Abstract
Selected from random pools of DNA or RNA molecules through systematic evolution of ligands by exponential enrichment (SELEX), aptamers can bind to target molecules with high affinity and specificity, which makes them ideal recognition elements in the development of biosensors. To date, aptamer-based biosensors have used a wide variety of detection techniques, which are briefly summarized in this article. The focus of this review is on the development of aptamer-based fluorescent biosensors, with emphasis on their design as well as properties such as sensitivity and specificity. These biosensors can be broadly divided into two categories: those using fluorescently-labeled aptamers and others that employ label-free aptamers. Within each category, they can be further divided into “signal-on” and “signal-off” sensors. A number of these aptamer-based fluorescent biosensors have shown promising results in biological samples such as urine and serum, suggesting their potential applications in biomedical research and disease diagnostics.
Keywords: Aptamers, biosensor, fluorescence, nanosensor, fluorescence resonance energy transfer (FRET)
INTRODUCTION
Biosensors for accurate and rapid detection of environmental pollutes, pathogens, disease markers, as well as various other chemicals/biologics/materials are highly desirable for a wide variety of applications such as environmental monitoring, waste management, drug discovery, disease diagnosis, clinical toxicology, forensics, among others [1,2]. Enzymes, antibodies, aptamers, etc. have all been investigated as the recognition elements for biosensors, with aptamers becoming increasingly popular over the last decade. Aptamers are typically single-strand (ss) nucleic acids screened out from combinatorial oligonucleotide (ODN) libraries of DNA or RNA, through a process termed “SELEX” (systematic evolution of ligands by exponential enrichment) [3,4]. Possessing comparable target binding affinity/selectivity to antibodies, aptamers can also offer advantages such as easier engineering and synthesis, lower batch-to-batch variability, better thermal stability, smaller size, lower immunogenicity, more versatile chemistry, among others [5]. Over the years, aptamers have been selected against a broad range of targets such as metal ions, small molecules, peptides, proteins, cells, etc. [5].
With these desirable characteristics, aptamer-based sensors have been developed to detect a diverse set of targets including adenosine [6,7], adenosine triphosphate (ATP) [8–11], cocaine [8,12], Hg2+ [13,14], toxins [15,16], thrombin [17–20], vascular endothelial growth factor (VEGF) [21,22], IgE [23,24], and many others. Besides the most commonly-used fluorescence detection method that is sensitive, robust, and cost-effective, various other detection techniques have also been coupled to aptamer-based sensors. For example, cocaine was detected in picomolar (pM) concentration with an electrochemiluminescence (ECL) sandwich biosensor, designed based on the understanding that a single aptamer could be split into two fragments which could form a folded, associated complex in the presence of its target [25]. Other chemiluminescence-coupled sensors based on rolling circle amplification of aptamers have also been reported [26,27].
One popular class of sensor design is based on electrochemical techniques [28,29], where redox-active aptamers are immobilized onto a conducting support to probe the features of electronic transfer. Upon binding to its target, the aptamer folds into a well-defined three-dimensional structure, either shielding or prompting the electron transfer between redox-active units and the electrode substrate. For example, various sensors of this type have been developed for sensing thrombin [20,30].
Field-effect transistor (FET) between aptamer and carbon nanotube or polypyrrole nanotube can be employed for sensor applications[31]. Generally, a gate voltage is applied to nanotubes with surface-bound aptamers. The association between the aptamer and its target results in charge transfer into the nanotube and changes in electrical properties between the source and electrode attached on the nanotube. Sensitivity as high as femtomolar (fM) detection has been achieved with this technique [32].
Target recognition by an aptamer can also be monitored by surface plasmon resonance (SPR). A thin layer of gold (the sensitive layer) on a glass surface with high refractive index can absorb laser light and produce surface plasmons on the gold surface, which occurs only at a specific angle and wavelength. When the target binds to the aptamer immobilized on the layer close to the sensitive layer, the refractive index of the layer and the SPR angle changes which can be employed to measure biological interactions with good sensitivity. Aptamer-based SPR sensing has been successfully used to detect various molecular targets in the nanomolar (nM) range [24,33,34].
Compared to optical/electrochemical detection, colorimetric measurement is directly visible and thus more convenient. However, its sensitivity is typically much lower (in the millimolar [mM] to micromolar [µM] range) [9,11,35–37]. Besides these traditional detection methods, other techniques such as quartz crystal microbalance (QCM) [21], acoustic wave [38,39], and micro-cantilever [40,41] have also been explored for aptamer-based sensing and several excellent review articles are available [28,42–49]. Herein we will focus on aptamer-based fluorescent biosensors, which can be divided into two major categories: those using fluorescently-labeled aptamers and others that employ label-free aptamers.
BIOSENSORS BASED ON FLUORESCENTLY-LABELED APTAMERS
Aptamers are well known to have distinctly different conformations/structures before and after binding with the targets. Through direct modification with fluorophores, the conformational and/or structural changes of aptamers can affect the fluorescence of a dye or the fluorescence resonance energy transfer (FRET) between two dyes. The signal change, either increase (i.e. the “signal-on” mode) or decrease (i.e. the “signal-off” mode), can reflect the extent of the binding process thereby allowing for quantitative measurement of target concentration.
The “Signal-On” Mode
FRET-based biosensors
In a typical “signal-on” biosensor based on fluorescently-labeled aptamers, one end of the aptamer is conjugated to a fluorophore with its fluorescence signal quenched by a quencher (which can also be another fluorophore) through FRET. Upon separation of the fluorophore and the quencher (through enzyme cleavage, aptamer binding, etc.), the fluorescence signal is recovered which can be used for detection and quantitative measurement of the target concentration.
In an early study, a pair of aptamers that can recognize two distinct epitopes of thrombin were each labeled with a different fluorophore through alkyl linkers [50]. The presence of thrombin induced association of the two aptamers, which annealed and brought the two fluorophores in close proximity for FRET. This strategy successfully detected thrombin with low detection limit of pM. In another report, sensors for ATP or adenosine diphosphate (ADP) based on a DNA enzyme (MgZ), which can cleave its RNA substrate in the presence of Mg2+, were shown to have low detection limit in the µM range [51].
Recently, a DNA aptamer against adenosine was immobilized onto a magnetic microparticle, which was able to sense adenosine in the uM range in 30% serum [52]. Initially, the 5’ end of the aptamer was modified with fluorescein and hybridized with a 12-mer complementary DNA, conjugated to a quencher at its 3’-end. The binding of adenosine induced a conformational change of the aptamer which displaced the quencher strand and recovered the fluorescence signal. Although this system worked well in aqueous buffers, the background was high and the signal was low in serum, possibly due to light absorption and/or scattering by serum. Upon immobilization of the aptamer onto magnetic nanoparticles, flow cytometry was able to detect individual particles, thereby effectively decreasing the light path and consequent interferences within serum.
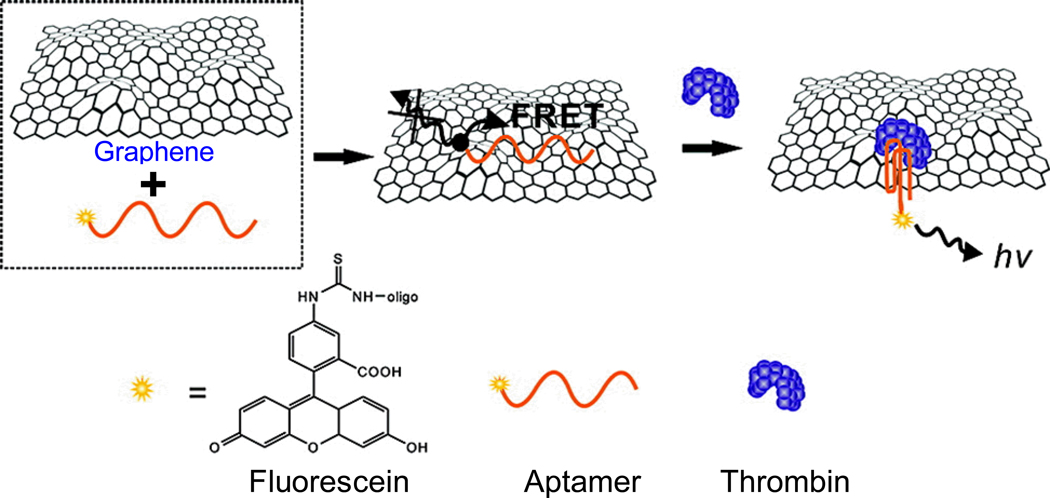
Graphene exhibits intriguing electronic properties which makes it an excellent energy acceptor, and hence an effective fluorescence quencher [53,54]. In one study, a fluorescein-labeled aptamer that binds to thrombin was non-covalently assembled onto graphene through π-π stacking (Fig. (1)) [55]. The fluorescence signal of fluorescein was initially quenched by graphene which was released upon thrombin binding, since fluorescein was too far away from graphene to be efficiently quenched. A detection limit of pM of thrombin, as well as good specificity in serum, was achieved [55].
