Abstract
As they emerge from the ground, seedlings adopt a photosynthetic lifestyle, which is accompanied by dramatic changes in morphology and global alterations in gene expression that optimizes the plant body plan for light capture. Phytochromes are red and far-red photoreceptors that play a major role during photomorphogenesis, a complex developmental program that seedlings initiate when they first encounter light. The earliest phytochrome signaling events after excitation by red light include their rapid translocation from the cytoplasm to subnuclear bodies (“photobodies”) that also contain other proteins involved in photomorphogenesis, including a number of transcription factors and E3 ligases. In the light, phytochromes and negatively acting transcriptional regulators that interact directly with phytochromes are destabilized, whereas positively acting transcriptional regulators are stabilized. Here, we discuss recent advances on the mechanisms linking phytochrome photoactivation in the cytoplasm and transcriptional regulation in the nucleus.
Phytochrome signaling and seedling development
A major difference between plant and animal development is that most of plant morphogenesis occurs post-embryonically, which confers plants with great developmental plasticity. Thus, even though they are rooted in the ground, plants can rapidly alter their growth and development in response to a wide spectrum of environmental cues. One of the most dramatic examples of this developmental plasticity occurs when seedlings emerge from soil [1]. Most seeds germinate in the ground, and seedlings must forage for light. These newly sprouted etiolated seedlings have a long primary stem and undeveloped embryonic leaves (called cotyledons), which are protected by an apical hook in the primary stem as the seedling pushes its way through the soil (Figure 1). In dicotyledonous plants, such as the reference plant, Arabidopsis thaliana, a differential growth program is initiated when the seedling emerges from the soil. Stem (hypocotyl) growth rate is rapidly inhibited and cotyledons and leaves expand, turn green as chloroplasts develop, and become photosynthetic, a process known as de-etiolation.
Figure 1. Two classes of photomorphogenetic mutants.
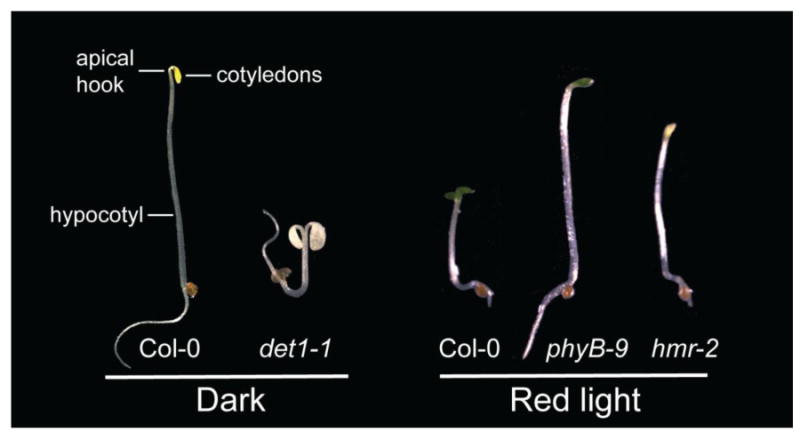
The left panel shows 4-day old wild-type Col-0 and det1-1 seedlings grown in the dark; the right panel shows 4-day old Col-0, phyB-9, and hmr-2 seedlings grown under 8 μmol m-2 s-1 of R light.
The morphological changes during de-etiolation are the result of a massive reprogramming of the transcriptome with 7-20% of the Arabidopsis genome differentially expressed between dark- and light-grown seedlings [2-6]. De-etiolation is first triggered by a suite of photoreceptors that perceive distinct colors of light [1]. The phytochrome (phy) family of receptors plays a major role in perceiving red (R) and far-red (FR) light, which carries information on the availability of photosynthetic energy and the proximity of neighboring plants.
Phys are bilin-containing proteins with two major domains: an N-terminal photosensory and signaling domain and a C-terminal dimerization and localization domain [7-10]. They exist in two relatively stable conformers: a R-light absorbing inactive Pr form (λmax=660) and a FR-absorbing active Pfr form (λmax=730) [8]. In Arabidopsis, the phy family consists of 5 members, phyA through phyE, that form homo- and hetero-dimers [11]; phyA and phyB are the most prominent phys and regulate almost every facet of plant development and growth [12, 13]. Due to differences in their photochemistry, stability, and rate of translocation to the nucleus, phyA and phyB are responsible for shared and distinct responses. PhyA proteins accumulate in the dark and in FR light and are rapidly degraded in R light [14]. PhyA plays a dominant role in FR light as well as during the dark-to-R transition. By contrast, phyB is relatively stable and plays a major role in R light. These different modes of action provide the molecular basis for distinct morphological responses of emerging etiolated seedlings to FR-rich shaded or R-rich open ambient light conditions.
