Abstract
Modification of proteins with ubiquitin chains is an essential regulatory event in cell cycle control. Differences in the connectivity of ubiquitin chains are believed to result in distinct functional consequences for the modified proteins. Among eight possible homogenous chain types, canonical Lys48-linked ubiquitin chains have long been recognized to drive the proteasomal degradation of cell cycle regulators, and Lys48 is the only essential lysine residue of ubiquitin in yeast. It thus came as a surprise that in higher eukaryotes atypical K11-linked ubiquitin chains regulate the substrates of the anaphase-promoting complex and control progression through mitosis. Here, we discuss recent findings that shed light on the assembly and function of K11-linked chains during cell division.
The ubiquitin code
Information can be transmitted in many ways, be it print media, television, internet, or social networks. Even the shortest notes relayed through these means rely on a code: the symbology of ☺ and ☹ or the more established written word. The more complex this code, the more information can be communicated, yet the response still depends on the recipient’s interpretation of the message. In eukaryotes, protein ubiquitination follows many of these principles.
Catalyzed by a cascade of E1 ubiquitin-activating, E2 ubiquitin-conjugating, and E3 ubiquitin-protein ligase enzymes, ubiquitin becomes covalently linked to Lys residues in proteins (Box 1; [1-3]). Modification with a single ubiquitin, referred to as monoubiquitination, often alters substrate localization or interactions [4]. This first ubiquitin can also function as the starting point for the synthesis of a polymeric chain, in which ubiquitin molecules are connected through isopeptide bonds between the C-terminus of one ubiquitin and the amino-group at one of seven Lys residues or the N-terminus of another ubiquitin [2]. Depending on the linkage between ubiquitin molecules, these chains can encode distinct information. For example, chains linked through Lys48 of ubiquitin (K48-linked chains) are a targeting device for protein degradation by the 26S proteasome [5, 6], whereas K63-linked chains act as molecular scaffolds, bringing together subunits of oligomeric kinase or DNA repair complexes [7, 8].
As K48- and K63-linked ubiquitin chains were discovered many years ago, much has been learned about their functions, and they are often referred to as “canonical” ubiquitin chains. By contrast, “non-canonical” or “atypical” chains remain incompletely characterized, leaving us with a poor understanding of the breadth of the ubiquitin code. Two atypical chain types, linear and K11-linked ubiquitin chains, were recently identified in cells, where they act in transcription factor activation and cell division, respectively [9, 10]. The important roles played by linear and K11-linked chains strongly support the notion that ubiquitination can constitute an elaborate code that cells use to control the activities of key signaling molecules. Here, we discuss insights into this system that have been gained from studying the assembly and function of K11-linked ubiquitin chains.
When are K11-linked ubiquitin chains detected in cells?
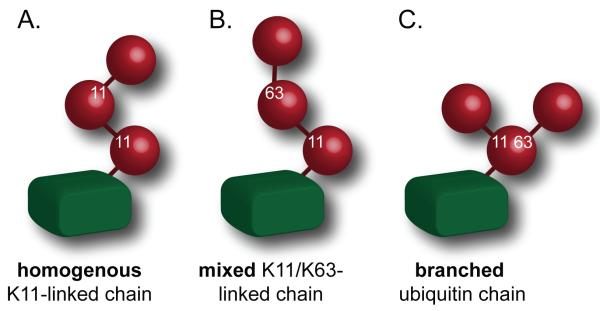
In homogenous chains, all ubiquitin molecules are connected through the same linkage (Figure 1A). For signaling purposes, chains that contain long stretches of uniform linkage might also be considered homogenous. If multiple linkages exist within a chain, these assemblies either have mixed or branched topologies (Figure 1B, C). K11-linkages have been detected in all chain types, and the different topologies might have consequences for their biological functions. For example, homogenous K11-linked chains mediate proteasomal degradation [9, 11], whereas mixed K11/K63-linked chains function non-proteolytically during endocytosis or NF-κB signaling [12, 13].
Figure 1. K11-linkages are found in chains of distinct topologies.
A. In homogenous K11-linked chains, all ubiquitin molecules are connected through K11-linkages. B. In mixed chains, K11- and other linkages are found, yet only one amino-group is modified per ubiquitin molecule. C. In branched ubiquitin chains, a single ubiquitin is connected to at least two other ubiquitin molecules.
The existence of K11-linkages was first suggested by experiments that analyzed the specificity of the E2 Ube2S in vitro [14], and proteomics later identified K11-linkages in cells with varying abundance [15-20]. An early analysis found comparable levels of K11- and K48-linkages in yeast [16], while a later study reported a lower abundance for K11-linkages in this organism [17]. In asynchronously dividing human cells, K11-linkages only represent ~2% of the ubiquitin conjugate pool [19, 20]. The differences in the levels of K11-linkages among these studies could be due to technical reasons, such as distinct purification or growth procedures, or they might reveal insight into the regulation of K11-linkage formation. For example, K11-linkages accumulate when cells are stressed by proteasome inhibition, heat shock, and formation of toxic aggregates, or when they passage through a specific cell cycle stage [11, 16, 19, 20].
