; [α]D23 -11.2 (c 0.50, MeOH; observed optical rotation was the average for all conformers); UV (MeOH) λmax 203 (4.5), 308 (3.9), 360 (3.1) nm; IR (neat) νmax 3317, 2960, 1745, 1638, 1518, 1451, 1237, 1095, 736, 698 cm−1; 1H NMR (500 MHz, CDCl3) and 13C NMR (300 MHz, CDCl3) data, see Table 1; ESIMS/MS m/z 879 [M + H]+, 901 [M + Na]+; HRESIMS m/z [M + Na]+ 901.4436 (calcd for C44H62N8O11Na 901.4430).
Table 1.
NMR spectroscopic data for fijimycin A (1) in CDCl3
| Amino Acid |
Position | Major Rotamer |
Minor Rotamer |
||
|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | ||
| d-PhSar | 1 | 169.2 | 169.5 | ||
| 2 | 6.65, m | 61.2 | 6.05, m | 62.5 | |
| 3 | 134.2 | 133.3 | |||
| 4 (8) | 7.35, m | 128.9 | 7.35, m | 128.9 | |
| 5 (7) | 7.33, m | 128.5 | 7.33, m | 128.5 | |
| 6 | 7.21, m | 128.2 | 7.21, m | 128.2 | |
| 9 | 2.99, s | 32.6 | 2.94, s | 33.4 | |
| l-Ala | 10 | 171.9 | 172.3 | ||
| 11 | 5.14, t (6.0) | 46.2 | 5.11, t (6.0) | 46.2 | |
| 12 | 1.37, d (6.6) | 18.3 | 1.29, d (6.0) | 18.5 | |
| 11-NH | 8.22, d (10.8) | 7.74, d (6.0) | |||
| l-DiMeLeu | 13 | 168.6 | 167.5 | ||
| 14 | 3.68, d (7.2) | 63.2 | 4.17, d (4.8) | 62.2 | |
| 15 | 2.11, t (7.8) | 36.6 | 2.23, m | 35.3 | |
| 16 | 0.62, d (6.6) | 8.6 | 0.65, d (6.6) | 8.4 | |
| 17 | 1.05, m | 27.2 | 1.05, m | 27.2 | |
| 18 | 0.51, brs | 14.7 | 0.51, brs | 15.9 | |
| 19 | 0.72, brs | 21.7 | 0.44, brs | 21.7 | |
| 20 | 2.96, s | 29.2 | 2.88, s | 28.5 | |
| Sar | 21 | 167.7 | 168.4 | ||
| 22 | 3.49, d (14.4) | 51.2 | 3.61, d (17.4) | 51.3 | |
| 5.21, d (14.4) | 6.47, d (15.6) | ||||
| 23 | 3.23, s | 31.2 | 3.09, s | 36.9 | |
| d-Hyp | 24 | 173.6 | 172.9 | ||
| 25 | 5.31, brs | 55.7 | 4.87, m | 56.5 | |
| 26 | 1.98, d (13.8) | 35.5 | 2.19, m | 35.3 | |
| 2.43, t (10.8) | |||||
| 27 | 4.47, brs | 71.9 | 4.55, m | 70.6 | |
| 28 | 3.74, d (10.8) | 56.2 | 3.68, d (10.8) | 58.5 | |
| 3.87, d (10.8) | 4.11, dd (6.0) | ||||
| 27-OH | 6.35, brd (9.6) | 6.66, brd (11.4) | |||
| d-Leu | 29 | 170.4 | 173.2 | ||
| 30 | 4.77, m | 48.7 | 4.93, dd (7.2) | 49.4 | |
| 31 | 1.35, m | 40.8 | 1.42, m | 43.1 | |
| 32 | 1.60, m | 24.5 | 1.53, m | 24.6 | |
| 33 | 1.06, d (6.6) | 21.2 | 0.97, d (6.0) | 21.9 | |
| 34 | 0.86, d (6.6) | 23.3 | 0.88, d (7.2) | 23.3 | |
| 30-NH | 6.76, d (9.6) | 7.20, m | |||
| d-Thr | 35 | 167.5 | 167.1 | ||
| 36 | 4.87, m | 55.4 | 4.87, m | 55.4 | |
| 37 | 5.85, brs | 71.7 | 5.85, brs | 70.8 | |
| 38 | 1.35, m | 17.2 | 1.21, d (2.4) | 16.2 | |
| 36-NH | 8.51, brs | 8.58, brs | |||
| 3HyPic | 39 | 167.0 | 166.5 | ||
| 40 | 130.1 | 130.1 | |||
| 41 | 157.9 | 157.9 | |||
| 42 | 7.40, m | 126.7 | 7.40, m | 126.7 | |
| 43 | 7.46, m | 129.5 | 7.46, m | 129.5 | |