Fig. (1).
Schematic illustration of a graphene-based biosensor for thrombin. The fluorescently-labeled aptamer is quenched when the aptamer binds to graphene, due to FRET between the dye and graphene. The fluorescence recovers when the aptamer binds to thrombin, which separates fluorescein from the graphene surface. Adapted from [55].
In another example, graphene oxide was used to quench a fluorescein-labeled aptamer (which binds to ochratoxin A [OTA]) in its free form but not the folded form after OTA binding [56]. Although excellent selectivity for OTA against other toxin analogs was observed, this system had poor sensitivity (with low detection limit in the µM range) which was attributed to analyte absorption by graphene oxide. After coating the graphene oxide with poly(vinyl pyrrolidone) (PVP), the detection limit was significantly improved into the nM range, suitable for measurements in real food samples such as the 1% red wine with various concentrations of OTA [56].
Fluorophore local environment change upon aptamer binding
In many cases, conformational change of an aptamer upon target binding can significantly affect the fluorescence of a conjugated dye. The finding that fluorescence of excimer and monomer pyrene is sensitive to local environment changes [57,58] has been employed for aptamer-based biosensing. In an early report, it was found that incorporation of a bis-pyrenyl fluorophore at a site adjacent to the ATP-binding residues within an aptamer exhibited a significant increase of the excimer fluorescence signal intensity upon ATP binding, which could be used to detect mM concentrations of ATP [59]. Based on the notion that fragments of an aptamer can restore its binding activity once brought together, the non-binding loop of a cocaine-binding aptamer was split into two pieces and target-induced self-assembly was monitored with UV spectroscopy [60]. With both fragments conjugated to pyrene molecules, the pyrenes become close enough to form a pyrene excimer upon assembly of the two pieces. This resulted in a wavelength shift of fluorescence and longer fluorescence life-time, which could be used for quantitative detection of cocaine at µM concentration in urine samples [60].
Other fluorophores were also used to modify the 2’-ribose at selected positions of a number of aptamers, which all exhibited increased fluorescence upon target binding of the aptamer [61]. These biosensors were found to have wide detection ranges in buffer solutions, many of which also performed well in biological samples such as urine and serum. Since fluorophore conjugation may reduce the target binding affinity of an aptamer, through either steric hindrance or interference with its folding, fluorescein-modified uridine was incorporated into the nucleoside triphosphate (NTP) library for in vitro SELEX of RNA aptamers that bind to ATP [62]. To avoid multiple fluorescent residues in one aptamer, which may lead to non-detectable change in fluorescence signal upon target binding, only a small percentage of the NTP library is fluorescein-modified uridine. The selected aptamers were able to detect ATP in the µM range, with the best one containing only one fluorescein-modified uridine. In addition, other fluorescent dyes could also be used to achieve similar signaling specificity and sensitivity [62].
Enzyme-linked immunosorbent assay (ELISA) with aptamers
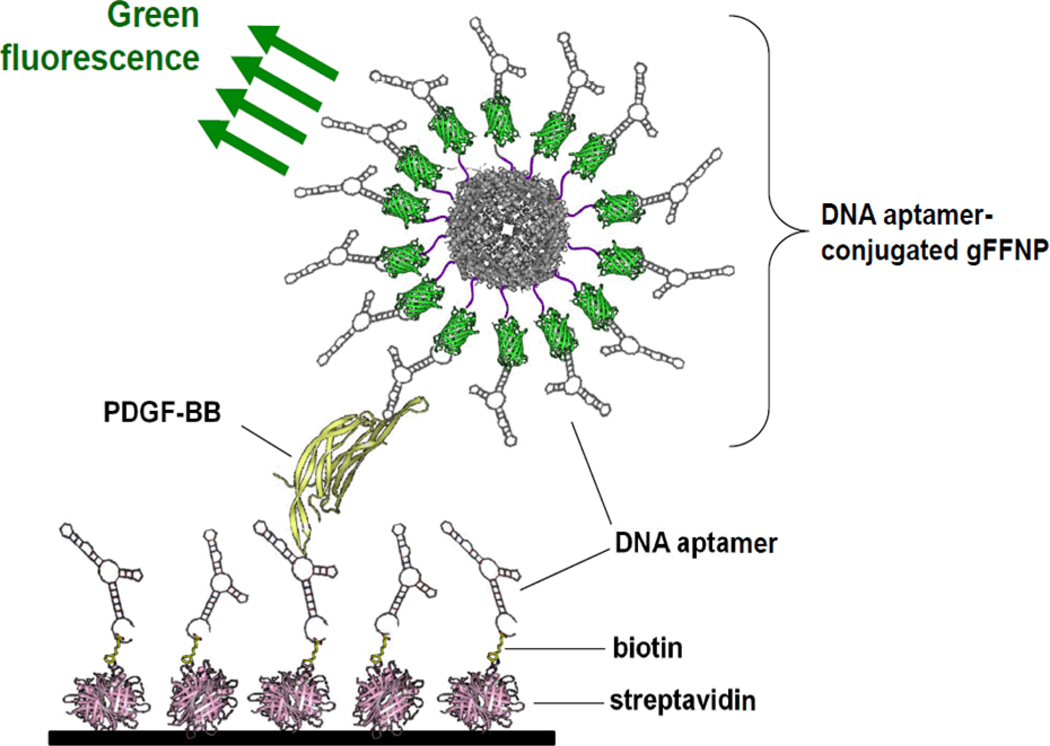
A pair of aptamers recognizing the same antigen can be used in a sandwich ELISA assay, similar to conventional antibody-based assays. Two aptamers that bind to platelet-derived growth factor B-chain homodimer (PDGF-BB) were conjugated to the surface of a green fluorescent ferritin nanoparticle (which functioned as a reporter probe) and the bottom of wells respectively for a sandwich ELISA (Fig. (2)) [63]. This aptamer-based ELISA was capable of detecting PDGF-BB at a concentration of as low as 100 fM, with higher sensitivity than antibody-based ELISA due to the multivalency effect of aptamer-conjugated nanoparticles.
Fig. (2).
A schematic illustration of an aptamer-based assay of platelet-derived growth factor B-chain homodimer (PDGF-BB) using aptamer-conjugated green fluorescent ferritin nanoparticles (gFFNP). Adapted from [63].
Fluorescence polarization (FP)-based biosensors
Changes of FP reflect the changes in tumbling rate of a fluorophore-modified molecule in the binding event. An FP-based competitive assay was developed, based on target-induced aptamer conformational change which affects the tumble rate of fluorophore-labeled aminoglycoside [64,65]. In another report, a thrombin-binding aptamer was labeled with fluorescein and immobilized onto a glass support [66]. When thrombin in solution bound to the immobilized aptamer, changes in the evanescent wave-induced fluorescence anisotropy could be detected quickly (within a few minutes) and sensitively (in nM range).
The “Signal-Off” Mode
Although sensors based on the “signal-off” mode are usually less sensitive than those based on the “signal-on” mode [67,68], they can sometimes lead to better detection of targets with low-affinity aptamers. More importantly, simpler designs with fewer steps are needed for “signal-off” sensors, which can be convenient and cost-effective [69,70].
FRET-based biosensors
Not only can FRET be employed to design “signal-on” sensors, it can also be used for “signal-off” sensors. In general, a fluorescence donor and a quencher are conjugated respectively to both ends of the aptamer. Upon target binding, conformational change of the aptamer drives the donor and quencher in close proximity which leads to fluorescence quenching. In one study, one stem of the three-way junction of a cocaine-binding aptamer was shortened, which adopted a free conformation in the absence of cocaine and closed upon cocaine binding [69]. The short stem was end-labeled with a fluorophore and a quencher respectively, where fluorescence quenching as a result of closed conformation quantitatively reflected the concentration of cocaine. Detection limit in the µM range was achieved, with high selectivity for cocaine over its metabolites [69]. However, the background signal was quite high when this sensor was used in serum.
