Abstract
Coral reefs are among the most productive marine ecosystems and are the source of a large group of structurally unique biosynthetic products. Annual reviews of marine natural products continue to illustrate that the most prolific source of bioactive compounds consist of coral reef invertebrates—sponges, ascidians, mollusks, and bryozoans. This account examines recent milestone developments pertaining to compounds from invertebrates designated as therapeutic leads for biomedical discovery. The focus is on the secondary metabolites, their inspirational structural scaffolds and the possible role of microorganism associants in their biosynthesis. Also important are the increasing concerns regarding the collection of reef invertebrates for the discovery process. The case examples considered here will be useful to insure that future research to unearth bioactive invertebrate-derived compounds will be carried out in a sustainable and environmentally conscious fashion.
Our account begins with some observations pertaining to the natural history of these organisms. Many still believe that a serious obstacle to the ultimate development of a marine natural product isolated from coral reef invertebrates is the problem of compound supply. Recent achievements through total synthesis can now be drawn on to forcefully cast this myth aside. The tools of semisynthesis of complex natural products or insights from SAR efforts to simplify an active pharmacophore are at hand and demand discussion. Equally exciting is the prospect that invertebrate-associated micro-organisms may represent the next frontier to accelerate the development of high priority therapeutic candidates.
Currently in the United States there are two FDA approved marine-derived therapeutic drugs and two others that are often cited as being marine-inspired. This record will be examined first followed by an analysis of a dozen of our favorite examples of coral reef invertebrate natural products having therapeutic potential. The record of using complex scaffolds of marine invertebrate products as the starting point for development will be reviewed by considering eight case examples. The potential promise of developing invertebrate-derived micro-organisms as the starting point for further exploration of therapeutically relevant structures is considered. Also significant is the circumstance that there are some 14 sponge-derived compounds that are available to facilitate fundamental biological investigations.
Keywords: Reef invertebrates, Biosynthetic products, Invertebrate-microbial associations, Bioactive small molecules, Marine-derived bacteria
1. Introduction
1.1. A glimpse into the past
It has taken almost three score years to fully achieve the intent of ocean-based research begun by organic chemists in 1950s. Initially, some of these pioneers sought to understand the small molecule constituents of marine invertebrates and most such efforts involved the isolation and characterization of steroids and toxins. After just a few years, it was also recognized that through further shaping and refining this focus could be accelerated through interdisciplinary meetings. The first such gathering, of what now seems to be a never-ending annual stream, was held in 1960, and included 32 papers focused on the topic Biochemistry and Pharmacology of Compounds Derived from Marine Organisms.1
The actual progress of translating the novel chemical constituents from marine organisms into substances of biomedical importance occurred, from 1950 to 1970, at a very slow pace. Nevertheless, international attention was drawn to this subject area. One measure of this development was that throughout the 1970s there were several strategic publications that appeared as peer-reviewed articles or monographs. One noteworthy example was the 1976 Science article Drugs from the Sea.2 Also during this decade, the emphasis on marine invertebrates, especially sponges, soft corals, and ascidians clearly emerged, in part because they were abundant in near shore habitats and were easy to collect.
In the early 1980s, the tunicate, Trididemnum solidum, derived compound, didemnin B3 was advanced by the NCI to a Phase I anticancer clinical trial.4 Even though the trial was subsequently discontinued this development represented a first for a marine natural product. Today, there are two compounds in clinical use based exactly on marine-derived invertebrate structures—ecteinascidin 743, a.k.a. Yondelis® from an Ascidian (EU approval 2007), and ziconotide, a.k.a. Prialt® from a Cone shell (US approval 2004); and both compounds will be further discussed later in this review. As another important milestone, the National Academy of Sciences published an important white paper ‘Marine Biotechnology in the 21st Century: Problems, Promise, and Products’.5 A central point of this report was ‘the search for new drugs and agrichemical compounds should be revitalized using innovative methods to gain a more fundamental understanding of the biosynthetic capabilities of marine organisms.’ In this review we will show that the collective efforts over many decades have brought us to the doorstep of realizing the promise envisioned in 1950s of using the small molecules from marine-derived invertebrates to benefit humankind.