Homogenous K11-linked chains were discovered as the product of the human E3 anaphase-promoting complex (APC/C), an essential regulator of cell division [9]. Drosophila and Xenopus APC/C also assemble K11-linked chains [21, 22]. When the APC/C is activated during mitosis, K11-linked chains rise dramatically in abundance [11], and a blockage of K11-linkage formation in Xenopus embryos resulted in cell division defects similar to those observed for APC/C-inhibition [9]. Conversely, when cells exit the cell cycle during differentiation, the levels of K11-linkages appear to decrease [19]. Together with the discovery that most known K11-specific enzymes are linked to mitotic control (see below), these observations raise the possibility that homogenous K11-linked ubiquitin chains are important regulators of cell division in higher eukaryotes.
How are K11-linked ubiquitin chains assembled during mitosis?
The APC/C is the only E3 known to assemble homogenous K11-linked ubiquitin chains. It recognizes its substrates via degenerate degron sequences, referred to as D- and KEN-boxes [23, 24], which are sandwiched between a cofactor, Cdc20/Cdh1, and a core APC/C-subunit, APC10 [25]. This mode of binding places substrates close to the RING-subunit APC11, which recruits the E2 Ube2C/UbcH10 to catalyze ubiquitin transfer. Substrate binding to the APC/C can be stabilized by additional means, such as Cks proteins that interact with substrates and phosphorylated APC/C-subunits or a C-terminal IR-appendix that is directly recognized by core subunits of the APC/C [26, 27].
Chain initiation
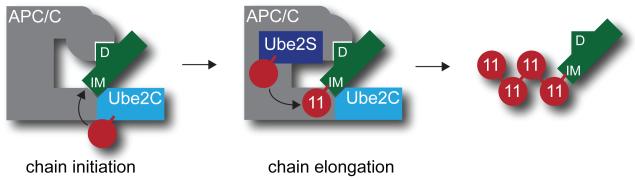
Following binding of substrates to the APC/C, formation of K11-linked chains is initiated by Ube2C (Figure 2; [9, 28-31]). Ube2C catalyzes the transfer of ubiquitin to a substrate lysine and also the formation of short, preferentially K11-linked chains [9, 32]. In vitro, E2s of the Ube2D/UbcH5 family also promote initiation, but several observations argue against a crucial role for these enzymes in K11-linked chain formation in cells. First, depletion of all Ube2D homologs did not affect human cell division [21, 33], while loss of Ube2C caused mitotic delay in several cell types [30, 34-38]. Second, as Ube2D is recognized by most of the >600 human E3 enzymes, only low concentrations of Ube2D are likely available for the APC/C. In contrast, Ube2C only acts with the APC/C, dependent upon an N-terminal APC/C-targeting motif that is absent in Ube2D [39, 40]. Third, whereas Ube2C preferentially assembles short K11-linked chains during initiation, Ube2D does not display any linkage specificity [9, 32]. Thus, Ube2C is the physiological chain-initiating E2 of mitotic APC/C.
Figure 2. Mechanism of K11-linked ubiquitin chain formation by the APC/C.
Substrates (green) are bound by the APC/C through degrons referred to as D-boxes (D) or KEN-boxes (not shown). Substrates also contain chain initiation motifs (IM) that are recognized by Ube2C, APC/C, or both, and that promote chain initiation by the E2 Ube2C. Following initiation, chains are extended by the K11-specific E2 Ube2S. Ube2C and Ube2S do not compete for binding to the APC/C.
Chain initiation by Ube2C is strongly promoted by conserved sequence motifs in substrates, referred to as initiation motifs [31]. These initiation motifs are patches of positively charged residues that are located in the proximity of a substrate’s D-box and include the preferentially modified lysines [31, 41]. The mutation of all positive charges in initiation motifs to alanine residues interferes with initiation. By contrast, if the positive charge is retained, yet all ubiquitin acceptor sites are removed, initiation can still occur at non-optimal lysine residues outside the motif [31]. This suggests that initiation motifs are recognized by an APC/C-component, but whether this is Ube2C, which contains a polar surface next to its active site [42], a core APC/C-subunit, or both, remains to be tested.
Kinetic studies with the SCF (Skp1/Cullin 1/F-box), another E3 required for cell cycle control, pointed to initiation as the rate-limiting step for chain assembly [43], an observation that applies to many substrates decorated with K11-linked chains. Biochemical analyses showed that the rate of initiation by Ube2C is slow compared to the processive chain elongation step [44, 45]. Accordingly, Ube2C levels are limiting for the degradation of many proteins modified with K11-linked chains [30, 34, 40], and the composition or accessibility of initiation motifs recognized by Ube2C can determine the timing of APC/C-substrate degradation without affecting the substrate’s affinity for the APC/C [31]. As the rate-limiting step, initiation has a profound impact on the processivity of chain formation, explaining why the processivity, and not the substrate’s affinity for the APC/C, correlates with the timing of APC/C-substrate degradation [44].
