Abstract
Objectives
Using a genome-wide association scan and DNA pooling, we previously identified 5 novel genetic susceptibility loci for Behçet’s disease. Herein, we establish the genetic effect within the UBAC2 gene, replicate this genetic association, and identify a functional variant within this locus.
Methods
A total of 676 Behçet’s disease patients and 1,096 controls were studied. The discovery set included 156 patients and 167 controls from Turkey, and the replication sets included 376 patients and 369 controls, and 144 patients and 560 controls, from Turkey and Italy, respectively. Genotyping of 14 SNPs within and around UBAC2 was performed using TaqMan SNP genotyping assays.
Results
The genetic association between Behçet’s disease and UBAC2 was established, replicated and confirmed (Meta-analysis OR= 1.84, meta-analysis P= 1.69X10−7). Haplotype analysis identified both a disease risk and a protective haplotype (P= 0.00014 and 0.0075, respectively). Using conditional haplotype analysis we identified the SNP rs7999348 (A/G) within UBAC2 as the most likely SNP with a genetic effect independent of the haplotypic effect formed by the remaining associated SNPs in this locus. Indeed, we demonstrate that rs7999348 tags a functional variant associated with increased mRNA expression of a UBAC2 transcript variant in PBMCs of individuals homozygous for the Behçet’s disease-associated “G” allele. Further, our data suggest the possibility of multiple genetic effects that increase susceptibility to Behçet’s disease in the UBAC2 locus.
Conclusion
We established and confirmed the genetic association between UBAC2 and Behçet’s disease in three independent sets of patients and controls. We identified the minor allele in rs7999348 as a disease-risk allele that tags altered UBAC2 expression.
Introduction
Behçet’s disease is a systemic inflammatory disease characterized by the presence of recurrent oro-genital ulceration, inflammatory eye disease, central nervous system involvement, skin involvement, and gastrointestinal involvement. Other disease manifestations include arthritis, arterial aneurysms, and recurrent deep venous thrombosis. The disease is most common along the ancient “silk road” route, and thus is most prevalent in East Asia, the Middle East, North Africa, and southern Europe. Both men and women are equally affected; however, younger patients and men tend to have a more severe disease with higher morbidity and mortality (1).
The pathogenesis of Behçet’s disease is poorly understood. Evidence for a genetic contribution to the disease etiology is largely derived from familial aggregation of the disease (2), disease incidence studies in immigrant populations(3), and the universally confirmed association between Behçet’s disease and HLA-B51 which is estimated to account for ~19% of the genetic risk for this disease (4). Evidence for involvement of environmental factors includes the association between poor oral health with Behçet’s disease (5–6). Evidence for possible infectious etiology contributing the Behçet’s disease comes from the high frequency of isolating S. aureus from acne lesions in Behçet’s disease patients compared to acne vulgaris patients (7), the presence of higher titers of anti-mycobaterial heat shock protein antibodies in patients’ sera (8), and the more frequent mouth colonization with S. mutans in patients compared to controls (9).
Recently, we performed a genome-wide association study (GWAS) in a set of Turkish Behçet’s disease patients and controls, and identified 5 novel candidate genetic susceptibility loci for the disease (10). These candidate loci include KIAA1529, CPVL, LOC100129342, UBASH3B, and UBAC2. Two subsequent GWAS studies in Behçet’s disease established and validated the genetic association between IL10 and IL23R and Behçet’s disease (11–12).
In this report, we genotype 14 SNPs in the UBAC2 locus, and identified a functional tag SNP within UBAC2 that increases susceptibility to Behçet’s disease. Further, we confirmed and replicated the genetic association in the UBAC2 enetic locus in a total of three independent sets of patients and controls.
Materials and Methods
Patients and controls
Three independent sets of Behçet’s disease patients and ethnically-matched controls from Turkey and Italy were included in this study. The first set included 156 patients and 167 controls from Turkey, the second set included 376 patients and 369 controls from Turkey, and the third set included 144 patients and 560 controls from Italy. All patients fulfilled the 1990 International Study Group classification criteria for Behçet’s disease (13). The study protocols were approved by the ethics committees and Institutional Review Boards at our institutions. All study participants signed an informed written consent. Buffy coat samples from normal blood donors were obtained from the Oklahoma Blood Institute and used to separate peripheral blood mononuclear cells (PBMCs) to measure UBAC2 transcript levels.