Using a fluorescently-labeled thrombin-binding aptamer which forms either a G-quadruplex (where G can interact with Pb2+) or a hairpin structure (which can allow for the interaction of T with Hg2+), selective detection of Pb2+ and Hg2+ in the nM range was achieved by measuring terminal fluorescence quenching upon ion binding [70]. In addition, measurements in soil and pond water samples were also performed to validate the practicality of this sensor. Similarly, a dual-labeled L-argininamide-binding aptamer was used for detection of L-argininamide in the µM range, which was specific for this molecule in the presence of L-arginine, glycine, L-lysine, and guanidine [71].
Other fluorescent biosensors
Quantum dot (QD)-labeled aptamer has been used as a fluorescent reagent for on-strip detection of OTA, with a sensitivity comparable to common immunoassays [16]. Two DNA probes, one containing the complete complementary sequence to the aptamer and the other which binds to the extra 18-mer poly A at the 5’-end of aptamer, were used in this design without the need of a quencher. The ratio of fluorescence was used to quantify OTA in sample solutions, which had low detection limit in the ng/mL range [16].
In some cases, the analyte can quench the fluorescence of certain dyes after binding to an aptamer. For example, an adenosine sensor was developed using an abasic site-containing triplex, which was modified with fluorescent furano-dU in the binding site [72]. The bound adenosine was stabilized by π-stacking of a flanked TAT sequence, and formed hydrogen-bonds with the two neighboring furano-dU. Such hydrogen-bonding led to a decrease in the fluorescence signal, which could be used to selectively detect adenosine over other bases, as well as ATP and adenosine monophosphate (AMP), at concentrations in the nM to µM range [72].
FLUORESCENT BIOSENSORS WITH LABEL-FREE APTAMERS
Despite the many interesting applications of biosensors based on fluorescently-labeled aptamers, covalent labeling of aptamers with fluorophores can be time-consuming and costly. In addition, the conjugated fluorophore may interfere with target binding of the aptamer. To overcome these challenges, a number of strategies have been developed to construct biosensors based on label-free aptamers.
The “Signal-On” Mode
Fluorophore displacement upon aptamer binding
One common strategy for “signal-on” aptamer-based biosensors involves the use of label-free aptamer which, upon target binding, can displace fluorophores that are either previously quenched or of little fluorescence. Aptamer-conjugated gold nanoparticles, with the fluorescence of N, N-dimethyl-2, 7-diazapyrenium dication (DMDAP) completely quenched by gold upon its intercalation onto the aptamer, were prepared for PDGF detection [73]. When both DMDAP and PDGF were added to the sensor, they competed for aptamer binding which left the unbound DMDAP highly fluorescent. This approach detected PDGF at as low as pM concentrations in buffer solutions, however the background fluorescence was very high in cell culture media. Subsequently, a similar strategy was used for ATP detection, which exhibited nM sensitivity in urine samples [74].
Besides gold nanoparticles, certain fluorophores can also be quenched by nucleic acid bases. An ssODN containing an abasic site, termed “dSpacer”, was constructed and hybridized to an adenosine-binding aptamer [7]. The resulting duplex was embedded with 2-amino-5, 6, 7-trimethyl-1, 8-naphthyridine (ATMND) at dSpacer which had its fluorescence quenched. Upon adenosine binding, the aptamer underwent structural changes and released the dSpacer-containing ssODN, leading to fluorescence recovery of ATMND. This sensor was able to detect adenosine in the µM range with high specificity over uridine, cytidine, as well as adenosine analogs.
In a follow-up study, a vacant site between DNA sequences was used to directly incorporate ATMND, which was stabilized through hydrogen-bonding, π-π stacking, and electrostatic forces by cytosine in the opposite strand and flanking guanines in the same strand [75]. The quenched fluorescence of ATMND could be recovered after release of one strand of the DNA duplex, caused by either DNAzyme-cleavage or analyte-dependent structural changes of aptamers. With this design, Hg2+ and adenosine were successfully detected in the nM to uM range, respectively. A similar biosensor from another group was also reported to have a detection limit of adenosine in the µM range [76].
Messenger activation upon aptamer binding
With rational engineering, target binding of aptamers can be used to activate certain sequences which may serve as messengers for further signaling. A recombinant protein was constructed which contained the enhanced yellow fluorescent protein (EYFP) and the enhanced cyan fluorescent protein (ECFP) [77]. The EYFP and ECFP were bridged by a Rev-peptide, which can bind to Rev-responsive element (RRE) RNA. Such binding resulted in a conformational change of the peptide which increased the FRET efficiency between EYFP and ECFP. With this signaling system, a theophylline-binding aptamer was inserted into the RRE RNA to generate a FRET-based biosensor with µM sensitivity in cell lysates [78].
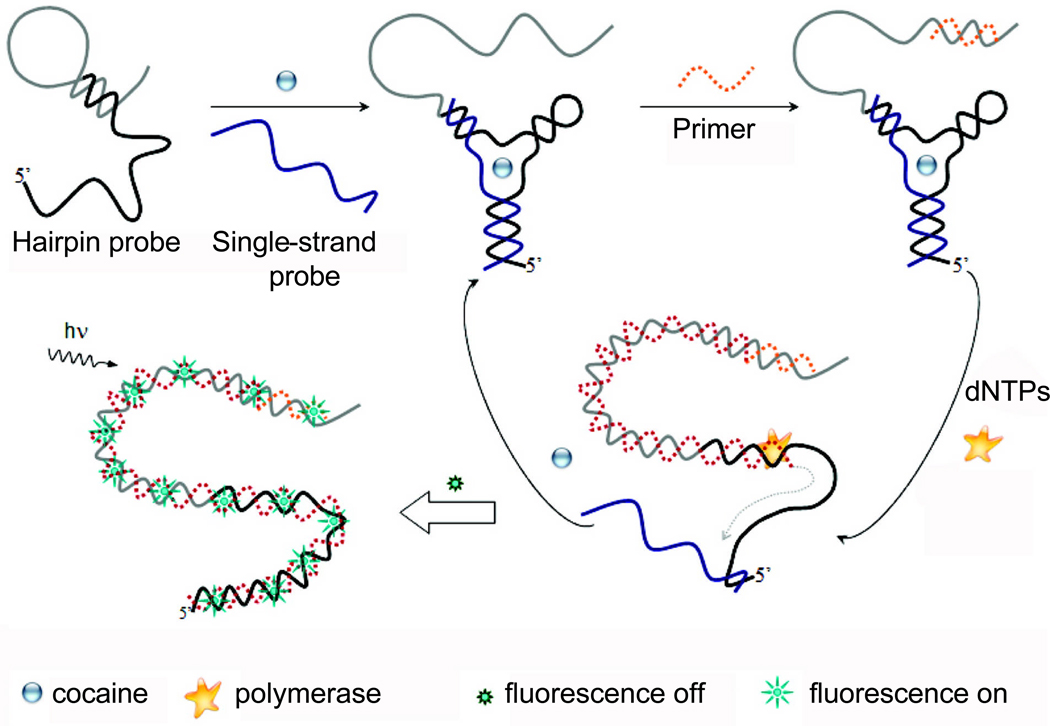
In another report, the messenger sequence was amplified by polymerase chain reaction (PCR) to achieve enhanced signal (Fig. (3)) [79]. The cocaine-binding aptamer was split into two recognition sequences which were incorporated into a hairpin DNA and an ssDNA respectively. Upon cocaine binding, the two DNAs were brought together which led to a structural change of the hairpin DNA. Its 3’ end became available for hybridization with a primer, which can then be amplified by PCR. When the hairpin DNA was converted to a duplex, cocaine and ssDNA were displaced which could bind to another hairpin DNA for a new cycle of amplification. The newly-synthesized double strand (ds) DNA in the system was then detected with a dye that was specific for ds DNA, which exhibited nM sensitivity for cocaine [79]. Recently, a thiol-containing cocaine-binding aptamer was immobilized onto gold nanoparticles which were further attached to magnetic beads [12]. Through rolling circle amplification and the use of molecular beacons, nM concentration of cocaine was detectable with a moderate linear dynamic range of 1–50 nM.
Fig. (3).
A schematic illustration of cocaine sensing strategy based on strand displacement amplification. Adapted from [79].