| 44 | 8.31, brs | 141.3 | 8.17, d (3.6) | 140.3 | |
| 41-OH | 11.62, s | 11.57, s | |||
4.6. Fijimycin B
Fijimycin B was isolated as an amorphous white powder; [α]D23 -18.2 (c 0.50, MeOH; observed optical rotation was the average for all conformers); UV (MeOH) λmax 203 (4.5), 308 (3.9), 360 (3.1) nm; IR (neat) νmax 3329, 2959, 1746, 1640, 1516, 1452, 1290, 1184, 1090 cm−1; 1H NMR (500 MHz, CDCl3) and 13C NMR (300 MHz, CDCl3) data, see Table 2; ESIMS/MS m/z 859 [M + H]+, 881 [M + Na]+; HRESIMS m/z [M + Na]+ 881.4737 (calcd for C42H66N8O11Na 881.4743).
Table 2.
NMR spectroscopic data for fijimycin B (2) in CDCl3
| Amino Acid |
Position | Major Rotamer |
Minor Rotamer |
||
|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | ||
| l-NMeLeu | 1 | 169.2 | 169.7 | ||
| 2 | 5.44, m | 54.3 | 5.18, m | 53.0 | |
| 3 | 1.79, m | 36.1 | 1.76, m | 38.0 | |
| 4 | 1.61, m | 24.5 | 1.60, m | 24.7 | |
| 5 | 0.83, m | 21.9 | 0.83, m | 21.9 | |
| 6 | 0.93, m | 24.3 | 0.93, m | 24.3 | |
| 7 | 3.04, s | 29.8 | 3.02, s | 36.8 | |
| l-Ala | 8 | 173.6 | 174.0 | ||
| 9 | 4.99, m | 45.8 | 4.96, m | 46.3 | |
| 10 | 1.39, d (6.6) | 17.8 | 1.46, d (7.2) | 15.1 | |
| 9-NH | 7.17, d (7.8) | 7.92, d (8.4) | |||
| l-DiMeLeu | 11 | 168.8 | 169.2 | ||
| 12 | 5.07, d (10.8) | 58.4 | 3.75, d (11.4) | 63.2 | |
| 13 | 2.18, m | 36.9 | 2.22, m | 37.3 | |
| 14 | 0.60, d, (10.8) | 8.7 | 0.73, d (7.2) | 8.6 | |
| 15 | 1.78, m | 28.0 | 1.63, m | 27.4 | |
| 16 | 0.94, m | 21.9 | 0.94, m | 21.9 | |
| 17 | 0.77 | 15.7 | 0.77, m | 15.7 | |
| 18 | 2.82, s | 29.6 | 2.91, s | 29.5 | |
| Sar | 19 | 167.5 | 167.9 | ||
| 20 | 3.87, d (16.0) | 52.3 | 3.38, d (13.8) | 50.6 | |
| 5.31, d (16.0) | 5.44, d (13.8) | ||||
| 21 | 2.92, s | 35.8 | 3.23, s | 35.5 | |
| d-Hyp | 22 | 174.0 | 173.6 | ||
| 23 | 5.16, m | 54.1 | 4.83, d (9.6) | 56.0 | |
| 24 | 2.05, d (13.8) | 37.6 | 1.93, d (14.4) | 35.6 | |
| 2.18, m | 2.22, m | ||||
| 25 | 4.53, t (5.4) | 71.2 | 4.45, brd, (9.6) | 71.7 | |
| 26 | 3.70, d (10.8) | 58.0 | 3.77, m | 56.2 | |
| 4.35, m | |||||
| 25-OH | 6.76, d (11.4) | 5.58, d (12.0) | |||
| d-Leu | 27 | 172.3 | 172.5 | ||
| 28 | 4.93, m | 49.3 | 4.79, m | 49.7 | |
| 29 | 2.42, m | 38.2 | 2.16, m | 38.7 | |
| 30 | 1.33, m | 24.8 | 1.39, m | 24.5 | |
| 31 | 0.98, m | 22.2 | 0.94, m | 22.2 | |
| 32 | 0.98, m | 23.4 | 0.95, m | 23.3 | |
| 28-NH | 7.44, d (9.0) | 7.55, d (8.4) | |||
| l-Thr | 33 | 166.4 | 166.2 | ||
| 34 | 4.40, m | 54.3 | 4.37, m | 54.9 | |
| 35 | 5.17, m | 69.3 | 5.22, m | 68.6 | |
| 36 | 1.20, d (6.6) | 13.8 | 1.22, d (6.6) | 13.8 | |
| 34-NH | 8.92, d (6.0) | 8.99, d (6.0) | |||
| 3HyPic | 37 | 167.9 | 168.2 | ||