Genotyping and data analysis
Genotyping of single nucleotide polymorphisms (SNP) within and around the UBAC2 gene was performed using TaqMan SNP Genotyping Assays (Applied Biosystems). A total of 14 SNPs were genotyped in this study. The SNPs selected for genotyping represent common genetic variants within the UBAC2 locus that showed a genetic association with Behçet’s disease in our pooled DNA GWAS. Our GWAS included 59 SNPs located in the LD block containing UBAC2 (Supplementary Table 1). 48 of these SNPs, also included in HapMap, capture 86% of variants in this LD block with a mean r2 value of 0.978 in CEU+TSI population. Only individuals with a genotyping success rate of >90% were used for subsequent analysis. All SNPs had a genotyping success rate of >90%. Allele frequencies in patients and controls were determined. A Chi2 test was used to examine genetic association between each of the genotyped SNPs and Behçet’s disease, and odds ratios were determined. Hardy-Weinberg equilibrium (HWE) p values were calculated in controls using Haploview 4.2 (14) and were not significant (>0.05). Haplotype analysis was performed using Haploview 4.2 and PLINK (14–15). Conditional haplotype analysis and meta-analysis were performed using PLINK (15). Each genotyped SNP was individually examined for an independent genetic effect against the haplotypic background formed by all remaining SNPs.
PBMC separation, RNA extraction, and real time RT-PCR
Peripheral blood mononuclear cells (PBMC) were separated from buffy coat samples obtained from normal healthy blood donors using density gradient centrifugation (Amersham Biosciences, Uppsala, Sweden). DNA was extracted using the DNeasy Kit (Qiagen, Valencia, CA), and RNA was extracted using a combination of Trizol (Invitrogen, Carlsbad, CA) and RNeasy kit (Qiagen, Valencia, CA), as previously described (16). RNA was treated with Turbo DNA-free (Ambion, Austin, TX) to digest any contaminating DNA. Real time RT-PCR was performed to measure the relative concentration of UBAC2 transcript variants 1 and 2, with normalization to the housekeeping gene beta-actin, using iScript One-Step RT-PCR Kit With SYBR Green (Bio-Rad, Hercules, CA) and the Rotor-Gene 3000 real-time thermocycler (Corbett Research, Australia). The PCR steps used are as follows: 50°C for 10 minutes, 95°C for 5 minutes, and 45 cycles of 95°C for 10 seconds followed by 55°C for 30 seconds. Primer sequences are: UBAC2 transcript variant 1 forward: 5′ GCTCCAGTGGGCTCTACAAG 3′, reverse: 5′ CCTCCAAATCTGGAAGTCGT 3′; UBAC2 transcript variant 2 forward: 5′ TGCTGGATGTTGCTGTTTTC 3′, reverse: 5′ CAGGCTGGAAGTCGTTCTTG 3′; beta-actin forward: 5′ GCACCACACCTTCTACAATGAGC 3′, reverse: 5′ GGATAGCACAGCCTGGATAGCAAC 3′.
Results
We have previously reported the candidate association between UBAC2 and Behçet’s disease. This association was initially discovered in our GWAS using pooled DNA samples from Behçet’s disease patients and controls, and was later validated with single-sample genotyping in the same set (P=5.8X10−3) (10). Herein, we genotyped 14 SNPs within and around UBAC2 (Figure 1). These experiments were performed using the 156 patients and 167 controls included in our discovery set. The SNPs selected for genotyping are SNPs that showed evidence for genetic association with Behçet’s disease in our GWAS using pooled DNA samples in the UBAC2 locus (Supplementary Table 1).