Quencher deactivation upon aptamer binding
Similar to the abovementioned “signal-on” biosensors using fluorescently-labeled aptamers [52,55,56], label-free aptamers can also activate fluorophores by either deactivation or removal of a quencher. For example, quencher-conjugated beads were used to screen out quencher-specific RNA aptamers which could separate a fluorophore-quencher pair, and/or lower the energy level of the quencher to reduce its FRET efficiency, upon binding thereby recovering the fluorescence [80,81]. Although µM concentrations of RNA aptamers could be detected and this interesting design may be incorporated into future SELEX of aptamers, potential applications of this strategy are limited since the target has to be an effective fluorescence quencher. Similar strategy was also adopted to design biosensors for ATP and hemin (an iron-containing porphyrin) with nM to µM detection limit using a fluorescent polymer, whose fluorescence could be quenched by hemin through either FRET or photo-induced electron transfer [82].
In another study, QD was linked to a thiol-containing thrombin-binding aptamer, which was hybridized with a complementary strand labeled with a quencher [83]. Although nM sensitivity was achieved, this design suffered from non-specific adsorption of DNA to the QD surface. In addition, it took a very long time (overnight) for displacement of the complementary DNA, which was also difficult to hybridize to the QD-conjugated aptamer due to multiple factors such as steric hindrance. Another similar QD-based biosensor was also reported to exhibit µM sensitivity for cocaine detection [84].
Enhancement of fluorophore binding upon aptamer binding
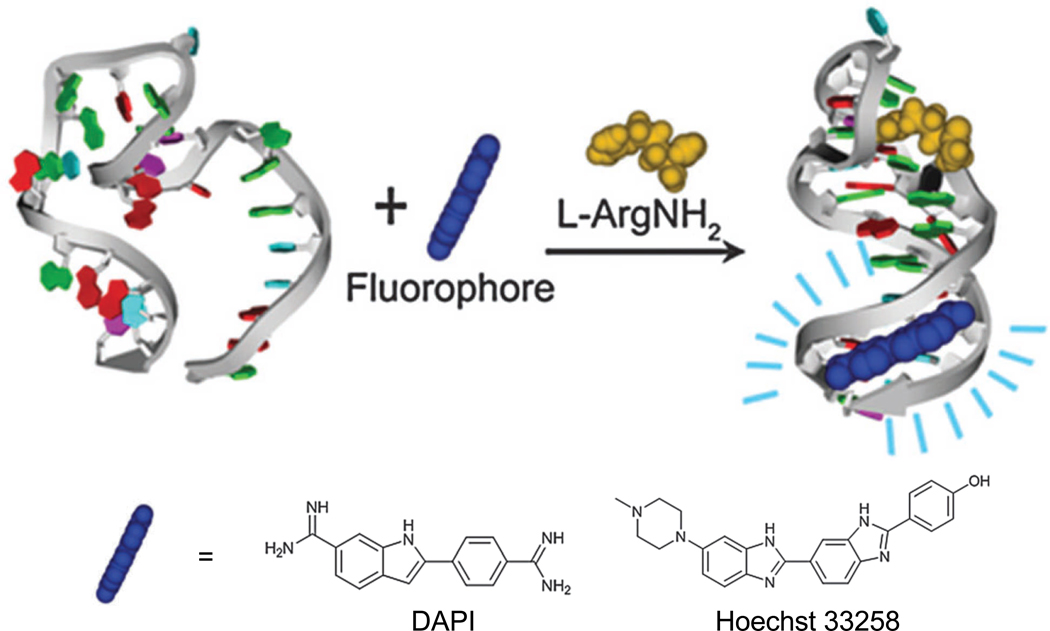
Some fluorescent dyes such as 4’, 6-diamidino-2-phenylindol (DAPI) [85], Hoechst 33258 [85], and malachite green (MG) [86–88] have little/weak fluorescence when they are free in solution, but exhibit enhanced fluorescence after binding with aptamers. Since the stem base pairs of a DNA aptamer (which is specific for L-argininamide) were not involved in target binding, the AT-rich sequences in the stem region were designed to serve as a dye-binding domain (Fig. (4)) [85]. AT-selective minor groove-targeting dyes (e.g. DAPI and Hoechst 33258) were incorporated in these sensors, which could detect L-argininamide in µM concentrations. In the absence of L-argininamide, the aptamer adopted equilibrated conformations between ss and folded hairpin, which has no stable stem formation for dye binding.
Fig. (4).
A schematic illustration of fluorescent detection of L-argininamide (L-ArgNH2) with DAPI and Hoechst 33258. Adapted from [85].
In an intriguing report, a MG-binding aptamer was connected with an adenosine-binding aptamer, which was partially hybridized with a short bridging strand that was more complementary to the MG-binding aptamer to completely suppress its MG-binding capability [86]. Since MG molecules in the solution could not bind to the aptamer, they emitted little fluorescence. However, addition of adenosine induced a structural change of the adenosine-binding aptamer, which in turn weakened the hybridization of the bridging strand. As a result, the MG-binding aptamer regained proper folding to recognize MG and yielded enhanced fluorescence. Detection limit of adenosine at µM concentrations was achieved. In another report, biosensors of a similar design were also developed for highly specific detection of ATP, flavin mono-nucleotide, and theophylline [89].
An aptamer was selected for high affinity binding to the Hoechst dye, which emitted strong fluorescence around 470 nm after binding [90]. Subsequently, it was identified that the 5’-AGG bulge and U4 loop of this aptamer were the key domains for lighting up the Hoechst dye [91]. When UUGG residues were added to its 3’ end, the aptamer adopted a different confirmation with no AGG bulge which led to a loss of fluorescence. An ADP biosensor was designed based on this finding, through connecting the aptamer with an ADP-binding RNA aptamer using UUGG as the communication module, which was able to detect ADP in the µM range within a few minutes. Good selectivity over AMP and cyclic AMP was also achieved [91].
The “Signal-Off” Mode
Fluorophore displacement upon aptamer binding
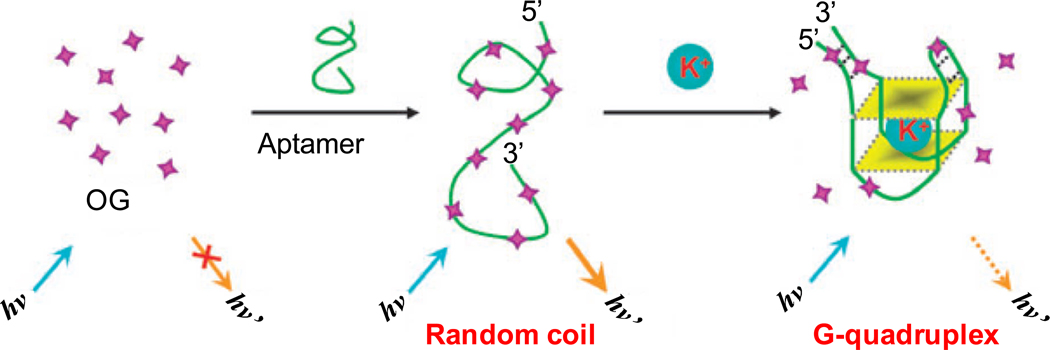
When complexed to reduced graphene oxide, the fluorescence of acridine orange (AO) is quenched. A hemin-binding aptamer, which formed a G-quadruplex in the presence of K+ or Na+, can compete with reduced graphene oxide for AO thereby regenerating its fluorescence [92]. Conversely, the presence of hemin in solution can substitute AO from the aptamer complex, which leads to fluorescence quenching. With this “signal-off” sensor, nM concentrations of hemin in buffer solution could be detected with high specificity [92]. In another study, the dye OliGreen (OG) which exhibits > 1000 fold enhanced fluorescence upon binding to ssDNA was used to indirectly monitor the presence of K+ (Fig. (5)) [93]. This sensor specifically detected K+ over other cations such as Na+, Li+, NH4+, Mg2+, and Ca2+ with a detection limit in the nM range in urine samples.
Fig. (5).
A schematic representation of a K+ sensor based on modulation of the fluorescence of the complex formed between the dye (OG) and an ATP -binding aptamer. Adapted from [93].
Using dsDNA as a template, CuSO4 was reported to form fluorescent copper nanoparticles that accumulated in the major groove of dsDNA [94]. Based on this phenomenon, an analyte-binding aptamer was hybridized with a part of its complementary strand to yield a dsDNA for association with Cu2+ to emit fluorescence [95]. Upon addition of analyte which bound to the aptamer, a reduced fluorescence was observed. These “signal-off” sensors were able to detect ATP and cocaine in the nM range in both cancer cells and human serum.