1.2. Natural history considerations
The well recognized ratio of 2.43 for the marine versus the terrestrial bionetwork is often cited as the stimulus for using Oce-ana as a starting point for marine natural products discovery. Not surprisingly this community of scientists has given a great deal of attention to the three largest of the six major zones defined by oceanographers (% of the world’s relative surface area) which are: Pacific (30.5), Atlantic (15.1), Indian (13.4), Southern (4.0), Arctic (2.8), and all others (2.3).6 The greatest invertebrate biodiversity is found in the oceans, and the Marine Life Project7 estimates the extent of known marine species at around 250,000, which can be extrapolated to a potential of at least a million marine species for chemical exploration. Many groups of invertebrates are exclusively (or almost exclusively) marine and this includes phyla such as Porifera, Coelenterata, Mollusca, Tunicata, and Annelida. These marine derived organisms continue to be rewarding sources of chemodiversity. By contrast, little attention has been given to the study of the natural products of freshwater aquatic zones because of their scant content of invertebrate species. For example, marine sponges represent greater than 95% of the phylum Porifera, as the representation of freshwater specimens is minor.8 Furthermore, the ratio of fresh water habitats versus marine is 1:300.9
Optimism expressed by organic chemists in 1950s that the exploration of Oceana’s invertebrates for novel secondary metabolites would be rewarding has now been realized. A majority of the approximately 30,000 marine natural products isolated from marine organisms and reported in approximately 7000 publications10 are obtained from invertebrates. However, there is still room for new discovery as only a fraction of the invertebrates from the ocean have been explored. Interestingly, the publication record from academic marine bioorganic labs indicates a phenomena we term ‘island-philic.’ This is based on the situation that many studies seem to be initiated by expeditions to habitats adjacent to islands. The Pacific Ocean, a favorite collection spot for the collection of invertebrate organisms, covers a surface area about 15 times the United States and contains about 25,000 islands.
1.3. Some comments on key invertebrate phyla
It is useful to highlight a few features of marine invertebrate organisms from biologically diverse habitats that have been both productive and unproductive as sources of natural products of biomedical importance. A trio of sponges are shown in Figure 1 which illustrate different outcomes. Almost all coral reefs contain massive sponges, such as Xestospongia muta (Fig. 1, panel A1), which can be as large in size as the individual engaged in taking its photo! Unfortunately, none of these huge specimens have been a source of significant biomolecules. Alternatively, specimens of less than 5 cm in diameter have been chemically prolific. One such example is Psammocinia aff. bulbosa (Fig. 1, panel A2), a source of the preclinical candidate psymberin (12). The bright yellow, deep water sponge, Lissondendoryx sp. (Fig. 1, panel A3) has attracted much interest because of its content of halichondrin B (27). However, it was only with great difficulty that more than 1 metric ton of this specimen was collected (dredging) to afford just 300 mg of this important cytotoxin. This forcefully illustrates the difficulty of relying on environmental collections for hit-to-lead development.
Figure 1.
Natural history of coral reef invertebrates with potential as a source of significant metabolites.
The production of secondary metabolites on a multi-mg scale by an ascidian of interest can also be quite variable. This is certainly true for the very photogenic tunicates Didemnum molle (Fig. 1, panel B1), whose symbiotic cyanobacterium, Prochloron didemni, always adds an attractive green hue to the creamy ectosome. Disappointingly, a rather thorough survey of this ascidian from Papua New Guinea and Palau showed that some colonies produce cyclic peptides but many others do not,11 and there does not appear to be a way by visual inspection to determine which colonies will be chemically rich. This circumstance is in contrast to other ascidian-prochloron assemblages which reliably are a source of an array of polypeptide cyanobactin metabolites.12 As another parallel example, a consistent biosynthetic process apparently operates to produce ET-743 (1) from the Atlantic tunicate Ecteinascidia turbinate (Fig. 1, panel B2). Both wild colonies and those maintained in aquaculture farms always produce this anticancer active metabolite, but the overall yields are low.13 Consequently, the timetable for its isolation and characterization was protracted, with the first significant report to stimulate the survey of E. turbinata for bioactive constituents occurring in 196914 and the full structure was only fully described many years later, in 1990.15,16
Nudibranchs are striking in their coloration and in their ability to concentrate bioactive metabolites by sequestering metabolites by dietary transfer.17 Two of our favorites, shown here, consist of the spongivorous Chromodoris lochi (Indo-Pacific) (Figure 1 panel C1) and Chromodoris quadricolor (Red Sea) (Fig. 1, panel C2). Both organisms sequester latrunculins, powerful actin inhibitor polyketide-peptides, produced by two different sponges that are the preferred resting spots for these nudibranchs.