Many phytochrome signaling components have been identified using forward genetic studies mainly in Arabidopsis [15]. The photomorphogenetic mutants can be divided into 2 general classes. The first class of mutants show de-etiolated or constitutively photomorphogenetic phenotypes, including short hypocotyls, expanded cotyledons, partial chloroplast differentiation, and de-repression of light-induced gene expression in the dark (Figure 1). This class of mutants includes the de-etiolated 1 (det1) mutants [16], constitutively photomorphogenic 1 (cop1) mutants [17], mutants in the components of the COP9 signalosome (CSN) [18], the quadruple mutant of the suppressor of phytochrome A-105 1-4 (spa1-4) [19], and the recently reported quadruple mutant phytochrome interacting factors 1,3,4,5 (pifq) [5, 6, 20]. Because mutations in these genes are recessive, these components most likely define negatively acting components in the light signaling pathways; they share similar phenotypes with a dominant constitutively active phyB allele, YHB [3, 21], suggesting that phys initiate photomorphogenesis by repressing these negative regulators.
The second class includes mutants that share similar phenotypes with phyA and/or phyB loss-of-function mutants with long hypocotyls and under-developed cotyledons in the light. They are defective in either phyA or phyB signaling or both. For example far-red elongated hypocotyl 1 (fhy1) [22, 23], far-red elongated hypocotyl 3 (fhy3) [22, 24], long after far-red light 1 (laf1) [25], far-red impaired response 1 (far1) [26], long hypocotyl in far-red 1 (hfr1) [27] are specific for phyA signaling, whereas the elongated hypocotyl 5 (hy5) mutant is defective in both phyA and phyB signaling [28, 29]. The recently reported hemera (hmr) mutant defines a new subclass of phy signaling mutants, which are not only impaired in phyA and phyB signaling with long hypocotyls under both R and FR light but also are defective in chloroplast development and die as albino seedlings [30] (Figure 1). The albino phenotype in hmr is most likely due to defects in chloroplast differentiation, in addition to phy signaling, because the quintuple phy mutant still makes chlorophyll and is pale-green in the light [31]. HMR may be a dual function protein, as it is localized in both chloroplasts and nuclei.
Many of the mutants from both classes define loci that encode transcriptional regulators of light responsive genes, including the positively acting HY5, LAF1, HFR1, FHY3, FAR1, as well as the negatively acting PIFs. A key mechanism by which phys regulate gene expression is to modulate the protein stability of these transcriptional regulators in the nucleus, where light directly regulates the affinity between phys and downstream signaling components. In addition, light regulates the translocation of phys to the nucleus into subnuclear foci called nuclear bodies, where phys and downstream signaling components are co-localized. This review focuses on the recent advances in our understanding of phy signaling mechanisms, with an emphasis placed on early events linking photoactivation of phys in the cytoplasm to transcriptional regulation in the nucleus.
Phytochrome nuclear accumulation is the earliest detectable light response
Akira Nagatani's group reported 15 years ago that Arabidopsis phyB contains putative nuclear localization signals (NLSs) and could be localized to the nucleus [32]. Subsequent studies using phytochromes fused with a fluorescent protein tag confirmed these earlier results and further demonstrated that the nuclear accumulation of all five phys in Arabidopsis is light-dependent [33-36]. In dark-grown seedlings, phys accumulate in the Pr form, and mainly localize to the cytoplasm. However, both phyA and phyB rapidly accumulate in the nucleus within minutes after R light exposure [37]. Regulation of phy nuclear accumulation is a key regulatory mechanism for phy signaling [10].
PhyB nuclear localization is regulated by light-dependent unmasking of the NLS. In R light when phyB is the in Pfr conformer, the N-terminal photosensory domain interacts less strongly with the C-terminal NLS domain than in the Pr conformer [38]. This led to a model in which photoactivation triggers a large movement of the two domains, exposing the NLS, and leading to nuclear accumulation.
The regulatory mechanism for phyA nuclear accumulation is more complex (Figure 2). No NLS has been identified in phyA. Rather, phyA nuclear localization requires binding to a pair of plant-specific proteins, FHY1 and its paralog FHL (FHy1-Like), both of which contain a conserved NLS and shuttle phyA to the nucleus [39-41]. FHY1 and FHL are required for both phyA nuclear accumulation and its subsequent signaling events [42, 43]. Although early reports suggested that FHY1 and FHL accumulate in the dark and interact stronger with the Pfr form of phyA in vitro [39, 40], more recently, it has been shown that FHY1 and FHL interact preferentially with the Pr form of phyA in vivo [42, 44, 45].
Figure 2. Regulatory mechanisms of phyA nuclear accumulation.
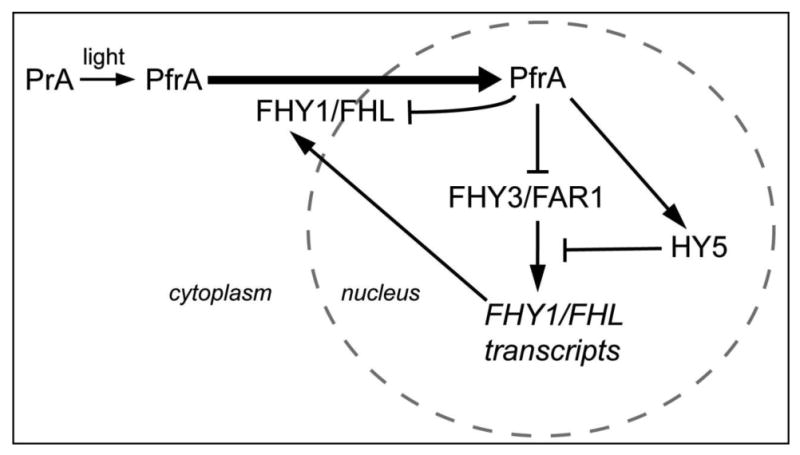
The nuclear accumulation of phyA is facilitated by FHY1 and FHL. FHY1/FHL are down-regulated by phyA signaling both at the transcriptional and post-translational levels.