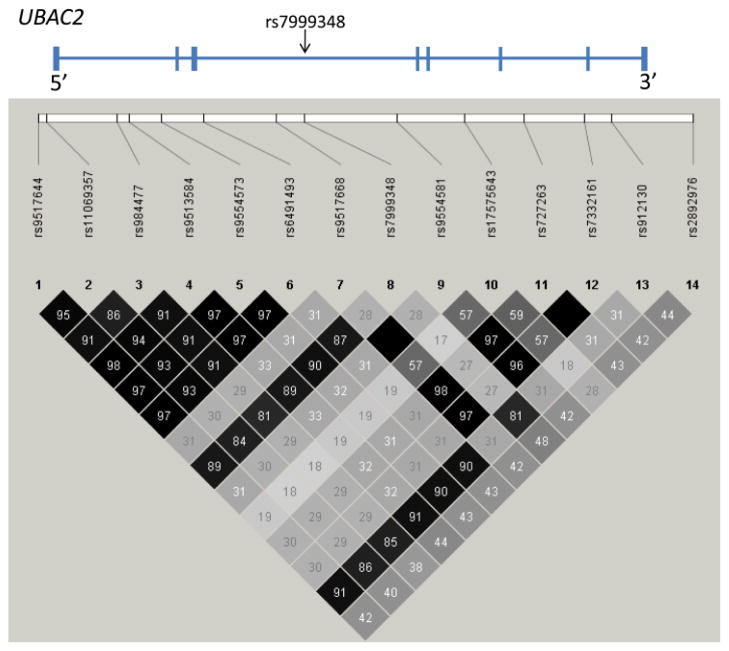
Figure 1.
Linkage disequilibrium (LD) plot showing the 14 SNPs genotyped in the UBAC2 locus and pair-wise correlation coefficient (r2) values. The location of the newly-identified functional SNP rs7999348 is depicted with an arrow.
We show genetic association between all the 14 SNPs genotyped in the UBAC2 locus and Behçet’s disease (Table 1). The SNP rs9517668 located in the third intron within UBAC2 shows the most significant association among the genetic variants tested (odds ratio= 2.62, p= 3.61X10−5).
Table 1.
Genetic association between UBAC2 and Behçet’s disease. Fourteen SNPs in the UBAC2 genetic locus were genotyped in a set of 156 patients and 167 normal healthy controls.
| SNP | Associated Allele | Frequency | OR | 95% Confidence Interval | P value | ||
|---|---|---|---|---|---|---|---|
| Cases | Controls | LL | UL | ||||
| rs9517644 | T | 0.44 | 0.32 | 1.72 | 1.23 | 2.41 | 0.0013 |
| rs11069357 | A | 0.45 | 0.33 | 1.68 | 1.21 | 2.35 | 0.002 |
| rs984477 | G | 0.46 | 0.34 | 1.65 | 1.17 | 2.32 | 0.0043 |
| rs9513584 | G | 0.45 | 0.32 | 1.74 | 1.23 | 2.46 | 0.0017 |
| rs9554573 | A | 0.44 | 0.32 | 1.73 | 1.24 | 2.43 | 0.0012 |
| rs6491493 | G | 0.44 | 0.32 | 1.74 | 1.25 | 2.43 | 0.0011 |
| rs9517668 | T | 0.23 | 0.10 | 2.62 | 1.64 | 4.18 | 3.61E-05 |
| rs7999348 | G | 0.48 | 0.34 | 1.78 | 1.28 | 2.48 | 0.00058 |
| rs9554581 | T | 0.22 | 0.10 | 2.48 | 1.56 | 3.95 | 8.53E-05 |
| rs17575643 | T | 0.15 | 0.06 | 2.91 | 1.63 | 5.20 | 0.00018 |
| rs727263 | A | 0.23 | 0.11 | 2.45 | 1.55 | 3.88 | 1.00E-04 |
| rs7332161 | A | 0.22 | 0.11 | 2.43 | 1.54 | 3.85 | 0.00011 |
| rs912130 | G | 0.44 | 0.33 | 1.58 | 1.13 | 2.21 | 0.0071 |
| rs2892976 | G | 0.36 | 0.23 | 1.96 | 1.37 | 2.80 | 0.00023 |
OR, odds ratio; LL, lower limit; UL, upper limit
The association between rs9517668 within UBAC2 and Behçet’s disease was then replicated and conformed in a second larger independent set consisting of 376 patients and 369 controls from Turkey (odds ratio= 1.82, 95% confidence interval= 1.31–2.53, P= 0.00034). This association was not detected in the Italian set of Behçet’s disease cases and controls (odds ratio= 1.41, 95% confidence interval= 0.91–2.18, P= 0.12). However, a meta-analysis for the association between rs9517668 and Behçet’s disease in the three independent sets included in this study revealed a meta-analysis odds ratio of 1.84 and a P value of 1.69X10−7 (Table 2).