| 38 | 131.0 | 130.8 | |||
| 39 | 157.3 | 157.3 | |||
| 40 | 7.24, m | 126.0 | 7.24, m | 126.0 | |
| 41 | 7.31, m | 128.8 | 7.31, m | 128.8 | |
| 42 | 8.10, brs | 139.9 | 8.05, m | 139.9 | |
| 39-OH | 11.63, s | 11.66, s | |||
4.7. Fijimycin C
Fijimycin C was isolated as an amorphous white powder; [α]D23 +2.0 (c 0.20, MeOH; observed optical rotation was the average for all conformers); UV (MeOH) λmax 203 (4.5), 308 (3.9), 360 (3.1) nm; IR (neat) νmax 3319, 2960, 1745, 1638, 1520, 1452, 1296, 1197 cm−1; 1H NMR (500 MHz, CDCl3) and 13C NMR (300 MHz, CDCl3) data, see Table 3; ESIMS/MS m/z 895 [M + H]+, 917 [M + Na]+; HRESIMS m/z [M + Na]+ 917.4365 (calcd for C44H62N8O12Na 917.4379).
Table 3.
NMR spectroscopic data for fijimycin C (3) in CDCl3
| Amino Acid |
Position | Major Rotamer |
Minor Rotamer |
||
|---|---|---|---|---|---|
| δH (J in Hz) | δC | δH (J in Hz) | δC | ||
| d-PhSar | 1 | 168.3 | 168.4 | ||
| 2 | 6.29, s | 60.2 | 6.37, s | 59.9 | |
| 3 | 131.2 | 131.4 | |||
| 4 (8) | 7.49, m | 129.5 | 7.49, m | 129.3 | |
| 5 (7) | 7.42, m | 129.0 | 7.42, m | 129.0 | |
| 6 | 7.31, m | 126.3 | 7.32, m | 126.3 | |
| 9 | 2.93, s | 31.4 | 2.98, s | 31.1 | |
| l-Ser | 10 | 170.5 | 171.6 | ||
| 11 | 5.08, m | 52.6 | 5.27, m | 52.5 | |
| 12 | 3.82, m | 62.5 | 3.78, m | 63.3 | |
| 3.93, m | 4.11, m | ||||
| 11-NH | 7.29, m | 7.74, d (9.0) | |||
| 12-OH | 4.89, m | 4.98, m | |||
| l-DiMeLeu | 13 | 169.2 | 168.6 | ||
| 14 | 5.10, d (10.8) | 58.7 | 3.87, m | 62.8 | |
| 15 | 2.20, m | 34.3 | 2.27, m | 36.5 | |
| 16 | 0.64, d (6.6) | 8.4 | 0.78, d (6.6) | 8.7 | |
| 17 | 2.02, m | 27.9 | 1.63, m | 27.4 | |
| 18 | 0.96, m | 21.7 | 0.95, m | 21.7 | |
| 19 | 0.80, d (6.6) | 15.6 | 0.81, m | 15.2 | |
| 20 | 2.87, s | 29.3 | 2.96, s | 29.6 | |
| Sar | 21 | 167.3 | 167.9 | ||
| 22 | 3.96, d (18.0) | 52.1 | 3.80, d (14.4) | 52.3 | |
| 5.09, m | 5.93, d (17.4) | ||||
| 23 | 2.93, s | 35.6 | 3.03, s | 36.9 | |
| d-Hyp | 24 | 173.8 | 173.8 | ||
| 25 | 5.17, d (9.0) | 54.1 | 4.81, m | 56.3 | |
| 26 | 2.03, d (14.4) | 37.6 | 2.14, m | 37.6 | |
| 2.16, m | 2.19, m | ||||
| 27 | 4.54, m | 71.3 | 4.49, m | 71.6 | |
| 28 | 3.75, d (11.4) | 58.0 | 3.68, d (12.0) | 58.1 | |
| 4.36, dd (6.0) | 3.95, m | ||||
| 27-OH | 6.44, brd (10.2) | 6.57, brd (10.8) | |||
| d-Leu | 29 | 173.4 | 174.0 | ||
| 30 | 4.90, m | 49.7 | 5.01, m | 49.3 | |
| 31 | 1.48, m | 39.7 | 2.43, t (12.0) | 38.7 | |
| 2.19, m | 1.41,m | ||||
| 32 | 1.86, m | 24.6 | 1.92, m | 27.1 | |
| 33 | 0.97, d (7.2) | 21.7 | 1.00, d (6.6) | 20.7 | |
| 34 | 1.05, d (7.2) | 23.3 | 1.03, d (7.2) | 23.3 | |
| 30-NH | 7.59, d (9.0) | 7.66, d (9.0) | |||
| l-Thr | 35 | 166.8 | 167.3 | ||