PhyA nuclear accumulation is dependent on FHY1/FHL; as such, it may not be a surprise that phyA nuclear accumulation is feed-back regulated through the regulation of FHY1 and FHL levels. FHY1 and FHL transcripts accumulate in the dark and are reduced in the light (Figure 2) [23, 46]. The expression of FHY1 and FHL is regulated by two transposase-derived transcription factors, FHY3 and FAR1 [46, 47]. Light reduces FHY1/FHL transcript levels through the down-regulation of FHY3/FAR1 transcription [46, 47]. In addition, light attenuates FHY3 and FAR1 function by promoting the accumulation of HY5, as HY5 binds to a cis-element next to a FHY3/FAR1 binding site in the FHY1/FHL promoters and negatively regulates the transcriptional activity of FHY3 or FAR1 [48].
In addition to regulation at the transcriptional level, phyA promotes the phosphorylation and proteosome-mediated degradation of FHY1 and FHL in R light [44, 45]. Together with the rapid degradation of phyA (discussed below), these mechanisms work in concert to attenuate phyA signaling in the light.
Are phytochrome-containing nuclear bodies (photobodies) the sites of phytochrome signaling?
When they enter the nucleus, phys are further compartmentalized to subnuclear foci, called phytochrome speckles or phytochrome nuclear bodies [49]. Because these are unique plant subnuclear domains that are regulated by light, we propose to name them “photobodies”. During the dark-to-light transition, photobodies containing both phyA and phyB are detected after 1-2 mins of R light [37, 50]. phyB's localization to photobodies is triggered by R light. In contrast, phyA photobody localization is triggered by R and FR light. These early photobodies are transient and disappear after 1 h of light exposure [37]. In addition to phyA and phyB, FHY1, FHL and the phytochrome interacting factors PIF3 and PIF7 have also been localized to the early photobodies [37, 51]. PhyB localization to the early photobodies is not dependent on phyA but dependent on PIF3 [37, 52]. However, it seems that phyB photobody localization does not require its interaction with PIF3 or other PIFs, as phyB mutations defective in PIF binding could still be localized to photobodies [52, 53]. By contrast, localization of PIF3 to early phy nuclear bodies requires its interaction with phys [50].
After 2 hours in R light, phy photobodies re-appear and remain present in the light [33, 37]. These late phy photobodies mainly contain phyB, as phyA is degraded rapidly during the dark-to-light transition (discussed below). Under continuous R light, the steady-state pattern (size and number) of photobodies is directly related to the amount of Pfr [54]. Under dim R light, phyB is evenly dispersed in the nucleoplasm, whereas under strong R light, phyB appears to be localized exclusively to a few large photobodies with diameters between 1μm to 2μm [30, 54].
Several models have been proposed for the function of photobodies. Photobodies could be storage depots or reservoirs for activated phys. In this model, photobodies are sites where activated phyB is stabilized, but are not sites of phy signaling events. Consistent with the model, the Pfr form of phyB associated with photobodies is more stable in vivo [55]. In addition, the N-terminus of phyB fused to a dimerization domain and an NLS is active in mediating light responses without localization to photobodies [56, 57].
A second model posits that the photobodies are sites of protein turnover. As will be elaborated below, Cullin-RING E3 ligases are master regulators of photomorphogenesis. COP1 and its target proteins LAF1, HY5, phyA, and HFR1 colocalize in nuclear bodies, leading to the proposal that photobodies are sites for protein degradation [58-61]. Also, phyA, FHY1 and PIF3 are localized to early photobodies prior to their degradation [37, 39, 44, 50]. Last year we reported a forward genetic screen for mislocalization of a phyB∷GFP fusion protein [30]. This screen identified a novel phy signaling component, HMR. In hmr mutants, phyB∷GFP localized to smaller photobodies than predicted based on the fluence rate of R light. In hmr mutants, phyA, PIF1, and PIF3 accumulated in the light, suggesting HMR either directly or indirectly controls the degradation of these proteins [30]. HMR is predicted to be structurally similar to RAD23, which is a multiubiquitin receptor that delivers multiubiquitylated proteins to the proteasome for degradation. HMR's localization to the periphery of nuclear photobodies, in combination with the other observations supports a model in which photobodies are sites for the degradation of photolabile proteins [30]. Further investigation of the biochemical functions of HMR will likely elucidate the link between photobodies and protein degradation.
Yet another model is that photobodies are sites for phytochrome signaling. This model is supported by a close correlation between the localization of phyB to large nuclear bodies under continuous R light and phyB-mediated responses [54]. The constitutively active phyB mutation, YHB, is localized to large phy nuclear bodies even in the dark [21]. The three models are not necessarily mutually exclusive as phy signaling involves the light-regulated proteolysis of PIF transcription factors (see below).