Table 2.
Meta-analysis for Behçet’s disease-associated alleles in rs9517668 and rs7999348 within the UBAC2 gene in three independent sets of Behçet’s disease patients and controls.
| SNP | Associated Allele | Turkish Set 1 | Turkish Set 2 | Italian Set | Meta-analysis | Meta-analysis | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P value | OR | P value | OR | P value | OR | P value | P value | ||
| rs9517668 | T | 2.62 | 3.61E-05 | 1.82 | 0.00034 | 1.41 | 0.12 | 1.84 | 1.69E-07 | 0.21 |
| rs7999348 | G | 1.78 | 0.00058 | 1.29 | 0.023 | 1.40 | 0.018 | 1.39 | 1.85E-05 | 0.49 |
OR, odds ratio
Haplotype analysis using all 14 SNPs genotyped in the discovery set identified 4 common haplotypes (frequency ≥ 5%) including a disease-risk haplotype with a frequency of 16% in patients and 6% in controls (P= 0.00014), and a protective haplotype with a frequency of 50% in patients and 62% in controls (P= 0.0075) (Table 3).
Table 3.
Haplotype analysis using 14 SNPs genotyped in the UBAC2 locus reveals both a disease-risk and a protective haplotype in Behçet’s disease.
| Haplotype | Frequency | P value | |
|---|---|---|---|
| Cases | Controls | ||
| CGCAGCAACCGGTA | 0.50 | 0.62 | 0.0075 |
| TAGGAGAGCCGGGG | 0.10 | 0.09 | 0.52 |
| TAGGAGTGTTAAGG | 0.16 | 0.06 | 0.00014 |
| TAGGAGAGCCGGGA | 0.11 | 0.12 | 0.6 |
Conditional haplotype analysis identified rs7999348 as the most likely SNP out of the 14 genotyped SNPs that might have an independent genetic effect againt the haplotypic effect formed by the remaining associated SNPs in this locus (likelihood ratio= 3.25, P= 0.071) (Supplementary Table 2). Genotype model association tests in rs7999348 suggest an additive model for this genetic effect, as the Cochran-Armitage test P value (P= 0.00078) was lower than the P values for the dominant or recessive models (P= 0.0059 and 0.0053, respectively). The genetic association between rs7999348 and Behçet’s disease was replicated in the second independent set of Turkish patients and controls (odds ratio= 1.29, 95% confidence interval= 1.04–1.60, P value= 0.023), and also in the Italian patients and controls (odds ratio= 1.40, 95% confidence interval=1.06–1.86, P value= 0.018). The results of a meta-analysis for the association with rs7999348 in Behçet’s disease in our three independent sets is presented in Table 2.