The initiation of K11-linked chain formation is tightly controlled. Similar to many cell cycle proteins, the transcription of Ube2C is regulated and peaks during mitosis [46], and overexpression of Ube2C as the result of an amplification of its genomic locus has been linked to cancer [34]. Higher levels of Ube2C destabilize the spindle checkpoint, a negative regulator of the APC/C that ensures accurate sister chromatid separation [40, 47, 48]. Accordingly, mice that overexpress Ube2C experience error-prone chromosome segregation, which can lead to tumorigenesis [49]. In addition, the stability of Ube2C is controlled by negative feedback centered on the APC/C [30, 37, 50]. Following the degradation of most of its substrates, the APC/C promotes autoubiquitination of Ube2C, which results in Ube2C degradation and downregulation of APC/C-activity. As the ubiquitination of Ube2C also depends on an initiation motif, APC/C substrates might delay Ube2C degradation by competing for initiation [31]. Nearby sequences in the N-terminus of Ube2C also mediate APC/C binding [40], and N-terminal epitope tags interfere with Ube2C-activity towards the APC/C [31], which explains previous results with GFPUbe2C that questioned an important role for Ube2C during mitosis [51]. The mechanism and timing of the APC/C-dependent degradation of Ube2C is conserved in higher eukaryotes [31, 37], underscoring the importance of keeping initiation in check.
Chain elongation
Following initiation, K11-linked chains are extended by a dedicated chain-elongating E2, Ube2S/E2-EPF (Figure 2; [21, 22, 52]). Ube2S interacts with the APC/C cofactor Cdc20 in early mitosis and with Cdh1 in late mitosis and G1 [21]. Additional experiments suggested that Ube2S is also recognized by a core subunit of the APC/C, the identity of which has not been clarified [21]. Apart from the APC/C, Ube2S has only been shown to bind the cullin-RING E3 subunit VHL [53].
While Ube2S has negligible activity towards Lys residues in APC/C substrates, it rapidly elongates K11-linked chains [21, 22, 52]. Ube2S displays an impressive specificity for K11-linkages (>95%), as determined by quantitative mass spectrometry, linkage-specific antibodies, and mutant ubiquitin [11, 14, 21, 22, 45, 54]. In vivo, Ube2S accounts for the large majority of K11-linked chain formation during cell division [11, 45].
The high specificity of Ube2S combined with established systems for assaying its activity make it a powerful model for dissecting the mechanism of linkage-specific chain formation [45]. A combination of NMR, computational docking and biochemical analysis revealed that Ube2S engages the thioester-linked donor ubiquitin in an additional non-covalent interaction, which restricts the conformational freedom of the donor and places it in an optimal position for nucleophilic attack by the acceptor Lys11 (Figure 3A; [45]). Tethering the donor ubiquitin increases the processivity of Ube2S, which can add up to 13 ubiquitin molecules to a chain in a single substrate-binding event to the APC/C. The processivity of chain elongation by Ube2S is comparable to that of the E3 SCF and its K48-specific E2 Ube2R1/Cdc34 [43]. Indeed, human Ube2R1 [45] and yeast Cdc34 [55] use a similar surface as Ube2S to bind donor ubiquitin, and a small-molecule inhibitor of Ube2R1 interferes with the integrity of the donor-binding site [56]. Interactions with the donor ubiquitin have also been observed with Ubc1 (another K48-specific E2 [57]), the HECT-E3 Nedd4L [58], and the SUMO-E2 Ubc9 [59], suggesting that donor tethering is of general importance for linkage formation.
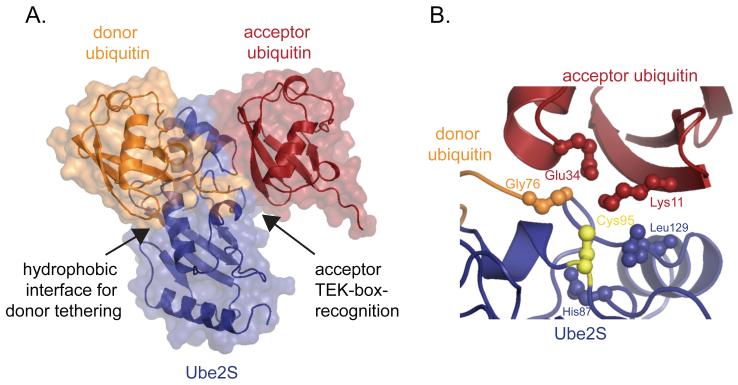
Figure 3. K11-linkage formation occurs through substrate-assisted catalysis.