| 36 | 4.85, m | 53.6 | 5.01, m | 53.2 | |
| 37 | 5.35, m | 69.7 | 5.45, m | 69.5 | |
| 38 | 1.21, d (6.6) | 13.6 | 1.23, d (6.6) | 13.6 | |
| 36-NH | 8.96, d (6.6) | 9.04, d (6.0) | |||
| 3HyPic | 39 | 167.1 | 167.3 | ||
| 40 | 131.0 | 131.0 | |||
| 41 | 157.5 | 157.5 | |||
| 42 | 7.27, m | 125.8 | 7.27, m | 125.8 | |
| 43 | 7.32, m | 128.6 | 7.33, m | 128.6 | |
| 44 | 8.08, d (3.6) | 139.9 | 8.11, d (3.6) | 140.0 | |
| 41-OH | 11.8, brs | 11.6, brs | |||
4.8. Acid hydrolysis and Marfey analysis
Hydrolysis of fijimycins A, B and C (0.5 mg each) was achieved, in separate experiments, by the addition of 0.5 mL of 6 N HCl at 115 °C for 12 h. The hydrolysate was evaporated to dryness and resuspended in water (1 mL) and again evaporated to dryness under a stream of N2 to remove traces of HCl. The solvent was removed under vacuum and 0.4 mL NaHCO3 (100 µL) and N-(3-fluoro-4, 6-dinitrophenyl)-l-leucinamide (FDLA, 50 µL [10 mg/mL solution in acetone]) were added, and the mixture was heated at 60 °C for 30 min. The reaction mixture was cooled, neutralized with 2 N HCl (200 µL), and diluted with CH3CN (300 µL). About 10 µL of the l-FDLA derivative was analyzed by LC-MS using a C18 column (Luna, 4.6 mm × 100 mm). Aqueous CH3CN containing 1% TFA was used as the mobile phase with a linear gradient elution (10–50% for 45 min) at a flow rate of 0.7 mL/min. l-FDLA-derivatized amino acids were detected by absorption at 340 nm. A Hewlett-Packard Series 1100 MSD mass spectrometer was used for detection in ESI (positive, mass range 100–1200 Da) mode. The retention times (min) of the derivatives were compared with those of authentic derivatized standards, with the exception of N,β-dimethylleucine (DiMeLeu, for which no standard was available): l-Thr (32.3), l-allo-Thr (33.3), l-Leu (45.2), l-Hyp (31.2), l-Ala (37.9), l- PhSar (46.5), l-NMeLeu (48.3), l-Ser (32.9); d-Thr (37.6), d-allo-Thr (35.2), d-Leu (52.5), d-Hyp (31.5), d-Ala (41.7), D-PhSar (48.4), D-NMeLeu (51.4), D-Ser (33.6) and Sar (36.0). The L-FDLA derivative of DiMeLeu fijimycins hydrolyses were detected at 50.7 min, while the L- and D-FDLA derivatives of DiMeLeu were detected at 50.7 and 54.7 min, respectively. No standards were available for DiMeLeu. However, the elution order was assumed to be the same as observed for Leu. The absolute configurations of DiMeLeu were assigned as S (l-DiMeLeu). It was shown that fijimycin A (1) was composed of d-PhSar, l-Ala, l-DiMeLeu, Sar, d-Hyp, d-Leu, and l-Thr; fijimycin B (2) of l-NMeLeu, l-Ala, l-DiMeLeu, Sar, d-Hyp, d-Leu, and l-Thr; and fijimycin C (3) of l-PhSar, l-Ser, l-DiMeLeu, Sar, d-HyP, d-Leu, and l-Thr.