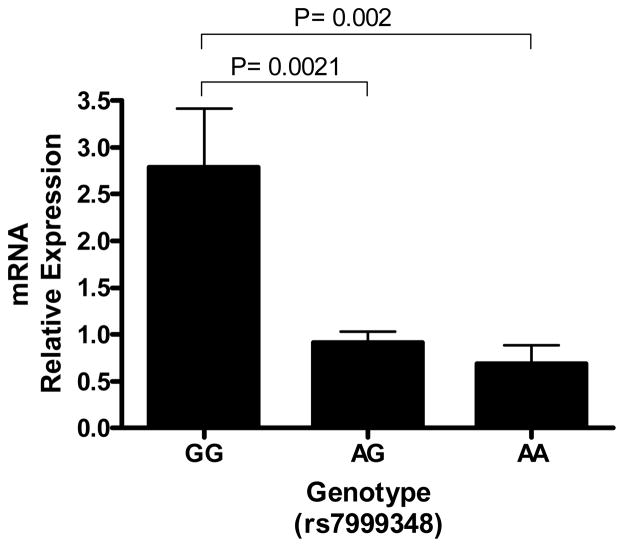
The SNP rs7999348 is an intronal SNP located within the UBAC2 gene. Using SNP function perdition algorithms incorporated within the FASTSNP software (17), it is predicted that rs7999348 changes transcription factor binding, with a small predicted probability of altering an intronic enhancer. Therefore, we hypothesized that disease-causing variants within the UBAC2 gene might alter the expression levels of UBAC2 transcripts. We tested mRNA expression of the two known protein-coding UBAC2 transcript variants in PBMCs collected from normal healthy donors that carry the homozygous risk genotype (GG), the homozygous protective genotype (AA), and the heterozygous genotype (AG) in rs7999348. We find that the expression of UBAC2 transcript variant 1 (NM_001144072.1) is significantly increased in the presence of the homozygous risk genotype in rs7999348 compared to both the heterozygous genotype and the homozygous protective genotype in this SNP (relative mRNA expression (mean ± SEM) is 2.79 ± 0.63 (n=4), 0.91 ± 0.12 (n=8), and 0.69 ± 0.19 (n=8) in GG, AG, and AA respectively (F=14.23, P= 0.0002 by one-way ANOVA) (Figure 2). Using either parametric (t-test) or non-parametric (Mann-Whitney test) analysis, there is significant difference in UBAC2 transcript variant 1 expression in individuals with the homozygous risk genotype compared to individuals with the homozygous protective genotype or heterozygous genotype (GG versus AA: t-test P= 0.002, Mann-Whitney P= 0.004; GG versus AG: t-test P= 0.0021, Mann-Whitney P= 0.004). We did not find a difference in the expression of UBAC2 transcript variant 2 (NM_177967.3) between the various genotypes (relative mRNA expression (mean ± SEM) is 0.95 ± 0.28 (n=4), 1.24 ± 0.81(n=8), and 2.68 ± 0.94 (n=8) in GG, AG, and AA respectively (F=1.13, P= 0.35 by one-way ANOVA). Transcript variant 2, which is shorter than transcript variant 1, is missing two consecutive in-frame coding exons and contains an alternate 5′ end exon leading to a different N-terminus compared to transcript variant 1.
Figure 2.
Relative mRNA expression of UBAC2 transcript variant 1 (NM_001144072.1) in PBMCs of healthy normal donors with homozygous risk (GG) (n= 4), heterozygous (AG) (n= 8), and homozygous protective (AA) (n= 8) rs7999348 genotypes.
To validate allele-specific UBAC2 expression in rs7999348, we used the GENe Expression VARiation (Genevar) expression quantitative trait loci database (18). While rs7999348 was not included in this database, we found a SNP that is in strong LD with rs7999348 (rs2181502, r2=0.88 with rs7999348). This SNP shows allele-specific gene expression profile similar to our findings. The minor allele in rs2181502 (which tags the minor and Behçet’s disease associated allele (G) in rs7999348) is associated with significant overexpression of UBAC2 in lymphoblastoid cell lines (P=0.03) (Supplementary Figure 1).
The most significant genetic effect detected in the UBAC2 locus is with the SNP rs9517668 located within UBAC2 (P= 1.69X10−7). There is relatively low linkage disequilibrium between rs9517668 and rs7999348 that tags a functional variant in UBAC2 (r2=0.28). This suggests that the genetic effect in rs9517668 might be independent of rs7999348. Indeed, a 2-SNP haplotype analysis reveals that the haplotypic genetic association P value for the 2-SNP haplotype formed by rs9517668 and rs7999348 (P= 0.00055) maintains significance independent of (when conditioning on) rs7999348 (P= 0.012). This highlights the possibility that rs9517668 tags a genetic effect that increases the susceptibility to Behçet’s disease independent of rs7999348, and that multiple independent genetic effects for Behçet’s disease might be present in this locus and will need to be further investigated.