A. Structural model of the ternary complex between Ube2S (blue), donor ubiquitin (orange), and acceptor ubiquitin (red), based on coordinates reported in [45]. The donor ubiquitin is tethered to the E2 by its thioester bond (not shown) and a non-covalent interaction around helix α2 (arrow). The acceptor ubiquitin is recognized through an electrostatic interaction that involves the TEK-box of ubiquitin (arrow). B. The catalytic center of K11-inkage formation consists of residues of the E2 Ube2S (blue) and the substrate acceptor ubiquitin (red). Ube2S contributes the active site cysteine (yellow), Leu129, and His87. The acceptor ubiquitin contributes Glu34 that helps deprotonate Lys11 and orients it towards the active site of Ube2S. The donor ubiquitin is shown in orange.
The acceptor ubiquitin is recognized by Ube2S with very low affinity through an electrostatic interface, which on ubiquitin consists of Lys6, Lys11, Glu34, Lys63, Thr12 and Thr14 [45]. This is the same motif, the “TEK-box”, that was previously found to be required for K11-linkage formation by Ube2C [9]. Non-specific E2s, such as Ube2D, also require the TEK-box of ubiquitin for formation of K11-linkages [60], revealing a key role for this surface in K11-linkage formation.
The low affinity for the acceptor ubiquitin suggested that the preferred recognition of a specific ubiquitin surface is unlikely to fully account for the high specificity of Ube2S, and indeed, an inspection of the ternary complex between Ube2S, donor, and acceptor ubiquitin revealed additional layers of regulation (Figure 3B). It was previously found that the active site of an E2 requires an acidic residue to suppress the pKa of the acceptor lysine, thereby turning it into a nucleophile ready for attack [61]. Ube2S lacks such an acidic residue and instead depends on a glutamate of the substrate, the acceptor ubiquitin, to activate the target lysine. Indeed, Glu34 of acceptor ubiquitin is positioned to support Lys11 deprotonation and help it orient towards the active site of Ube2S [45]. As other ubiquitin Lys residues lack an appropriately positioned acidic residue, only Lys11 is used for modification. These studies showed that K11-linkage formation proceeds through a mechanism of substrate-assisted catalysis.
Similar to initiation, the elongation of K11-linked ubiquitin chains appears to be under tight control. The expression of Ube2S is cell cycle-regulated [46], and increased levels of Ube2S lead to tumorigenesis [53, 62]. Further regulation of Ube2S is achieved by its binding to Cdc20 and Cdh1 at defined cell cycle stages and by its APC/C-dependent ubiquitination and degradation in late G1 [21]. As the degradation of Ube2S depends on Ube2C activity, and vice versa, the APC/C can compensate to some extent for a reduction in the levels of either E2 enzyme. This might explain why partial depletion of Ube2C has weak effects on mitosis [51], whereas complete depletion of Ube2C or co-depletion of Ube2C and Ube2S cause mitotic arrest [21, 38]. Thus, homogenous K11-linked ubiquitin chains are assembled through tightly regulated coordination between an initiating E2, Ube2C, and an elongating E2, Ube2S.
How are K11-linked ubiquitin chains disassembled during the cell cycle?
Deubiquitinating enzymes (DUBs) cleave isopeptide bonds between ubiquitin molecules to oppose signaling through ubiquitin chains. While some of the ~95 human DUBs display linkage-specificity, the majority of these enzymes act on most linkages with only minor preferences [63]. K11-linkages could, therefore, be disassembled by a K11-specific DUB, or alternatively, by a non-specific DUB that is targeted to substrates modified with K11-linked chains through an interaction with the APC/C or the substrate.
Linkage specificity was observed for various DUBs that contain a catalytic OTU (ovarian tumor) domain [63]. Among these OTU-DUBs, Cezanne preferentially cleaves K11-linkages in vitro [54], but its depletion is not associated with proliferation defects [64]. Cezanne might regulate NF-κB activation by deubiquitinating the signaling protein RIP, which is modified with K11/K63-linked mixed chains [12, 65]. Moreover, proteomic analysis identified the transcription factor HIF1α as a binding partner of Cezanne [66]. HIF1α is targeted for degradation by an E3 ligase that uses VHL as substrate adaptor [67], and VHL appears to bind Ube2S [53]. However, strong candidates for Cezanne-dependent deubiquitination still await discovery.