Down-regulation of phyA and phyB in the light
Both phyA and phyB levels are repressed in the light to desensitize or regulate phy signaling [61]. During the dark-to-R-light transition, phyA levels are rapidly reduced by 50 to 100 fold, whereas phyB levels are reduced gradually by 4 to 5 fold [62]. phyA accumulation is negatively regulated both at the transcriptional and post-translational levels. PHYA transcripts are strongly reduced by both R and FR light [63]. This is correlated with histone modification changes at the PHYA locus. Histone marks for gene activation, such as H3K9/14ac, H3K27ac and H3K4me3 are enhanced during phyA activation in the dark; in contrast, histone marks for gene silencing, such as H3K24me3, are enriched at the PHYA locus in the light [64].
The Pfr form of phyA is subject to ubiquitin-proteasome-dependent protein degradation [14, 61, 65, 66]. PhyA degradation is mediated by the E3 ubiquitin ligase Cullin4 (CUL4)-Damaged DNA Binding Protein1(DDB1)-COP1-SPA complex, where the RING protein COP1 is associated with members of the SPA protein family to form the substrate receptor for the E3 ubiquitin ligase [42, 61, 67, 68]. Recognition of phyA by a COP1-SPA complex is enhanced by phyA phosphorylation in the light, because unphosphorylated phyA preferentially interacts with signaling components FHY1/FHY3, preventing phyA from binding to COP1-SPA [42]. Independent of the CUL4-DDB1-COP1-SPA complex, other E3 ubiquitin ligases may also be involved in phyA degradation [42]. One possible candidate is a Cullin1 (CUL1) based E3 ubiquition ligase [69], although no substrate receptor F-box protein has been identified. PhyA degradation also requires HMR, which is structurally similar to the multiubiquitin receptor RAD23 and could possibly act like RAD23 in delivering phyA to the proteasome [30]. PhyA degradation occurs in both the cytoplasm and the nucleus, with faster degradation kinetics in the nucleus [70-73]. Within the nucleus, phyA degradation likely occurs on photobodies, because phyA and COP1 were co-localized to photobodies [37, 61]. This notion was further supported by the identification of the hmr mutant, where defects in photobodies led to phyA accumulation in the light [30].
Down-regulation of phyB occurs mainly at the post-translational level, as PHYB transcripts are relatively unaffected by light in Arabidopsis [74]. Degradation of phyB is mediated by COP1 [65], most likely through the CUL4-DDB1-COP1-SPA complex. In addition, phyB's protein level in the light is negatively regulated by PIFs [51, 65, 75], which could promote COP1-mediated phyB ubiquitylation, possibly by enhancing the interaction between phyB and COP1 [65]. This serves as one of the mechanisms for PIFs to regulate phyB signaling [76].
Repression of photomorphogenesis by Cullin4-RING ubiquitin ligases
Our understanding of how photomorphogenesis is repressed in dark-grown seedlings began from genetic screens for Arabidopsis mutants exhibiting de-etiolated (DET) or constitutive photomorphogenic (COP) phenotypes in the absence of light. These genetic screens identified a group of master repressors for photomorphogenesis, including DET1 [77], COP1 [78], COP10 [79], SPA1-4 [19, 80], and subunits of the COP9 signalosome (CSN) [18]. Characterization of the DET1 protein complex identified a WD-repeat protein DDB1 (Damaged DNA Binding Protein 1), a key component for regulating photomorphogenesis [81]. Recent biochemical analyses revealed that these master repressor proteins form three protein complexes involved in ubiquitin-proteasome mediated protein degradation, including two CUL4 based E3 ubiquitin ligases and the COP9 signalosome, which modulates the activity of Cullin-based E3 ubiquitin ligases [18, 67, 82].
CUL4 is part of the multisubunit Cullin-RING ubiquitin ligase superfamily [83]. The Cullin-RING ubiquitin ligases share a similar modular design. The C-terminus of CUL binds a catalytic RING subunit, such as Rbx1, and Rbx1-associated E2 ubiquitin conjugating enzyme; the N-terminus of CUL associates with an adaptor protein and a substrate receptor that recognizes target proteins. Cullin-RING ubiquitin ligases facilitate the transfer of ubiquitin from E2 to target proteins. CUL4-based E3s utilize DDB1 as adaptor proteins and DDB1-binding WD40 proteins as substrate receptors [84]. Recent biochemical analyses suggested that the master repressors for photomorphogenesis form two distinct E3 ubiquitin ligases: the CUL4-DDB1-DET1-COP10 complex and the CUL4-DDB1-COP1-SPA complex [67, 82]. Cullin-based E3s are regulated by the cyclic attachment and removal of the ubiquitin-related protein: RUB or NEDD8. The CSN is a highly conserved protein complex, which resembles the 19S lid of the 26S proteasome and catalyzes the deneddylation reaction for Cullins. Taken together, all three complexes are involved in ubiquitin-proteasome-mediated protein degradation.