One of the most chemically important bryozoans is Bugula neri-tina (Fig. 1, panel D), the source of bryostatin 1 (65), putatively produced by a type I polyketide synthase (PKS). This common fouling organism, consisting of brown-red tufts, cosmopolitan to warm water habitats in the Atlantic and Pacific oceans,13 has been used as prolific source to supply clinical trial material. There have been more than 80 anticancer clinical trials of bryostatin 1 and current ones have been shifted to focus on combination therapy.4
1.4. Invertebrate-derived secondary metabolites with therapeutic potential
The vivid colors of invertebrates and reef-building corals are among the most conspicuous elements of a living coral reef. The chromic molecules near the surface structure of coral reef creatures range between yellow, green, blue, brown, orange, red, purple and black. Little is known about the identity and role of color in reef-associated organisms.18,19 However, in the past some investigators have speculated that the discovery of bioactive substances ought to begin with examination of highly pigmented invertebrates because of their potential to use chemical defense for protection against other grazing organisms. Some clarifying insights on this point are now evident from the extensive chemical defense investigations of western Antarctic Peninsula sponges. These studies have shown that sponges exhibit chemical defense responses irrespective of the presence or absence of pigments.20
By the 1980’s emphasis began to be placed on adding value to a newly discovered natural product by establishing a relationship between structure, biological activity and its ability to target specific proteins in the human cell. Thus, a number of privileged scaffolds were discovered. Summarized in Table 1 are 14 examples selected from a much longer list4,21–23 of marine invertebrate compounds in clinical, preclinical trials or used for bioassay positive controls. As another enduring theme, when examining the optimum sources for chemical diversity it is clear that natural products occupy a special position and the structures derived from marine organism do not overlap with those amassed from decades of exploring terrestrial plants and insects. Finally, the chemodiversity represented in this short list is immense versus that present in most other organic structures created through synthesis.24
Table 1.
Our 14 favorite examples of invertebrate-derived natural products in clinical use or of therapeutic potential
| Entry | Compound name | Invertebrate source | Biosynthetic class |
Target | Comments |
|---|---|---|---|---|---|
| 1 | Ecteinascidin 743 (1) (ET-743) (Yondelis®) |
Ascidian Ecteinascidia turbinata | Alkaloid | Cancer; clinical use EU |
Enantiopure clinical compound via semisynthesis (see Fig. 1) |
| 2 | Manzamine A (2) | Sponge Haliclona sp.; Pellina; Pachypellina; Xestospongia; Ircinia; Amphimedon |
Alkaloid | Antimalarial; assay positive control |
Isolation from diverse sponges and a marine-derived bacterium providing a way for sustainable supply |
| 3 | Crambescidin 800 (3) |
Sponge Crambe crambe; Monanchora unguifera |
Alkaloid | Antimalarial; Cytotoxic |
Subject of total syntheses |
| 4 | Variolin A (4) | Sponge Kirkpatrickia varialosa (Antarctica) |
Alkaloid | CDK | Awesome structural challenge: core ratio H/C < 1 (C15H13N7O2) |
| 5 | Dehydrodidemnin B (5) (Aplidin®) |
Ascidian Aplidium albicans | Depsipeptide | Phase II cancer | An encouraging analogue replacing didemnin B, dropped from clinical trials (1995) |
| 6 | Discodermolide (6) | Sponge Discodermia dissoluta | Polyketide | Cancer | Advanced to Phase II, but discontinued 2004; total synthesis by A. Smith yielded one gram! |
| 7 | Dictyostatin-1 (7) | Sponge Spongia sp. (Indian Ocean); Corallistidae (Caribbean deep water) |
Polyketide | Cancer | Isolated from two different sponges; potent tubulin agent; active in cells with the P-glycoprotein MDR pump |
| 8 | Bryostatin-1 (8) | Bryozoan Bugula neritina | Polyketide- macrolide |
Phase II Cancer | Scale-up isolation for Phase II via aquaculture (see Fig. 1) |
| 9 | Fijianolide B (9) (Laulimalide) |
Sponge Cacospongia mycofijiensis | Polyketide- macrolide |
Cancer | Many analogues evaluated to develop SAR against micro- tubulin target. In vivo activity shown |
| 10 | Spongistatin 1 (10a) Altohyrtin (10b) Cinachyrolide A (10c) |