4.9. Methanolysis of fijimycin A (1)
To 1.5 mg of 1 was added 1.5 mL of a 0.5 N NaOMe solution, and the reaction was stirred at room temperature for 2 h under nitrogen. LC-MS analysis indicated the complete conversion of the starting material to the desired methyl ester. The mixture was neutralized by adding 1 mL saturated NH4Cl, and the resulting mixture was extracted with EtOAc. The solvent removed under reduced pressure, and the methanolysis product obtained (5) was purified by HPLC (semipreparative Phenomenex Luna C18, 10 mm × 250 mm, 5 µm) with a gradient solvent system (0–5 min; 40% aqueous CH3CN, 5–35 min; 40%–70% aqueous CH3CN, 35–45 min; 70%–100% CH3CN) at 2.5 mL/min flow rate using UV detection at 254 nm, to give 0.4 mg of the ring-opened methyl ester derivative of fijimycin A.
Supplementary Material
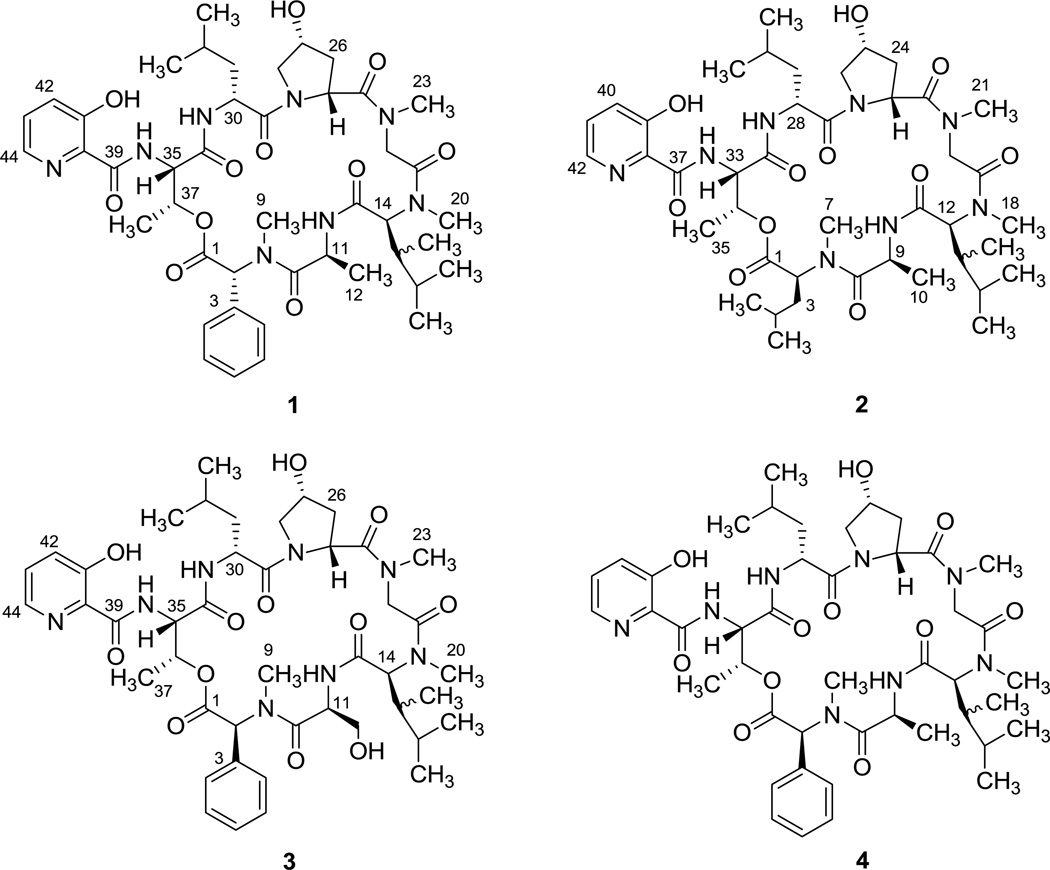
Figure 1.