Discussion
Behçet’s disease is a multi-systemic inflammatory disease of unclear etiology. The repeatedly confirmed genetic association between the disease and the HLA locus leaves no doubt that genetic factors play an important role in the pathogenesis of the disease. Despite being the only confirmed genetic association in all ethnicities, there is no established mechanistic explanation to date for how HLA-B51 increases the susceptibility for developing Behçet’s disease. Further, a lack of association between disease severity and HLA-B51 has been observed (19). This frustration is equally shared with the well established association between HLA-B27 and ankylosing spondylitis, which is one of the most robust genetic associations in complex diseases known to man, yet with incomplete insight into how this HLA allele predisposes to disease.
It became apparent in Behçet’s disease, as is the case in a number of other rheumatic and inflammatory diseases, that the association with the HLA region does not fully account for the disease genetic risk. Indeed, GWAS studies in the last few years have provided a large number of candidate genes for various immune-mediated diseases, some highlighted novel therapeutic targets, such as the association with IL23R in inflammatory bowel disease (20).
We have previously performed the first GWAS in Behçet’s disease, and identified at least 5 novel candidate susceptibility loci for the disease outside of the HLA region (10). We have also confirmed the known association with HLA-B51 in our sample set (Unpublished data).
Herein, we have established the genetic association in the UBAC2 locus and confirmed this association in two additional sets of patients and controls representing two ethnic groups. Using conditional haplotype analysis, rs7999348 was the most likely SNP among the genotyped SNPs to have a genetic effect independent of the haplotypic effect formed by the remaining associated SNPs in this locus. Indeed, We find that the expression of a UBAC2 transcript variant is significantly increased in individuals with the Behçet’s disease associated homozygous genotype in this variant compared to heterozygotes (P=0.0021) and compared to individuals with the protective genotype (P= 0.002). These findings are also consistent with UBAC2 expression profiles in the an expression quantitative trait loci database. Further, our data suggest that rs9517668 increases the susceptibility to Behçet’s disease independent of rs7999348, raising the possibility of multiple genetic effects in the UBAC2 locus. Re-sequencing will be needed to identify disease causal variants in this locus. Future efforts should also focus on determining the effect of increased UBAC2 expression upon the pathogenesis of Behçet’s disease. While our data support the genetic association between UBAC2 and Behçet’s disease in two Caucasian populations, a similar finding in other ethnicities remains to be determined.
The function of the protein encoded by UBAC2 is not known. However, the presence of a ubiquitin-associated domain in this gene product predicts involvement in ubiquitination pathways. Indeed, UBAC2 mRNA is ubiquitously expressed (BioGPS). Of interest, we have previously reported the genetic association between another ubiquitination-related gene with Behçet’s disease (UBASH3B) (10). The genetic association with a third ubiquitination-related gene (SUMO4) has also been reported in Behçet’s disease (21). These data suggest that ubiquitination defects might be involved in the pathogenesis of Behçet’s disease.
In summary, we establish and replicate a genetic association between Behçet’s disease and UBAC2 gene. Indeed, we identify a functional variant-tagging SNP that increases the risk of Behçet’s disease and that is associated with higher mRNA expression of a common UBAC2 splice variant in PBMCs.
Supplementary Material
Acknowledgments
Funding
This work was supported by the American College of Rheumatology ACR REF Rheumatology Investigator award, and the National Institutes of Health grants numbers R03AI076729, P20RR020143, and P30AR053483.
We would like to thank Dr. Sandra D’Alfonso, Dr. Raffaella Scorza, Dr. Angela Tincani, and Dr. Andrea Doria for providing control samples for our study.
Footnotes
Conflict of Interest Statement
None of the authors has any financial conflict of interest with the work and results presented herein.