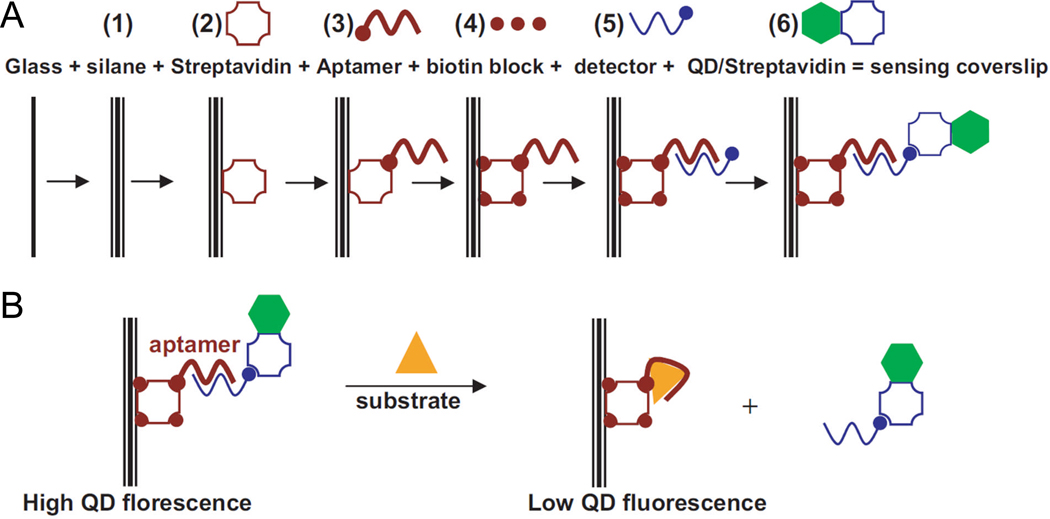
Following the aforementioned success of QD-based “signal-on” biosensors [84], a “signal-off” biosensor was also developed by hybridizing a Cy5-labeled short ODN to the aptamer which served as a FRET acceptor [84]. Similar sensitivity (at µM level) as the “signal-on” sensor was achieved with this design. QD has also been used as a fluorophore to develop a real-time fluorescent flow sensor with an adenosine-binding aptamer (Fig. (6)) [96]. However, this sensor has very poor sensitivity in the mM range.
Fig. (6).
A QD-based biosensor. A. Preparation of the sensing coverslip with both sides modified. B. ATP sensing with the coverslip. Adapted from [96].
In another report, a Hg2+ sensor was constructed by immobilizing to the fiber optical sensor surface a DNA aptamer containing a 5’ short sequence, which can be hybridized to a Cy5.5-labeled cDNA [97]. For each detection, a Hg2+-containing sample was mixed with a fixed concentration of Cy5.5-labeled cDNA and then added to the sensor for competitive binding to the DNA aptamer, which could specifically detect Hg2+ at nM concentration.
Fluorescence-based competitive assay
Among the various “signal-off” sensor designs, competitive assay is simple and straightforward, in which a limited amount of fluorescently-labeled analyte is directly replaced by unlabeled analyte from the sample which results in a decrease in fluorescence. Thiazole orange (TO), a dye with weak fluorescence in solution, becomes highly fluorescent after its non-specific binding to double helical nucleic acids. Taking advantage of this property, TO was conjugated to guanidine monophosphate (GMP) and AMP and tested for their ability to “light up” their corresponding aptamers [98]. The GMP and AMP sensors based on this strategy exhibited high specificity over other NTPs.
FP-based displacement assay
Direct labeling of an aptamer in FP-based assays is feasible for targets larger than aptamers such as proteins [66]. For measurement of aptamer binding with small molecules, a displacement assay was designed where a fluorescein-labeled ODN was pre-hybridized with an aptamer, which was subsequently displaced by small molecules that can be recognized by the aptamer [99]. In a model assay with an OTA-binding aptamer, FP-based sensing could detect OTA in nM concentrations. More importantly, such FP-based assay could allow for immediate measurement of target concentration, since equilibrium is typically reached in a few seconds.
Repositioning of quencher upon aptamer binding
Different from the aforementioned studies where aptamer-conjugated donor and quencher were brought together upon target binding [69–71], in one study QD was conjugated to a thrombin-binding aptamer which was hybridized with a complementary strand, labeled with a redox-active metal complex at the 5’ end [100]. The QD fluorescence can be quenched when the aptamer bound to thrombin, which unraveled the duplex and brought the metal complex in close proximity to the QD. Though the sensitivity of this sensor was not clearly stated in the report, it can be estimated to be in the nM range based on the dose-response curve.
CONCLUSION
In this review article, we have comprehensively summarized the development of aptamer-based fluorescent biosensors for proteins, small molecules, metal ions, among others (Table 1). Although aptamer-based biosensors can have enormous potential impact in many areas such as disease diagnosis, to date very few of them have been demonstrated useful in biological samples. Most studies on aptamer-based biosensors have been performed in model systems (e.g. aqueous buffer) under well-controlled laboratory settings for proof-of-concept, with few clinically relevant targets been investigated. It will be ideal if different aptamer-based sensors can be compared side-by-side using the same model system, which may significantly help in deciding which sensors are the best candidates for potential clinical/commercial development. The National Cancer Institute (NCI) has required each of its funded nanotechnology centers to test their newly developed nanosensors using the same standard samples, which is expected to readily identify which new sensors truly stand out from the large pool of candidates. Extending a similar standard to a much broader range of research laboratories across the country would be highly beneficial. Choosing the right candidate(s) at an early stage not only saves precious research time, but can also significantly reduce the cost for new sensor development.
Table 1.
A tabulated summary of aptamer-based fluorescent biosensors.
| Target | Labeling method |
Mode | Detection limit |
Biological sample? |
Ref. |
|---|---|---|---|---|---|
| Adenosine | Covalent | Signal-on | µM | 30% serum | [52] |
| Signal-off | nM | No | [72] | ||
| Label-free | Signal-on | µM | No | [7] | |
| µM | No | [75] | |||
| µM | No | [76] | |||
| µM | No | [86] | |||
| ATP | Covalent | Signal-on | µM | No | [51] |
| mM | No | [59] | |||
| µM | No | [62] | |||
| Label-free | Signal-on | nM | No | [74] | |
| nM | No | [82] | |||
| Signal-off | nM | Cells, serum | [95] | ||
| µM | No | [96] | |||
| ADP | Covalent | Signal-on | µM | No | [51] |
| Label-free | Signal-on | µM | No | [91] | |
| AMP | Covalent | Signal-on | mM | Urine, serum | [61] |
| Tyrosinamid | Covalent | Signal-on | mM | Urine, serum | [61] |
| L-argininamide | Label-free | Signal-on | µM | No | [85] |
| Thrombin | Covalent | Signal-on | pM | Serum | [55] |
| pM | Cell extract, plasma | [50] | |||
| nM | No | [66] | |||
| Label-free | Signal-on | nM | No | [83] | |
| Signal-off | nM | No | [100] | ||
| Theophylline | Label-free | Signal-on | µM | Cell lysate | [78] |
| OTA | Covalent | Signal-on | nM | 1% Red wine | [56] |
| Signal-off | nM | Red wine | [16] | ||
| Label-free | Signal-off | nM | No | [99] | |
| Cocaine | Covalent | Signal-on | µM | Urine | [60] |
| Signal-off | µM | Serum | [69] | ||
| Label-free | Signal-on | nM | No | [79] | |
| nM | No | [12] | |||
| µM | No | [84] | |||
| Signal-off | nM | No | [95] | ||
| µM | No | [84] | |||
| PDGF | Covalent | Signal-on | fM | No | [63] |
| Label-free | Signal-on | pM | No | [73] | |
| Pb2+ | Covalent | Signal-off | pM | Soil, pond water | [70] |
| Hg2+ | Covalent | Signal-off | nM | Soil, pond water | [70] |
| Label-free | Signal-on | nM | No | [75] | |
| Signal-off | nM | No | [97] | ||
| K+ | Label-free | Signal-off | nM | Urine | [93] |
| Hemin | Label-free | Signal-off | nM | No | [92] |
Although aptamers can be selected to target any analyte of choice, the sensing and diagnostic market is still largely dominated by antibodies. Few aptamer-based sensors are commercially available. One of the most promising areas for aptamer-based sensors is for detection of small molecules (e.g. metal ions, metabolites, etc.) in biological systems and in the environment, since it is very difficult to generate highly specific antibodies for small molecules due to challenges in eliciting immune responses to small molecules. For potential commercial applications, it is essential to design easy-to-use, accurate, and cost-effective sensors or kits. Once a sufficient number of highly sensitive and practical sensors are obtained, the next step is to develop sensor arrays for more sophisticated target analysis. To date, most aptamer-based sensors were developed for in vitro detection while significant future effort is expected for in vivo biosensing. The future of aptamer-based biosensors looks increasingly brighter, yet many hurdles remain to be overcome.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the University of South Florida start-up fund, the University of Wisconsin Carbone Cancer Center, NCRR 1UL1RR025011, a DOD PCRP IDEA Award, and the support from TE Connectivity.