Structures of fijimycins A–C (1–3) and etamycin A (4).
Acknowledgments
This research is a result of financial support from the National Institutes of Health/Fogarty Center International Cooperative Biodiversity Groups program (grant U01-TW007401-01) and NIH award RO1 GM084350-01. We thank the people of Fiji for their hospitality and the government and local resource owners for permission to collect samples in their local offshore waters. We thank C. A. Kauffman for fermentation assistance and S. Kelly for performing the 16S RNA analysis. We also thank A. Mrse (UCSD) for assistance with NMR experiments and Y. Su (UCSD) for HRMS and MS/MS data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting Information Available
The 1H, 13C, gCOSY, gHSQC, gHMBC and TOCSY spectra of 1–3 are available on line at: http://www.sciencedirect.com.
Reference and Notes
- 1.Hanson MR, Chung CL. J. Drugs Dermatol. 2009;8:281. [PubMed] [Google Scholar]
- 2.Blunt JW, Copp BR, Hu WP, Munro MH, Northcote PT, Prinsep MR. Nat. Prod. Rep. 2008;25:35. doi: 10.1039/b701534h. [DOI] [PubMed] [Google Scholar]
- 3.Fenical W, Jensen PR. Nat. Chem. Biol. 2006;2:666. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 4.Jensen PR, Williams PG, Oh DC, Zeigler L, Fenical W. Appl. Environ. Microbiol. 2007;73:1146. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. Int. Ed. Engl. 2003;42:355. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 6.Asolkar RN, Freel KC, Jensen PR, Fenical W, Kondratyuk TP, Park EJ, Pezzuto JM. J. Nat. Prod. 2009;72:396. doi: 10.1021/np800617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii K, Ikai Y, Mayumi T, Oka H, Suzuki M, Harada K. Anal. Chem. 1997;69:3346. [Google Scholar]
- 8.Fujii K, Shimoya T, Ikai Y, Oka H, Harada K. Tetrahedron. Lett. 1998;39:2579. [Google Scholar]
- 9.Sheehan JC, Zachau HG, Lawson WB. J. Am. Chem. Soc. 1958;80:3349. [Google Scholar]
- 10.Mukhtar TA, Wright GD. Chem. Rev. 2005;105:529. doi: 10.1021/cr030110z. [DOI] [PubMed] [Google Scholar]
- 11.Haste NM, Perera VR, Maloney KN, Tran DN, Jensen PR, Fenical W, Nizet V, Hensler ME. J.Antibiot. 2010;63:219. doi: 10.1038/ja.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dugave C, Demange L. Chem. Rev. 2003;103:2475. doi: 10.1021/cr0104375. [DOI] [PubMed] [Google Scholar]
- 13.Mongkolvisut W, Sutthivaiyakit S, Leutbecher H, Mika S, Klaiber I, Möller W, Rösner H, Beifuss U, Conrad J. J. Nat. Prod. 2006;69:1435. doi: 10.1021/np0602012. [DOI] [PubMed] [Google Scholar]
- 14.Williams DE, Yu K, Behrisch HW, Soest RV, Andersen RJ. J. Nat. Prod. 2009;72:1253. doi: 10.1021/np900121m. [DOI] [PubMed] [Google Scholar]
- 15.Gontang EA, Fenical W, Jensen PR. Appl. Environ. Microbiol. 2007;73:3272. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.