Based on our current knowledge, it appears more likely that K11-linked chains are disassembled by DUBs that are targeted to their substrates through an interaction with the APC/C. Initial analyses showed that APC/C-dependent ubiquitination is opposed by DUBs present in cell extracts [44]. These and later studies also found that deubiquitination activity co-purified with the APC/C [44, 68], and a proteomic survey of the DUB-interactome provided evidence for DUB-binding to the APC/C [66]. Functional interactions with the APC/C are now known for two DUBs, Usp44 and Usp37. Usp44 opposes APC/C-dependent inactivation of the spindle checkpoint in mitosis [69]. One of its substrates is the APC/C activator Cdc20, which is ubiquitinated and degraded by the APC/C during spindle checkpoint inactivation [48, 70]; Usp44 also protects Cdc20 from ubiquitination by the APC/C in postmitotic neurons [71], and Usp44-overexpression delays APC/C-activation and chromosome segregation in mouse fibroblasts [72]. Usp37 binds APC/CCdh1 in G1 and deubiquitinates the APC/C substrate cyclin A [73]. Together with its kinase partner Cdk2, cyclin A inactivates APC/CCdh1 in late G1, a reaction that is facilitated by its Usp37-dependent stabilization. Thus, Usp44 and Usp37 can restrict the activity of the major K11-specific ligase, the APC/C, in addition to deubiquitinating substrates modified with K11-linked chains.
As expected for DUBs with catalytic USP domains, Usp44 and Usp37 display little linkage-specificity in vitro ([73], and M.R., unpublished results), and they are primarily targeted to K11-linked chains through binding the APC/C. Similar interactions between DUBs and E3s occur with high frequency, which might result in dynamic regulation of ubiquitination [66]. The case of Usp37 shows that the interplay between ubiquitination and deubiquitination can be even more intricate. During exit from mitosis, Usp37 is degraded in an APC/C-dependent manner after being modified with K11-linked ubiquitin chains itself [73]. During G1, several mechanisms turn Usp37 from an APC/C substrate into a stable and abundant APC/C inhibitor, including transcription by E2F, activation by Cdk2, and stabilization as a result of Ube2C- and Ube2S-degradation [21, 30, 73]. The close connections between the E3 ligase responsible for K11-linked chain formation and DUBs underscore that the assembly of K11-linked chains is a tightly regulated and highly dynamic process, as expected for a key component of cell cycle control.
How are K11-linked ubiquitin chains recognized?
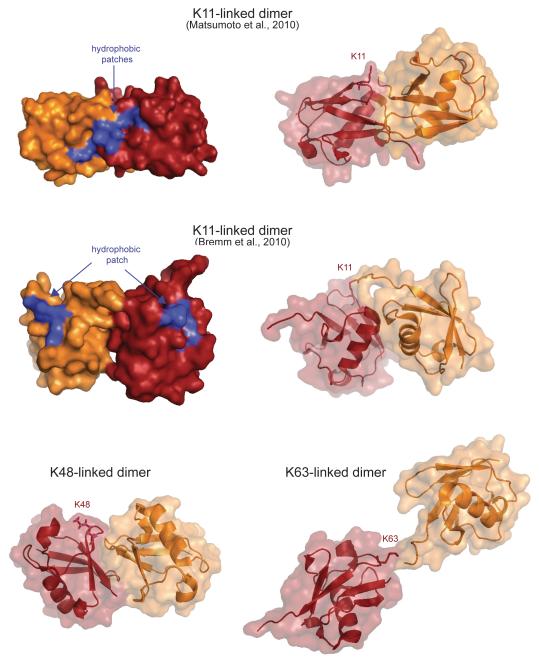
Proteins that specifically interact with K11-linked ubiquitin chains and not with chains of other topologies are likely to exploit structural features that are unique to K11-linkages, a hypothesis that was in principle validated with the development of K11-linkage specific antibodies [11]. Indeed, K11-linked ubiquitin dimers adopt compact conformations that are distinct from K48- or K63-linked dimers, and differences between two reported structures of K11-linked ubiquitin dimers and an NMR-analysis indicated that K11-linked chains could populate at least two conformations (Figure 4; [11, 54]). Importantly, in each conformation, the hydrophobic patch of ubiquitin, the major surface recognized by ubiquitin-binding proteins, is exposed and ready to engage in interactions.
Figure 4. K11-linked ubiquitin dimers have a unique structure.
Two structures of K11-linked ubiquitin dimers, as well as comparable structures of K48- and K63-linked dimers are displayed (based on coordinates reported in pdb-files 3NOB, 2XEW, 2PEA, 2JF5). The acceptor ubiquitin containing the modified lysine residue is shown in red, the donor ubiquitin is depicted in orange. Both K11-linked ubiquitin dimers show compact structures with the hydrophobic patch of ubiquitin being exposed for potential interactions.
While the unique structures of K11-linked ubiquitin dimers suggest that proteins specifically interacting with chains of this topology could exist, the nature of such binding partners remains elusive. K11-linked chains are recognized by proteasomal receptors for substrate degradation, but the same receptors can also interact with K48-linked chains [9, 11, 14]. K11-linkages can also be bound by ubiquitin-binding domains in proteins, such as NEMO, which were previously shown to interact with ubiquitin chains of different topologies [12]. However, it is interesting to note that an analysis of substrate adaptors of p97/Cdc48, a segregase that mobilizes ubiquitinated proteins from complexes [74, 75], found some enrichment for K11-linkages [75]. Whether this is caused by specific binding to K11-linked chains, and whether it is biologically meaningful, needs to be tested in future experiments. How K11-linked ubiquitin chains are decoded remains, unfortunately, mostly elusive.