The COP1/SPA1 E3 also mediates the degradation of positively acting factors for photomorphogenesis [67]. Besides phyA, its substrates include 3 transcriptional regulators, the bZIP protein HY5 [85], the MYB protein LAF1 [60] and the helix-loop-helix protein HFR1 [58, 86, 87]. The selective degradation of these transcriptional regulators in the dark is a key mechanism to repress photomorphogenesis. Consistent with this notion, expressing truncations of HY5 or HFR1 without their corresponding COP1 binding motifs stabilizes HY5, leading to enhanced photomorphogenetic phenotypes [58, 59, 87, 88]. COP1/SPA-mediated protein degradation could occur on photobodies, as COP1 colocalizes with phyA, HY5, LAF1, and HFR1 in photobodies and all SPA proteins are localized to nuclear bodies [58, 60, 61, 87, 89]. Although DET1 and COP1/SPA form distinct E3 ubiquitin ligases, genetic studies suggest that DET1 works in concert with COP1 in plant development [90]. One possible mechanism is that the DET1 E3 is involved in the regulation of components of the COP1/SPA E3, such as COP1 [82].
It has been proposed that phys inhibit the function of the DET/COP master repressor proteins in the light; however the mechanism is still not totally clear. Some possibilities include: i) Light regulation of the nuclear/cytoplasmic partitioning of COP1 [91]. COP1 accumulates in the nucleus in the dark to repress photomorphogenesis and are mainly localized to the cytoplasm in the light. The nuclear accumulation of COP1 is dependent on DET1, COP10, and direct interaction with the CSN1 subunit of CSN [92-94]. Therefore, phys could either directly regulate COP1 localization or indirectly regulate its localization through DET1 or CSN. However, it should be mentioned that the kinetics of this regulation is slow during the dark-to-light transition and there are always COP1 proteins remaining in the nucleus in the light [65]; ii) Phys could negatively regulate the amount of COP1/SPA complex by down-regulating both SPA1 and SPA2 protein levels [95]; iii) Phys could regulate COP1/SPA activity, as COP1/SPA1 interactions are weakened in R, FR, and blue light [89]. Recently, it has been shown that the blue light photoreceptors cryptochromes directly regulate either the formation of the substrate receptor COP1/SPA1 complex or the interaction between the substrate receptor COP1/SPA complex and its target proteins [96-98]. Phys could utilize a similar mechanism to regulate the activity of COP1.
Initiation of photomorphogenesis by removal of phytochrome interacting factors
Phytochrome interacting factors (PIFs) are bHLH transcription factors that negatively regulate photomorphogenesis [76]. Since the founding member, PIF3, was identified by Peter Quail's laboratory in 1998 [99], great advances have been made by several labs to understand the roles of PIFs in photomorphogenesis. The PIFs belong to a subfamily of bHLH transcription factors including 15 members. All of the characterized PIFs, including PIF3, PIF1/PIL5, PIF4, PIF5/PIL6, and PIF7, have been shown to bind to the G-box motif (CACGTG) in light-regulated genes, where they act as either transcriptional activators or repressors [5, 51, 100-105]. PIFs are involved in shared and distinct phy-mediated responses. During de-etiolation, PIF1, PIF3, PIF4, PIF5 and PIF7 are all involved in hypocotyl growth inhibition [75, 100, 101, 106-109]. PIF1, PIF3, and PIF5 are also repressors for chloroplast development, down-regulating the expression of genes encoding key chlorophyll biosynthetic enzymes [5, 6, 101, 103, 110]. Moreover, PIF1, down-regulates carotenoid biosynthesis by repressing the expression of a key carotenoid biosynthetic enzyme, phytoene synthase [111]. Therefore, PIFs link phy signaling to downstream light-mediated morphological responses.
All of the PIF proteins contain phytochrome interacting domains in the N-terminal region and bHLH DNA binding and dimerization domains at their C-terminus. PIFs preferentially bind to the active Pfr form of phy [76]. PIF1 and PIF3 are capable of interacting with both photoactivated phyA and phyB, however, PIF4, PIF5, PIF6, and PIF7 bind only to phyB [51, 99, 101, 108]. Upon light activation, accumulation of phys in the nucleus triggers phosphorylation of PIFs and rapid degradation of PIFs by the ubiquitin-proteosome system [50]. It is still not completely clear which kinase is responsible for PIF phosphorylation. It was recently suggested that PIF1 is phosphorylated by CK2 and possibly other unidentified kinases [112]. The E3 ubiquitin ligase for PIF degradation is also unknown. With the exception of PIF7, most PIFs have half-lives between 5 to 20 mins in the light [113-117]. The significance of PIF degradation was demonstrated by the characterization of the pif1pif3pif4pif5 quadruple (pifq) mutant [5, 6]. The pifq mutant showed de-etiolated phenotypes in the dark similar to those of a constitutively active phyB YHB allele and the det/cop mutants [3, 5]. Thus, PIFs may be required to promote etiolation (or stem growth); phys may regulate photomorphogenesis by promoting PIF degradation [5, 6].
A key question now is to understand the relationship between PIFs and DET/COP proteins (Figure 3). COP1/SPA does not appear to be the E3 for PIF degradation but rather is required for at least PIF3 accumulation [20, 37, 65]. However, the mechanism linking COP1/SPA and PIF3 stability is still unknown (Figure 3). COP1/SPA could regulate the function of PIFs indirectly through the HLH protein HFR1, which forms heterodimers with PIFs and prevents PIF binding to DNA [102].