REFERENCES
- 1.Cooper MA. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002;1:515–528. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- 2.Yeom SH, Kang BH, Kim KJ, Kang SW. Nanostructures in biosensor--a review. Front. Biosci. 2011;16:997–1023. doi: 10.2741/3731. [DOI] [PubMed] [Google Scholar]
- 3.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 4.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 5.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Xia J, Li W, Zhang S. Multianalyte electrochemical biosensor based on aptamer- and nanoparticle-integrated bio-barcode amplification. Chem. Asian J. 2010;5:294–300. doi: 10.1002/asia.200900217. [DOI] [PubMed] [Google Scholar]
- 7.Xiang Y, Tong A, Lu Y. Abasic site-containing DNAzyme and aptamer for label-free fluorescent detection of Pb2+ and adenosine with high sensitivity, selectivity, and tunable dynamic range. J. Am. Chem. Soc. 2009;131:15352–15357. doi: 10.1021/ja905854a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Y, Chen C, Zhou M, Dong S, Wang E. Microfluidic electrochemical aptameric assay integrated on-chip: A potentially convenient sensing platform for the amplified and multiplex analysis of small molecules. Anal. Chem. 2011 doi: 10.1021/ac101988n. Epub. [DOI] [PubMed] [Google Scholar]
- 9.Liu F, Zhang J, Chen R, Chen L, Deng L. Highly effective colorimetric and visual detection of ATP by a DNAzyme-aptamer sensor. Chem. Biodivers. 2011;8:311–316. doi: 10.1002/cbdv.201000130. [DOI] [PubMed] [Google Scholar]
- 10.Nakano S, Mashima T, Matsugami A, Inoue M, Katahira M, Morii T. Structural aspects for the recognition of ATP by ribonucleopeptide receptors. J. Am. Chem. Soc. 2011;133:4567–4579. doi: 10.1021/ja110725d. [DOI] [PubMed] [Google Scholar]
- 11.Wang W, Wu WY, Zhong X, Wang W, Miao Q, Zhu JJ. Aptamer-based PDMS-gold nanoparticle composite as a platform for visual detection of biomolecules with silver enhancement. Biosens. Bioelectron. 2011;26:3110–3114. doi: 10.1016/j.bios.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Ma C, Wang W, Yang Q, Shi C, Cao L. Cocaine detection via rolling circle amplification of short DNA strand separated by magnetic beads. Biosens. Bioelectron. 2011;26:3309–3312. doi: 10.1016/j.bios.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Kang T, Yoo SM, Yoon I, Lee S, Choo J, Lee SY, Kim B. Au nanowire-on-film SERRS sensor for ultrasensitive Hg2+ detection. Chemistry. 2011;17:2211–2214. doi: 10.1002/chem.201001663. [DOI] [PubMed] [Google Scholar]
- 14.Xu JP, Song ZG, Fang Y, Mei J, Jia L, Qin AJ, Sun JZ, Ji J, Tang BZ. Label-free fluorescence detection of mercury(II) and glutathione based on Hg2+-DNA complexes stimulating aggregation-induced emission of a tetraphenylethene derivative. Analyst. 2010;135:3002–3007. doi: 10.1039/c0an00554a. [DOI] [PubMed] [Google Scholar]
- 15.Kuang H, Chen W, Xu D, Xu L, Zhu Y, Liu L, Chu H, Peng C, Xu C, Zhu S. Fabricated aptamer-based electrochemical "signal-off" sensor of ochratoxin A. Biosens. Bioelectron. 2010;26:710–716. doi: 10.1016/j.bios.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 16.Wang L, Chen W, Ma W, Liu L, Ma W, Zhao Y, Zhu Y, Xu L, Kuang H, Xu C. Fluorescent strip sensor for rapid determination of toxins. Chem. Commun. 2011;47:1574–1576. doi: 10.1039/c0cc04032k. [DOI] [PubMed] [Google Scholar]
- 17.Bai L, Yuan R, Chai Y, Yuan Y, Mao L, Zhuo Y. Highly sensitive electrochemical label-free aptasensor based on dual electrocatalytic amplification of Pt-AuNPs and HRP. Analyst. 2011;136:1840–1845. doi: 10.1039/c0an00755b. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Zhang J, Li J, Yang HH, Fu F, Chen G. An ultrasensitive signal-on electrochemical aptasensor via target-induced conjunction of split aptamer fragments. Biosens. Bioelectron. 2010;25:996–1000. doi: 10.1016/j.bios.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Li L, Zhao H, Chen Z, Mu X, Guo L. Aptamer-based electrochemical approach to the detection of thrombin by modification of gold nanoparticles. Anal. Bioanal. Chem. 2010;398:563–570. doi: 10.1007/s00216-010-3922-2. [DOI] [PubMed] [Google Scholar]
- 20.Rahman MA, Son JI, Won MS, Shim YB. Gold nanoparticles doped conducting polymer nanorod electrodes: ferrocene catalyzed aptamer-based thrombin immunosensor. Anal. Chem. 2009;81:6604–6611. doi: 10.1021/ac900285v. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yadavalli VK. Surface immobilization of DNA aptamers for biosensing and protein interaction analysis. Biosens. Bioelectron. 2011;26:3142–3147. doi: 10.1016/j.bios.2010.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhao S, Yang W, Lai RY. A folding-based electrochemical aptasensor for detection of vascular endothelial growth factor in human whole blood. Biosens. Bioelectron. 2011;26:2442–2447. doi: 10.1016/j.bios.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 23.Ohno Y, Maehashi K, Matsumoto K. Label-free biosensors based on aptamer-modified graphene field-effect transistors. J. Am. Chem. Soc. 2010;132:18012–18013. doi: 10.1021/ja108127r. [DOI] [PubMed] [Google Scholar]
- 24.Pollet J, Delport F, Janssen KP, Jans K, Maes G, Pfeiffer H, Wevers M, Lammertyn J. Fiber optic SPR biosensing of DNA hybridization and DNA-protein interactions. Biosens. Bioelectron. 2009;25:864–869. doi: 10.1016/j.bios.2009.08.045. [DOI] [PubMed] [Google Scholar]
- 25.Cai Q, Chen L, Luo F, Qiu B, Lin Z, Chen G. Determination of cocaine on banknotes through an aptamer-based electrochemiluminescence biosensor. Anal. Bioanal. Chem. 2011;400:289–294. doi: 10.1007/s00216-011-4739-3. [DOI] [PubMed] [Google Scholar]
- 26.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, Ostman A, Landegren U. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 2002;20:473–477. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 27.Di Giusto DA, Wlassoff WA, Gooding JJ, Messerle BA, King GC. Proximity extension of circular DNA aptamers with real-time protein detection. Nucleic Acids Res. 2005;33:e64. doi: 10.1093/nar/gni063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng AK, Sen D, Yu HZ. Design and testing of aptamer-based electrochemical biosensors for proteins and small molecules. Bioelectrochemistry. 2009;77:1–12. doi: 10.1016/j.bioelechem.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Willner I, Zayats M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. Engl. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 30.Swensen JS, Xiao Y, Ferguson BS, Lubin AA, Lai RY, Heeger AJ, Plaxco KW, Soh HT. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J. Am. Chem. Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoon H, Kim JH, Lee N, Kim BG, Jang J. A novel sensor platform based on aptamer-conjugated polypyrrole nanotubes for label-free electrochemical protein detection. Chembiochem. 2008;9:634–641. doi: 10.1002/cbic.200700660. [DOI] [PubMed] [Google Scholar]
- 32.Lee J, Jo M, Kim TH, Ahn JY, Lee DK, Kim S, Hong S. Aptamer sandwich-based carbon nanotube sensors for single-carbon-atomic-resolution detection of non-polar small molecular species. Lab Chip. 2011;11:52–56. doi: 10.1039/c0lc00259c. [DOI] [PubMed] [Google Scholar]