Why does the APC/C assemble K11-linked ubiquitin chains?
Without knowing specific binding partners for K11-linkages or highly K11-specific DUBs, we can only speculate about the properties of K11-linked chains that serve their function as essential mitotic regulators. One potential clue to understanding the role of K11-linked chains in mitosis comes from studying the conservation of the E2s Ube2C and Ube2S [76]. Both K11-specific enzymes are absent in budding yeast, and in this organism the APC/C assembles K48-linked chains [77]. It is an attractive hypothesis that K11-linked, but not K48-linked, ubiquitin chains are able to control reactions that are required for the more complex mitotic regulation of higher eukaryotes. For example, higher eukaryotes have many more APC/C-substrates than yeast, and modification with K11-linked chains might be a more efficient means of sending proteins off to degradation [11]. Moreover, spindle assembly, a process very tightly connected to APC/C-regulation, is more complex in higher eukaryotes than in yeast, as the former undergo nuclear envelope breakdown, utilize the Ran-GTPase for microtubule nucleation, and attach multiple microtubules to each kinetochore [78]. K11-linked chains might be more suited to allow the coordinated assembly of mitotic spindles.
Given these observations, it is interesting to note that cells unable to assemble K11-linked chains accumulate in mitosis with defective spindles [21]. While the aberrant spindle structures could be a consequence of aborted or dysfunctional spindle formation, an alternative explanation is provided by cohesion fatigue [79]. Cohesion fatigue (i.e. the loss of sister chromatid cohesion during a prolonged metaphase arrest) was observed by live imaging of cells that could not assemble K11-linked chains due to depletion of the APC/C-activator Cdc20 or that did not turn over K11-ubiquitin modified substrates as a result of sustained proteasome inhibition [79]. Events reminiscent of cohesion fatigue were also seen in studies that interfered with APC/C activity using siRNAs or small molecules [38, 80]. As cohesion fatigue depends on continuous kinetochore attachment to the spindle, K11-linked chains might indeed control spindle dynamics or stability.
The connections between K11-linked chains and the spindle were strengthened by the discovery that the APC/C controls Ran-dependent spindle assembly [81]. GTP-charged Ran activates spindle assembly factors by releasing them from inhibitors of the importin family [82, 83]. For two spindle assembly factors, HURP and NuSAP, activation by Ran also results in exposure of their APC/C-binding sites, which leads to their modification with K11-linked chains and their degradation by the proteasome [81]. However, as mechanisms that coordinate spindle assembly factor activation and degradation remain poorly understood, it is not known whether other chain types would also achieve the proper regulation of Ran-dependent spindle assembly.
There is, of course, the possibility that K11-linked ubiquitin chains are not better suited than other chain types to fulfill the many tasks of the APC/C. Instead, it might be properties of the enzymes that catalyze K11-linked chain assembly, rather than chain topology per se, that have been conserved during evolution. For example, the similarity in the composition of initiation motifs in substrates and the TEK-box in ubiquitin might increase the efficiency of K11-linked chain formation by allowing Ube2C to assemble short chains during initiation [9]. In addition, as Ube2S does not appear to interact with the RING-domain of APC11 [45], Ube2S does not compete with Ube2C for APC/C-binding [21, 45], and initiating and elongating E2s can be present on the same APC/C-molecule; such a close collaboration might also be advantageous to target many substrates during the short time span of mitosis. Future work is needed to distinguish between these possibilities and to find a molecular explanation for the role of K11-linked chains in cell cycle regulation.
Concluding remarks
Recent studies taught us much about the function and assembly of K11-linked chains. The striking accumulation of K11-linked chains in mitosis and the strong mitotic defects in cells lacking the K11-specific enzymes have firmly linked this modification to cell cycle control. We know less about the factors that specifically bind to and read K11-linked chains, either in mitosis or at other cell cycle stages. Moreover, we lack strong evidence that homogenous K11-linked chains function outside of mitosis, possibly because we have yet to identify mechanisms that lead to their postmitotic induction. Hence, while it is established that K11-linked chains are able to trigger degradation, it remains unclear whether they also provide more idiosyncratic, non-proteolytic functions that set them apart from canonical K48-linked chains. The discovery of specific functions of K11-linked chains would be exciting and important, as it would deepen our insight into the breadth of the ubiquitin code – cracking a code that is an elaborate device for the tight and complex regulation of cell division.
Box 1: The enzymatics of ubiquitin chain formation.