Figure 3. Phy signaling pathways to turn on photomorphogenesis.
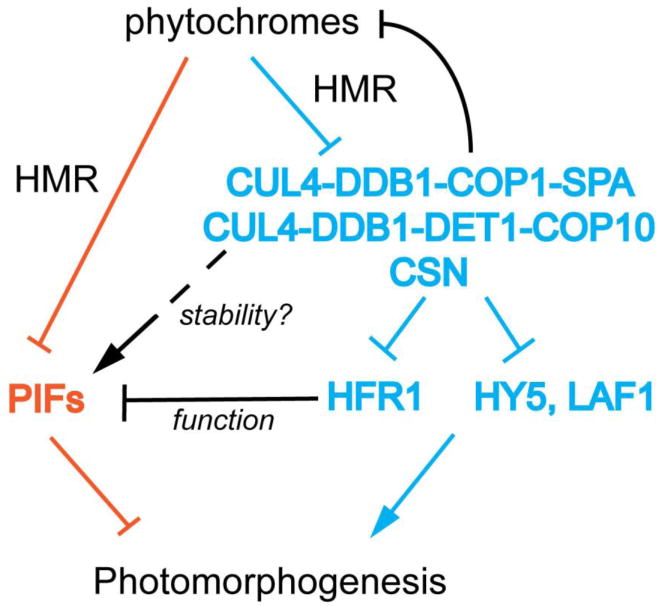
There are two major signaling pathways: first, in the “blue” pathway, phys de-repress the positively acting transcriptional regulators, including HY5, LAF1 and HFR1, by inhibiting the COP1 and DET1 containing CUL4 E3 ubiquitin ligases; Second, in the “orange” pathway, phys trigger the degradation of the negatively acting transcriptional regulators PIFs. The activity of PIFs could be enhanced by the COP1/DET1 through HFR1 repression; the protein stability of at least PIF3 is promoted by COP1 through an unknown mechanism. The recently identified HMR acts early in the pathways and is involved in the degradation of PIFs and phyA as well as the inhibition of DET1.
At the cellular level, because PIF3 is localized to early photobodies prior to its turnover, it has been proposed that photobodies are sites for PIF degradation [37]. The characterization of the hmr mutants that are defective in the late photobodies supported this hypothesis, as both PIF1 and PIF3 accumulate to higher levels in the hmr mutants in the light [30]. This suggests that photobodies are sites for PIF degradation. Because HMR is also required for phyA degradation in the light, HMR could be involved in PIF degradation directly or indirectly through the regulation of phys [30]. Further characterization of HMR function will be able to distinguish between these possibilities.
Concluding remarks
Numerous recent studies have established the main framework of phy signaling during seedling emergence. First, phys trigger the rapid degradation of the PIF family of bHLH transcription factors that promote the dark program. Second, phys negatively regulate two CUL4-based E3 ubiquitin ligases, CUL4-DDB1-COP1-SPA and CUL4-DDB1-DET1-COP10, to stabilize a group of transcriptional regulators, including HY5, LAF1, and HFR1 that promote photomorphogenesis. Both phyA and phyB are turned-over by the same mechanism to desensitize phy signaling in the light. Third, at the cellular level, phys rapidly accumulate in the nucleus and are compartmentalized to photobodies in the light. The translocation of phys is regulated by both conformational changes of phys, as well as by other signaling components. Photobodies are likely sites for light-regulated proteolytic events. Despite these great advances, many questions remain unanswered. It is still not clear how phys inhibit the activity of DET1, COP1/SPA, and CSN activities in the light. The mechanism of PIF degradation is also unknown. Finally, how photobodies are assembled and regulated, as well as their precise function in phy signaling will be an active area of future investigation.