- 33.Polonschii C, David S, Tombelli S, Mascini M, Gheorghiu M. A novel low-cost and easy to develop functionalization platform. Case study: aptamer-based detection of thrombin by surface plasmon resonance. Talanta. 2010;80:2157–2164. doi: 10.1016/j.talanta.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Zhou HS. Aptamer-based Au nanoparticles-enhanced surface plasmon resonance detection of small molecules. Anal. Chem. 2008;80:7174–7178. doi: 10.1021/ac801281c. [DOI] [PubMed] [Google Scholar]
- 35.Xia F, Zuo X, Yang R, Xiao Y, Kang D, Vallee-Belisle A, Gong X, Yuen JD, Hsu BB, Heeger AJ, Plaxco KW. Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes. Proc. Natl. Acad. Sci. USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elbaz J, Moshe M, Shlyahovsky B, Willner I. Cooperative multicomponent selfassembly of nucleic acid structures for the activation of DNAzyme cascades: a paradigm for DNA sensors and aptasensors. Chemistry. 2009;15:3411–3418. doi: 10.1002/chem.200802004. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Lu Y. Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angew. Chem. Int. Ed. Engl. 2005;45:90–94. doi: 10.1002/anie.200502589. [DOI] [PubMed] [Google Scholar]
- 38.Gronewold TM, Baumgartner A, Weckmann A, Knekties J, Egler C. Selection process generating peptide aptamers and analysis of their binding to the TiO2 surface of a surface acoustic wave sensor. Acta Biomater. 2009;5:794–800. doi: 10.1016/j.actbio.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Treitz G, Gronewold TM, Quandt E, Zabe-Kuhn M. Combination of a SAW-biosensor with MALDI mass spectrometric analysis. Biosens. Bioelectron. 2008;23:1496–1502. doi: 10.1016/j.bios.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 40.Shu W, Laurenson S, Knowles TP, Ko Ferrigno P, Seshia AA. Highly specific label-free protein detection from lysed cells using internally referenced microcantilever sensors. Biosens. Bioelectron. 2008;24:233–237. doi: 10.1016/j.bios.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Savran CA, Knudsen SM, Ellington AD, Manalis SR. Micromechanical detection of proteins using aptamer-based receptor molecules. Anal. Chem. 2004;76:3194–3198. doi: 10.1021/ac049859f. [DOI] [PubMed] [Google Scholar]
- 42.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cho EJ, Lee JW, Ellington AD. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 44.Danielsson B. Artificial receptors. Adv. Biochem. Eng. Biotechnol. 2008;109:97–122. doi: 10.1007/10_2007_088. [DOI] [PubMed] [Google Scholar]
- 45.Holthoff EL, Bright FV. Molecularly imprinted xerogels as platforms for sensing. Acc. Chem. Res. 2007;40:756–767. doi: 10.1021/ar700087t. [DOI] [PubMed] [Google Scholar]
- 46.Lee JH, Yigit MV, Mazumdar D, Lu Y. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv. Drug Deliv. Rev. 2010;62:592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JO, So HM, Jeon EK, Chang H, Won K, Kim YH. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2008;390:1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sadik OA, Aluoch AO, Zhou A. Status of biomolecular recognition using electrochemical techniques. Biosens. Bioelectron. 2009;24:2749–2765. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang G, Wang Y, Chen L, Choo J. Nanomaterial-assisted aptamers for optical sensing. Biosens. Bioelectron. 2010;25:1859–1868. doi: 10.1016/j.bios.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Heyduk E, Heyduk T. Nucleic acid-based fluorescence sensors for detecting proteins. Anal. Chem. 2005;77:1147–1156. doi: 10.1021/ac0487449. [DOI] [PubMed] [Google Scholar]
- 51.Chiuman W, Li Y. Simple fluorescent sensors engineered with catalytic DNA 'MgZ' based on a non-classic allosteric design. PLoS One. 2007;2:e1224. doi: 10.1371/journal.pone.0001224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang PJ, Liu J. Flow cytometry-assisted detection of adenosine in serum with an immobilized aptamer sensor. Anal. Chem. 2010;82:4020–4026. doi: 10.1021/ac9028505. [DOI] [PubMed] [Google Scholar]
- 53.Swathi RS, Sebastian KL. Long range resonance energy transfer from a dye molecule to graphene has (distance)(−4) dependence. J. Chem. Phys. 2009;130 doi: 10.1063/1.3077292. 086101. [DOI] [PubMed] [Google Scholar]
- 54.Swathi RS, Sebastian KL. Resonance energy transfer from a dye molecule to graphene. J. Chem. Phys. 2008;129 doi: 10.1063/1.2956498. 054703. [DOI] [PubMed] [Google Scholar]
- 55.Chang H, Tang L, Wang Y, Jiang J, Li J. Graphene fluorescence resonance energy transfer aptasensor for the thrombin detection. Anal. Chem. 2010;82:2341–2346. doi: 10.1021/ac9025384. [DOI] [PubMed] [Google Scholar]
- 56.Sheng L, Ren J, Miao Y, Wang J, Wang E. PVP-coated graphene oxide for selective determination of ochratoxin A via quenching fluorescence of free aptamer. Biosens. Bioelectron. 2011;26:3494–3499. doi: 10.1016/j.bios.2011.01.032. [DOI] [PubMed] [Google Scholar]
- 57.Wu C, Wang C, Yan L, Yang CJ. Pyrene excimer nucleic acid probes for biomolecule signaling. J. Biomed. Nanotechnol. 2009;5:495–504. doi: 10.1166/jbn.2009.1074. [DOI] [PubMed] [Google Scholar]
- 58.Lehrer SS. Intramolecular pyrene excimer fluorescence: a probe of proximity and protein conformational change. Methods Enzymol. 1997;278:286–295. doi: 10.1016/s0076-6879(97)78015-7. [DOI] [PubMed] [Google Scholar]
- 59.Yamana K, Ohtani Y, Nakano H, Saito I. Bis-pyrene labeled DNA aptamer as an intelligent fluorescent biosensor. Bioorg. Med. Chem. Lett. 2003;13:3429–3431. doi: 10.1016/s0960-894x(03)00799-6. [DOI] [PubMed] [Google Scholar]
- 60.Wu C, Yan L, Wang C, Lin H, Wang C, Chen X, Yang CJ. A general excimer signaling approach for aptamer sensors. Biosens. Bioelectron. 2010;25:2232–2237. doi: 10.1016/j.bios.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 61.Merino EJ, Weeks KM. Facile conversion of aptamers into sensors using a 2'-riboselinked fluorophore. J. Am. Chem. Soc. 2005;127:12766–12767. doi: 10.1021/ja053189t. [DOI] [PubMed] [Google Scholar]
- 62.Jhaveri S, Rajendran M, Ellington AD. In vitro selection of signaling aptamers. Nat. Biotechnol. 2000;18:1293–1297. doi: 10.1038/82414. [DOI] [PubMed] [Google Scholar]
- 63.Kim SE, Ahn KY, Park JS, Kim KR, Lee KE, Han SS, Lee J. Fluorescent Ferritin Nanoparticles and Application to the Aptamer Sensor. Anal. Chem. 2011 doi: 10.1021/ac200657s. Epub. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Killian J, Hamasaki K, Rando RR. RNA molecules that specifically and stoichiometrically bind aminoglycoside antibiotics with high affinities. Biochemistry. 1996;35:12338–12346. doi: 10.1021/bi960878w. [DOI] [PubMed] [Google Scholar]
- 65.Cho J, Hamasaki K, Rando RR. The binding site of a specific aminoglycoside binding RNA molecule. Biochemistry. 1998;37:4985–4992. doi: 10.1021/bi972757h. [DOI] [PubMed] [Google Scholar]
- 66.Potyrailo RA, Conrad RC, Ellington AD, Hieftje GM. Adapting selected nucleic acid ligands (aptamers) to biosensors. Anal. Chem. 1998;70:3419–3425. doi: 10.1021/ac9802325. [DOI] [PubMed] [Google Scholar]