Ubiquitin chain formation starts with ubiquitin being activated in an ATP-dependent manner by one of two E1 ubiquitin-activating enzymes [1]. In this reaction, a thioester bond is formed between the carboxy-terminus of ubiquitin and the active site cysteine of the E1 [84]. This ubiquitin is then transferred to a cysteine in the active site of an E2 ubiquitin conjugating enzyme [2]. Human cells contain 38 different E2s. Finally, ubiquitin is transferred to the ε-amino group of a substrate lysine with the help of an E3 ubiquitin-protein ligase. Human cells possess >600 different E3s, which fall into three different classes: i) HECT-E3s contain an active site cysteine in their HECT (homologous to E6AP C-terminus) domain [85] and the E2 transfers ubiquitin to this cysteine before the charged HECT modifies the substrate; ii) RING (really interesting new gene)-E3s bind at the same time to the substrate and the charged E2 and promote transfer of ubiquitin directly from the E2 to the substrate [3]; iii) finally, the hybrid RBR (RING-in between RING-RING)-E3s use a RING-domain to promote transfer of ubiquitin from the E2 to a cysteine in their second, RING-like domain [86]. The charged RBR-E3 then modifies the substrate. The linkage specificity of ubiquitin chain formation is likely determined by the HECT-E3, the E2 for RING-E3s, and the RBR-E3.
Acknowledgements
We thank Julia Schaletzky for many discussions and for critically reading the manuscript. We are also grateful to all other members of the Rape lab for their discussions and suggestions. Work in our lab is funded by a grant from the NIH, the NIH New Innovator Award, and a March of Dimes Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10(5):319–31. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10(11):755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 4.Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol. 2010;17(6):666–72. doi: 10.1038/nsmb.1842. [DOI] [PubMed] [Google Scholar]
- 5.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243(4898):1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 6.Thrower JS, et al. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19(1):94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng L, et al. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 8.VanDemark AP, et al. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell. 2001;105(6):711–20. doi: 10.1016/s0092-8674(01)00387-7. [DOI] [PubMed] [Google Scholar]
- 9.Jin L, et al. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133(4):653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10(7):706–13. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto ML, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39(3):477–84. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 12.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29(24):4198–209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boname JM, et al. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic. 2010;11(2):210–20. doi: 10.1111/j.1600-0854.2009.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271(5):2823–31. doi: 10.1074/jbc.271.5.2823. [DOI] [PubMed] [Google Scholar]
- 15.Peng J, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21(8):921–6. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 16.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziv I, et al. A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol Cell Proteomics. 2011;10(5):M111 009753. doi: 10.1074/mcp.M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett EJ, et al. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448(7154):704–8. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 19.Dammer EB, et al. Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J Biol Chem. 2011;286(12):10457–65. doi: 10.1074/jbc.M110.149633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaiser SE, et al. Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods. 2011 doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williamson A, et al. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2009;106(43):18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu T, et al. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107(4):1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349(6305):132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 24.Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14(6):655–65. [PMC free article] [PubMed] [Google Scholar]
- 25.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470(7333):274–8. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolthuis R, et al. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30(3):290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 27.Hayes MJ, et al. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat Cell Biol. 2006;8(6):607–14. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- 28.Yu H, et al. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr Biol. 1996;6(4):455–66. doi: 10.1016/s0960-9822(02)00513-4. [DOI] [PubMed] [Google Scholar]
- 29.Townsley FM, et al. Dominant-negative cyclin-selective ubiquitin carrier protein E2-C/UbcH10 blocks cells in metaphase. Proc Natl Acad Sci U S A. 1997;94(6):2362–7. doi: 10.1073/pnas.94.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rape M, Kirschner MW. Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature. 2004;432(7017):588–95. doi: 10.1038/nature03023. [DOI] [PubMed] [Google Scholar]
- 31.Williamson A. Regulation of ubiquitin chain initiation to determine the timing of substrate degradation. Mol Cell. 2011;42(6):744–757. doi: 10.1016/j.molcel.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick DS, et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8(7):700–10. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 33.Machida YJ, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589–96. doi: 10.1016/j.molcel.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 34.Wagner KW, et al. Overexpression, genomic amplification and therapeutic potential of inhibiting the UbcH10 ubiquitin conjugase in human carcinomas of diverse anatomic origin. Oncogene. 2004;23(39):6621–9. doi: 10.1038/sj.onc.1207861. [DOI] [PubMed] [Google Scholar]
- 35.Fujita T, et al. Clinicopathological relevance of UbcH10 in breast cancer. Cancer Sci. 2008 doi: 10.1111/j.1349-7006.2008.01026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berlingieri MT, et al. UbcH10 expression may be a useful tool in the prognosis of ovarian carcinomas. Oncogene. 2007;26(14):2136–40. doi: 10.1038/sj.onc.1210010. [DOI] [PubMed] [Google Scholar]