Acknowledgments
Our work on phytochromes and photomorphogenesis is supported by grants from the NIH GM52413 to J.C., and by NIH GM87388 and NSF IOS1051602 to M.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Chen M, et al. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 2.Jiao Y, et al. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–230. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 3.Hu W, et al. A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol Plant. 2009;2:166–182. doi: 10.1093/mp/ssn086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tepperman JM, et al. phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 2006;48:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- 5.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagatani A. Phytochrome: structural basis for its functions. Curr Opin Plant Biol. 2010;13:565–570. doi: 10.1016/j.pbi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Rockwell NC, et al. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–858. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulijasz AT, et al. Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature. 2010;463:250–254. doi: 10.1038/nature08671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fankhauser C, Chen M. Transposing phytochrome into the nucleus. Trends Plant Sci. 2008;13:596–601. doi: 10.1016/j.tplants.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Clack T, et al. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. Plant Cell. 2009;21:786–799. doi: 10.1105/tpc.108.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kami C, et al. Light-regulated plant growth and development. Curr Top Dev Biol. 2010;91:29–66. doi: 10.1016/S0070-2153(10)91002-8. [DOI] [PubMed] [Google Scholar]
- 13.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clough R, Vierstra R. Phytochrome degradation. Plant Cell Environ. 1997;20:713–721. [Google Scholar]
- 15.Chory J. Light signal transduction: an infinite spectrum of possibilities. Plant J. 2010;61:982–991. doi: 10.1111/j.1365-313X.2009.04105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chory J, et al. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989;58:991–999. doi: 10.1016/0092-8674(89)90950-1. [DOI] [PubMed] [Google Scholar]
- 17.Deng XW, et al. cop1: a regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 1991;5:1172–1182. doi: 10.1101/gad.5.7.1172. [DOI] [PubMed] [Google Scholar]
- 18.Serino G, Deng XW. The COP9 signalosome: regulating plant development through the control of proteolysis. Annu Rev Plant Biol. 2003;54:165–182. doi: 10.1146/annurev.arplant.54.031902.134847. [DOI] [PubMed] [Google Scholar]
- 19.Laubinger S, et al. The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell. 2004;16:2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su YS, Lagarias JC. Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell. 2007;19:2124–2139. doi: 10.1105/tpc.107.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitelam GC, et al. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desnos T, et al. FHY1: a phytochrome A-specific signal transducer. Genes Dev. 2001;15:2980–2990. doi: 10.1101/gad.205401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Deng XW. Arabidopsis FHY3 defines a key phytochrome A signaling component directly interacting with its homologous partner FAR1. Embo J. 2002;21:1339–1349. doi: 10.1093/emboj/21.6.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ballesteros ML, et al. LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 2001;15:2613–2625. doi: 10.1101/gad.915001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson M, et al. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fairchild CD, et al. HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 2000;14:2377–2391. [PMC free article] [PubMed] [Google Scholar]
- 28.Koornneef M, et al. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 29.Oyama T, et al. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997;11:2983–2995. doi: 10.1101/gad.11.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen M, et al. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell. 2010;141:1230–1240. doi: 10.1016/j.cell.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strasser B, et al. Arabidopsis thaliana life without phytochromes. Proc Natl Acad Sci U S A. 2010;107:4776–4781. doi: 10.1073/pnas.0910446107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. Plant J. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi R, et al. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999;145:437–445. doi: 10.1083/jcb.145.3.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kircher S, et al. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kircher S, et al. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell. 2002;14:1541–1555. doi: 10.1105/tpc.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim L, et al. Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 2000;22:125–133. doi: 10.1046/j.1365-313x.2000.00729.x. [DOI] [PubMed] [Google Scholar]
- 37.Bauer D, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen M, et al. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005;15:637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 39.Hiltbrunner A, et al. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15:2125–2130. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 40.Hiltbrunner A, et al. FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 2006;47:1023–1034. doi: 10.1093/pcp/pcj087. [DOI] [PubMed] [Google Scholar]
- 41.Genoud T, et al. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 2008;4:e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saijo Y, et al. Arabidopsis COP1/SPA1 complex and FHY1/FHY3 associate with distinct phosphorylated forms of phytochrome A in balancing light signaling. Mol Cell. 2008;31:607–613. doi: 10.1016/j.molcel.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang SW, et al. FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell. 2009;21:1341–1359. doi: 10.1105/tpc.109.067215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen Y, et al. Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 2005;139:1234–1243. doi: 10.1104/pp.105.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen Y, et al. Phytochrome A mediates rapid red light-induced phosphorylation of Arabidopsis FAR-RED ELONGATED HYPOCOTYL1 in a low fluence response. Plant Cell. 2009;21:494–506. doi: 10.1105/tpc.108.061259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin R, et al. Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science. 2007;318:1302–1305. doi: 10.1126/science.1146281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin R, et al. Discrete and essential roles of the multiple domains of Arabidopsis FHY3 in mediating phytochrome A signal transduction. Plant Physiol. 2008;148:981–992. doi: 10.1104/pp.108.120436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome a signaling. Plant Cell. 2010;22:3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M. Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling. Curr Opin Plant Biol. 2008;11:503–508. doi: 10.1016/j.pbi.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 50.Al-Sady B, et al. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oka Y, et al. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 2008;4:e1000158. doi: 10.1371/journal.pgen.1000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kikis EA, et al. Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 2009;5:e1000352. doi: 10.1371/journal.pgen.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen M, et al. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci U S A. 2003;100:14493–14498. doi: 10.1073/pnas.1935989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rausenberger J, et al. An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS One. 2010;5:e10721. doi: 10.1371/journal.pone.0010721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsushita T, et al. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003;424:571–574. doi: 10.1038/nature01837. [DOI] [PubMed] [Google Scholar]