- 67.Li N, Ho CM. Aptamer-based optical probes with separated molecular recognition and signal transduction modules. J. Am. Chem. Soc. 2008;130:2380–2381. doi: 10.1021/ja076787b. [DOI] [PubMed] [Google Scholar]
- 68.Xiao Y, Lubin AA, Baker BR, Plaxco KW, Heeger AJ. Single-step electronic detection of femtomolar DNA by target-induced strand displacement in an electrodebound duplex. Proc. Natl. Acad. Sci. USA. 2006;103:16677–16680. doi: 10.1073/pnas.0607693103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 70.Liu CW, Huang CC, Chang HT. Highly selective DNA-based sensor for lead(II) and mercury(II) ions. Anal. Chem. 2009;81:2383–2387. doi: 10.1021/ac8022185. [DOI] [PubMed] [Google Scholar]
- 71.Ozaki H, Nishihira A, Wakabayashi M, Kuwahara M, Sawai H. Biomolecular sensor based on fluorescence-labeled aptamer. Bioorg. Med. Chem. Lett. 2006;16:4381–4384. doi: 10.1016/j.bmcl.2006.05.054. [DOI] [PubMed] [Google Scholar]
- 72.Patel M, Dutta A, Huang H. A selective adenosine sensor derived from a triplex DNA aptamer. Anal. Bioanal. Chem. 2011;400:3035–3040. doi: 10.1007/s00216-011-4996-1. [DOI] [PubMed] [Google Scholar]
- 73.Huang CC, Chiu SH, Huang YF, Chang HT. Aptamer-functionalized gold nanoparticles for turn-on light switch detection of platelet-derived growth factor. Anal. Chem. 2007;79:4798–4804. doi: 10.1021/ac0707075. [DOI] [PubMed] [Google Scholar]
- 74.Chen SJ, Huang CC, Chang HT. Enrichment and fluorescence enhancement of adenosine using aptamer-gold nanoparticles, PDGF aptamer, and Oligreen. Talanta. 2010;81:493–498. doi: 10.1016/j.talanta.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 75.Xiang Y, Wang Z, Xing H, Wong NY, Lu Y. Label-free fluorescent functional DNA sensors using unmodified DNA: a vacant site approach. Anal. Chem. 2010;82:4122–4129. doi: 10.1021/ac100244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu Z, Morita K, Sato Y, Dai Q, Nishizawa S, Teramae N. Label-free aptamer-based sensor using abasic site-containing DNA and a nucleobase-specific fluorescent ligand. Chem. Commun. 2009;42:6445–6447. doi: 10.1039/b908345f. [DOI] [PubMed] [Google Scholar]
- 77.Endoh T, Funabashi H, Mie M, Kobatake E. Method for detection of specific nucleic acids by recombinant protein with fluorescent resonance energy transfer. Anal. Chem. 2005;77:4308–4314. doi: 10.1021/ac048491j. [DOI] [PubMed] [Google Scholar]
- 78.Endoh T, Shintani R, Mie M, Kobatake E, Ohtsuki T, Sisido M. Detection of bioactive small molecules by fluorescent resonance energy transfer (FRET) in RNA-protein conjugates. Bioconjug. Chem. 2009;20:2242–2246. doi: 10.1021/bc9002184. [DOI] [PubMed] [Google Scholar]
- 79.He JL, Wu ZS, Zhou H, Wang HQ, Jiang JH, Shen GL, Yu RQ. Fluorescence aptameric sensor for strand displacement amplification detection of cocaine. Anal. Chem. 2010;82:1358–1364. doi: 10.1021/ac902416u. [DOI] [PubMed] [Google Scholar]
- 80.Sparano BA, Koide K. A strategy for the development of small-molecule-based sensors that strongly fluoresce when bound to a specific RNA. J. Am. Chem. Soc. 2005;127:14954–14955. doi: 10.1021/ja0530319. [DOI] [PubMed] [Google Scholar]
- 81.Sparano BA, Koide K. Fluorescent sensors for specific RNA: a general paradigm using chemistry and combinatorial biology. J. Am. Chem. Soc. 2007;129:4785–4794. doi: 10.1021/ja070111z. [DOI] [PubMed] [Google Scholar]
- 82.Li B, Qin C, Li T, Wang L, Dong S. Fluorescent switch constructed based on hemin-sensitive anionic conjugated polymer and its applications in DNA-related sensors. Anal. Chem. 2009;81:3544–3550. doi: 10.1021/ac900110a. [DOI] [PubMed] [Google Scholar]
- 83.Zhang H, Stockley PG, Zhou D. Development of smart nanoparticle-aptamer sensing technology. Faraday Discuss. 2011;149:319–332. doi: 10.1039/c005373b. [DOI] [PubMed] [Google Scholar]
- 84.Zhang CY, Johnson LW. Single quantum-dot-based aptameric nanosensor for cocaine. Anal. Chem. 2009;81:3051–3055. doi: 10.1021/ac802737b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu Z, Yang C, Zhou X, Qin J. Label-free aptamer-based sensors for L-argininamide by using nucleic acid minor groove binding dyes. Chem. Commun. 2011;47:3192–3194. doi: 10.1039/c0cc04844e. [DOI] [PubMed] [Google Scholar]
- 86.Xu W, Lu Y. Label-free fluorescent aptamer sensor based on regulation of malachite green fluorescence. Anal. Chem. 2010;82:574–578. doi: 10.1021/ac9018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 88.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc. Natl. Acad. Sci. USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 90.Sando S, Narita A, Hayami M, Aoyama Y. Transcription monitoring using fused RNA with a dye-binding light-up aptamer as a tag: a blue fluorescent RNA. Chem. Commun. 2008;33:3858–3860. doi: 10.1039/b808449a. [DOI] [PubMed] [Google Scholar]
- 91.Furutani C, Shinomiya K, Aoyama Y, Yamada K, Sando S. Modular blue fluorescent RNA sensors for label-free detection of target molecules. Mol. Biosyst. 2010;6:1569–1571. doi: 10.1039/c001230k. [DOI] [PubMed] [Google Scholar]
- 92.Shi Y, Huang WT, Luo HQ, Li NB. A label-free DNA reduced graphene oxide-based fluorescent sensor for highly sensitive and selective detection of hemin. Chem. Commun. 2011;47:4676–4678. doi: 10.1039/c0cc05518b. [DOI] [PubMed] [Google Scholar]
- 93.Huang CC, Chang HT. Aptamer-based fluorescence sensor for rapid detection of potassium ions in urine. Chem. Commun. 2008;12:1461–1463. doi: 10.1039/b718752a. [DOI] [PubMed] [Google Scholar]
- 94.Rotaru A, Dutta S, Jentzsch E, Gothelf K, Mokhir A. Selective dsDNA-templated formation of copper nanoparticles in solution. Angew. Chem. Int. Ed. Engl. 2010;49:5665–5667. doi: 10.1002/anie.200907256. [DOI] [PubMed] [Google Scholar]
- 95.Zhou Z, Du Y, Dong S. Double-strand DNA-templated formation of copper nanoparticles as fluorescent probe for label-free aptamer sensor. Anal. Chem. 2011;83:5122–5127. doi: 10.1021/ac200120g. [DOI] [PubMed] [Google Scholar]
- 96.Bogomolova A, Aldissi M. Real-time aptamer quantum dot fluorescent flow sensor. Biosens. Bioelectron. 2011;26:4099–4103. doi: 10.1016/j.bios.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 97.Long F, Gao C, Shi HC, He M, Zhu AN, Klibanov AM, Gu AZ. Reusable evanescent wave DNA biosensor for rapid, highly sensitive, and selective detection of mercury ions. Biosens. Bioelectron. 2011;26:4018–4023. doi: 10.1016/j.bios.2011.03.022. [DOI] [PubMed] [Google Scholar]
- 98.Pei R, Rothman J, Xie Y, Stojanovic MN. Light-up properties of complexes between thiazole orange-small molecule conjugates and aptamers. Nucleic Acids Res. 2009;37:e59. doi: 10.1093/nar/gkp154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cruz-Aguado JA, Penner G. Fluorescence polarization based displacement assay for the determination of small molecules with aptamers. Anal. Chem. 2008;80:8853–8855. doi: 10.1021/ac8017058. [DOI] [PubMed] [Google Scholar]
- 100.Swain MD, Octain J, Benson DE. Unimolecular, soluble semiconductor nanoparticle-based biosensors for thrombin using charge/electron transfer. Bioconjug. Chem. 2008;19:2520–2526. doi: 10.1021/bc8003952. [DOI] [PubMed] [Google Scholar]