- 37.Mathe E, et al. The E2-C vihar is required for the correct spatiotemporal proteolysis of cyclin B and itself undergoes cyclical degradation. Curr Biol. 2004;14(19):1723–33. doi: 10.1016/j.cub.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 38.Zeng X, et al. Pharmacologic inhibition of the anaphase-promoting complex induces a spindle checkpoint-dependent mitotic arrest in the absence of spindle damage. Cancer Cell. 2010;18(4):382–95. doi: 10.1016/j.ccr.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, et al. APC2 Cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell. 2001;12(12):3839–51. doi: 10.1091/mbc.12.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Summers MK, et al. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31(4):544–56. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King RW, Glotzer M, Kirschner MW. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol Biol Cell. 1996;7(9):1343–57. doi: 10.1091/mbc.7.9.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin Y, Hwang WC, Basavappa R. Structural and functional analysis of the human mitotic-specific ubiquitin-conjugating enzyme, UbcH10. J Biol Chem. 2002;277(24):21913–21. doi: 10.1074/jbc.M109398200. [DOI] [PubMed] [Google Scholar]
- 43.Pierce NW, et al. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462(7273):615–9. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124(1):89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 45.Wickliffe KE, et al. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit e2. Cell. 2011;144(5):769–81. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miniowitz-Shemtov S, et al. ATP is required for the release of the anaphase-promoting complex/cyclosome from inhibition by the mitotic checkpoint. Proc Natl Acad Sci U S A. 2010;107(12):5351–6. doi: 10.1073/pnas.1001875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy SK, et al. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446(7138):921–5. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 49.van Ree JH, et al. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188(1):83–100. doi: 10.1083/jcb.200906147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka A, et al. Cell cycle-dependent expression of mammalian E2-C regulated by the anaphase-promoting complex/cyclosome. Mol Biol Cell. 2000;11(8):2821–31. doi: 10.1091/mbc.11.8.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker A, et al. UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. J Cell Sci. 2008;121(Pt 14):2319–26. doi: 10.1242/jcs.031591. [DOI] [PubMed] [Google Scholar]
- 52.Garnett MJ, et al. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11(11):1363–9. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung CR, et al. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12(7):809–16. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- 54.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17(8):939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saha A, et al. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42(1):75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceccarelli DF, et al. An allosteric inhibitor of the human cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145(7):1075–87. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 57.Hamilton KS, et al. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9(10):897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 58.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36(6):1095–102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bosanac I, et al. Modulation of K11-linkage formation by variable loop residues within UbcH5A. J Mol Biol. 2011;408(3):420–31. doi: 10.1016/j.jmb.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 61.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13(6):491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 62.Tedesco D, et al. The ubiquitin-conjugating enzyme E2-EPF is overexpressed in primary breast cancer and modulates sensitivity to topoisomerase II inhibition. Neoplasia. 2007;9(7):601–13. doi: 10.1593/neo.07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–63. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 64.Neumann B, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464(7289):721–7. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Enesa K, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283(11):7036–45. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 66.Sowa ME, et al. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaelin WG. Von Hippel-Lindau disease. Annu Rev Pathol. 2007;2:145–73. doi: 10.1146/annurev.pathol.2.010506.092049. [DOI] [PubMed] [Google Scholar]
- 68.Matyskiela ME, Morgan DO. Analysis of activator-binding sites on the APC/C supports a cooperative substrate-binding mechanism. Mol Cell. 2009;34(1):68–80. doi: 10.1016/j.molcel.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stegmeier F, et al. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature. 2007;446(7138):876–81. doi: 10.1038/nature05694. [DOI] [PubMed] [Google Scholar]
- 70.Nilsson J, et al. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10(12):1411–20. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Y, et al. A Cdc20-APC ubiquitin signaling pathway regulates presynaptic differentiation. Science. 2009;326(5952):575–8. doi: 10.1126/science.1177087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, van Deursen J, Galardy PJ. Overexpression of Ubiquitin Specific Protease 44 (USP44) induces chromosomal instability and is frequently observed in human T-cell leukemia. PLoS One. 2011;6(8):e23389. doi: 10.1371/journal.pone.0023389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang X, et al. Deubiquitinase USP37 is activated by CDK2 to antagonize APC(CDH1) and promote S phase entry. Mol Cell. 2011;42(4):511–23. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 74.Rape M, et al. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107(5):667–77. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 75.Alexandru G, et al. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134(5):804–16. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 2010;24(4):981–93. doi: 10.1096/fj.09-136259. [DOI] [PubMed] [Google Scholar]
- 77.Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39(4):548–59. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J Cell Sci. 2008;121(Pt 10):1577–86. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Daum JR, et al. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21(12):1018–24. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang HC, et al. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell. 2009;16(4):347–58. doi: 10.1016/j.ccr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Mol Cell. 2010;38(3):369–82. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ribbeck K, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol Biol Cell. 2006;17(6):2646–60. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sillje HH, et al. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol. 2006;16(8):731–42. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 84.Olsen SK, et al. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature. 2010;463(7283):906–12. doi: 10.1038/nature08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10(6):398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 86.Wenzel DM, et al. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474(7349):105–8. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]