- 57.Palagyi A, et al. Functional analysis of amino-terminal domains of the photoreceptor phytochrome B. Plant Physiol. 2010;153:1834–1845. doi: 10.1104/pp.110.153031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jang IC, et al. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005;19:593–602. doi: 10.1101/gad.1247205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ang LH, et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 60.Seo HS, et al. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003;423:995–999. doi: 10.1038/nature01696. [DOI] [PubMed] [Google Scholar]
- 61.Seo HS, et al. Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 2004;18:617–622. doi: 10.1101/gad.1187804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Canton FR, Quail PH. Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 1999;121:1207–1216. doi: 10.1104/pp.121.4.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jang IC, et al. Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell. 2011;23:459–470. doi: 10.1105/tpc.110.080481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jang IC, et al. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–2383. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quail PH. The phytochromes: a biochemical mechanism of signaling in sight? Bioessays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 67.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20:2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quint M, et al. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rosler J, et al. Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci U S A. 2007;104:10737–10742. doi: 10.1073/pnas.0703855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toledo-Ortiz G, et al. Subcellular sites of the signal transduction and degradation of phytochrome A. Plant Cell Physiol. 2010;51:1648–1660. doi: 10.1093/pcp/pcq121. [DOI] [PubMed] [Google Scholar]
- 72.Debrieux D, Fankhauser C. Light-induced degradation of phyA is promoted by transfer of the photoreceptor into the nucleus. Plant Mol Biol. 2010;73:687–695. doi: 10.1007/s11103-010-9649-9. [DOI] [PubMed] [Google Scholar]
- 73.Wolf I, et al. Light-regulated nuclear import and degradation of Arabidopsis phytochrome-A N-terminal fragments. Plant Cell Physiol. 2011;52:361–372. doi: 10.1093/pcp/pcq194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clack T, et al. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- 75.Al-Sady B, et al. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci U S A. 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pepper A, et al. DET1, a negative regulator of light-mediated development and gene expression in Arabidopsis, encodes a novel nuclear-localized protein. Cell. 1994;78:109–116. doi: 10.1016/0092-8674(94)90577-0. [DOI] [PubMed] [Google Scholar]
- 78.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki G, et al. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes Dev. 2002;16:554–559. doi: 10.1101/gad.964602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hoecker U, et al. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- 81.Schroeder DF, et al. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol. 2002;12:1462–1472. doi: 10.1016/s0960-9822(02)01106-5. [DOI] [PubMed] [Google Scholar]
- 82.Chen H, et al. Arabidopsis CULLIN4 Forms an E3 Ubiquitin Ligase with RBX1 and the CDD Complex in Mediating Light Control of Development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Biedermann S, Hellmann H. WD40 and CUL4-based E3 ligases: lubricating all aspects of life. Trends Plant Sci. 2011;16:38–46. doi: 10.1016/j.tplants.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 84.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Osterlund MT, et al. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 86.Duek PD, et al. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol. 2004;14:2296–2301. doi: 10.1016/j.cub.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 87.Yang J, et al. Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell. 2005;17:804–821. doi: 10.1105/tpc.104.030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang KY, et al. Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-deltaN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiol. 2003;133:1630–1642. doi: 10.1104/pp.103.029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saijo Y, et al. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 2003;17:2642–2647. doi: 10.1101/gad.1122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nixdorf M, Hoecker U. SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta. 2010;231:825–833. doi: 10.1007/s00425-009-1088-y. [DOI] [PubMed] [Google Scholar]
- 91.von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79:1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 92.Chamovitz DA, et al. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86:115–121. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- 93.von Arnim AG, et al. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 1997;114:779–788. doi: 10.1104/pp.114.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang X, et al. Regulation of COP1 nuclear localization by the COP9 signalosome via direct interaction with CSN1. Plant J. 2009;58:655–667. doi: 10.1111/j.1365-313X.2009.03805.x. [DOI] [PubMed] [Google Scholar]
- 95.Balcerowicz M, et al. Light exposure of Arabidopsis seedlings causes rapid destabilization as well as selective post-translational inactivation of the repressor of photomorphogenesis SPA2. Plant J. 2011;65:712–723. doi: 10.1111/j.1365-313X.2010.04456.x. [DOI] [PubMed] [Google Scholar]
- 96.Zuo Z, et al. Blue Light-Dependent Interaction of CRY2 with SPA1 Regulates COP1 activity and Floral Initiation in Arabidopsis. Curr Biol. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu B, et al. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lian HL, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ni M, et al. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 100.Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 102.Hornitschek P, et al. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. Embo J. 2009;28:3893–3902. doi: 10.1038/emboj.2009.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moon J, et al. PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc Natl Acad Sci U S A. 2008;105:9433–9438. doi: 10.1073/pnas.0803611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 105.Oh E, et al. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Oh E, et al. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fujimori T, et al. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004;45:1078–1086. doi: 10.1093/pcp/pch124. [DOI] [PubMed] [Google Scholar]
- 108.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lorrain S, et al. Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J. 2009;60:449–461. doi: 10.1111/j.1365-313X.2009.03971.x. [DOI] [PubMed] [Google Scholar]
- 110.Stephenson PG, et al. PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A. 2009;106:7654–7659. doi: 10.1073/pnas.0811684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Toledo-Ortiz G, et al. Direct regulation of phytoene synthase gene expression and carotenoid biosynthesis by phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2010;107:11626–11631. doi: 10.1073/pnas.0914428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bu Q, et al. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biol Chem. 2011;286:12066–12074. doi: 10.1074/jbc.M110.186882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shen H, et al. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008;20:1586–1602. doi: 10.1105/tpc.108.060020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lorrain S, et al. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 115.Nozue K, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 116.Shen Y, et al. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shen H, et al. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]