References
- 1.Yazici H, Tuzun Y, Pazarli H, Yurdakul S, Ozyazgan Y, Ozdogan H, et al. Influence of age of onset and patient’s sex on the prevalence and severity of manifestations of Behcet’s syndrome. Ann Rheum Dis. 1984;43(6):783–9. doi: 10.1136/ard.43.6.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gul A, Inanc M, Ocal L, Aral O, Konice M. Familial aggregation of Behcet’s disease in Turkey. Ann Rheum Dis. 2000;59(8):622–5. doi: 10.1136/ard.59.8.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mahr A, Belarbi L, Wechsler B, Jeanneret D, Dhote R, Fain O, et al. Population-based prevalence study of Behcet’s disease: differences by ethnic origin and low variation by age at immigration. Arthritis Rheum. 2008;58(12):3951–9. doi: 10.1002/art.24149. [DOI] [PubMed] [Google Scholar]
- 4.Hirohata S, Kikuchi H. Behcet’s disease. Arthritis Res Ther. 2003;5(3):139–46. doi: 10.1186/ar757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akman A, Kacaroglu H, Donmez L, Bacanli A, Alpsoy E. Relationship between periodontal findings and Behcet’s disease: a controlled study. J Clin Periodontol. 2007;34(6):485–91. doi: 10.1111/j.1600-051X.2007.01085.x. [DOI] [PubMed] [Google Scholar]
- 6.Mumcu G, Ergun T, Inanc N, Fresko I, Atalay T, Hayran O, et al. Oral health is impaired in Behcet’s disease and is associated with disease severity. Rheumatology (Oxford) 2004;43(8):1028–33. doi: 10.1093/rheumatology/keh236. [DOI] [PubMed] [Google Scholar]
- 7.Hatemi G, Bahar H, Uysal S, Mat C, Gogus F, Masatlioglu S, et al. The pustular skin lesions in Behcet’s syndrome are not sterile. Ann Rheum Dis. 2004;63(11):1450–2. doi: 10.1136/ard.2003.017467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Direskeneli H, Hasan A, Shinnick T, Mizushima R, van der Zee R, Fortune F, et al. Recognition of B-cell epitopes of the 65 kDa HSP in Behcet’s disease. Scand J Immunol. 1996;43(4):464–71. doi: 10.1046/j.1365-3083.1996.d01-53.x. [DOI] [PubMed] [Google Scholar]
- 9.Mumcu G, Inanc N, Aydin SZ, Ergun T, Direskeneli H. Association of salivary S. mutans colonisation and mannose-binding lectin deficiency with gender in Behcet’s disease. Clin Exp Rheumatol. 2009;27(2 Suppl 53):S32–6. [PubMed] [Google Scholar]
- 10.Fei Y, Webb R, Cobb BL, Direskeneli H, Saruhan-Direskeneli G, Sawalha AH. Identification of novel genetic susceptibility loci for Behcet’s disease using a genome-wide association study. Arthritis Res Ther. 2009;11(3):R66. doi: 10.1186/ar2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Remmers EF, Cosan F, Kirino Y, Ombrello MJ, Abaci N, Satorius C, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behcet’s disease. Nat Genet. 2010;42(8):698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuki N, Meguro A, Ota M, Ohno S, Shiota T, Kawagoe T, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behcet’s disease susceptibility loci. Nat Genet. 2010;42(8):703–6. doi: 10.1038/ng.624. [DOI] [PubMed] [Google Scholar]
- 13.Criteria for diagnosis of Behcet’s disease. International Study Group for Behcet’s Disease. Lancet. 1990;335(8697):1078–80. [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60(4):1076–84. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34(Web Server issue):W635–41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26(19):2474–6. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gul A, Uyar FA, Inanc M, Ocal L, Tugal-Tutkun I, Aral O, et al. Lack of association of HLA-B*51 with a severe disease course in Behcet’s disease. Rheumatology (Oxford) 2001;40(6):668–72. doi: 10.1093/rheumatology/40.6.668. [DOI] [PubMed] [Google Scholar]
- 20.Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314(5804):1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou S, Yang P, Du L, Zhou H, Lin X, Liu X, et al. SUMO4 gene polymorphisms in Chinese Han patients with Behcet’s disease. Clin Immunol. 2008;129(1):170–5. doi: 10.1016/j.clim.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.