Sponge 10a Spongia sp., Spirastrella spinispirulifera; 10b Hyrtios alatum, Haliclona sp.; 10c Cinachyra sp. |
Polyketide- macrolide |
Cancer | Isolated in low yields, 0.003–0.17 mg/kg from five different sponges possessing nM in vitro activity versus cancer cells. Synthesis: 10a = 10b |
| 11 | Aplyronine A (11) | Sea Hare Aplysia kurodai | Polyketide- macrolide |
Antitumor | It inhibits of G-actin polymerization & F-actin depolymerizing. X-Ray reveals interactions within the actin–aplyronine A complex |
| 12 | Psymberin (12) (Irciniastatin A) |
Sponge Ircinia ramosa, Psammocinia aff. bulbosa |
Polyketide- peptide |
Cancer | Many analogues evaluated against solid tumors. Target unknown. |
| 13 | Mycothiazole (13) | Sponge Cacospongia mycofijiensis | Polyketide- peptide |
Mitochondrial complex I inhibitor |
A potent cancer cell cytotoxin with efficacy as a molecular probe for mitochondrial biology and HIF-mediated hypoxic signaling |
| 14 | Pseudopterosin A (14) |
Soft coral Pseudopterogorgia elisabethae |
Diterpene glycoside |
Antiinflammatory; wound healing |
First discovered in 1986, its 25 analogues are of continuing interest |
There are steady streams of annual reviews4,21,25–27 that have provided a chronicle of the ever changing pipeline of small molecules justifiably designated as ‘marine pharmaceuticals.’ A selected list of such invertebrate-derived compounds plus a few others that are of significance are presented in Table 1. These entries can be divided into five biosynthetic structural classes. These frameworks embedded in the compounds of this list include: alkaloid, depsipeptide, polyketide, polyketide-peptide, or terpenoid. A very recent special issue (December 2010)28 was devoted to pharmaceutical biotechnology and it included 16 perspectives on this subject. One other valuable treatise from 2010 summarized the 13 natural products in current phases of clinical development as potential therapeutics against cancer, antiviral, pain, wound healing and schizophrenia.21 While some of these substances are listed in Table 1, the reader should scan these works for comprehensive information, additional perspectives, and the confounding synonyms that exist for these high priority compounds.
One of the most inspirational molecules of Table 1 is ecteinascidin 743 (1) also known as ET-743, Trabectedin, and now marketed as Yondelis®. This alkaloid, comprised of three fused tetrahydro-quinolines linked to a carbinolamine moiety is in current use in the EU to treat soft tissue sarcoma and platinum-sensitive ovarian cancers. The low isolation yield of 1 from E. turbinata (1 mg/Kg) has been replaced by an effective kilogram semisynthesis in 27 steps as shown in Scheme 1, from cyanosafracin B,29 obtained from fermentation of Pseudomonas fluorescens, as the strategy to provide the clinical material.30,31 Also important to this discussion, and alluded to earlier in this review, is that 1 was the first clinically approved anticancer agent ‘based on an actual marine natural product.’32 The reader is directed to a section in our review ‘Setting the Ara A and Ara C story straight’ for a view about the place in history occupied by such arabinose compounds, often designated as being marine-derived drugs.32
Scheme 1.
Semisynthetic route beginning with cyanosafracin B, obtained from fermentation to ecteinascidin 743 (1).
Each of the other 13 compounds listed in Table 1 have been the subjects of numerous fundamental studies. The comments included in Table 1 provide short vignettes for each and will guide the reader to a list of interesting publications for further information. Each structure shown stands as the lead compound for a host of analogues and there are a variety of total syntheses for one or more of their members.33–37 A brief summary to underscore their importance follows next. Extensive networks of fused nitrogen-containing rings are present in the trio of sponge-derived alkaloids, manzamine A (2) (Indo-Pacific), cram-bescidin-800 (3) (Indo-Pacific, and Caribbean), and variolin (4) (Antarctica). The very complex ascidian-derived structure, aplidine (5) [syn: dehydrodidemnin B, Aplidin®] differs from other members of the didemnin series by changes in the tripeptide B side chain. Synthetic material of 5 is being used in the Phase II trials for cancers such as pancreatic, colorectal, and lung. In 2003, 5 was approved in the EU as an orphan drug for the treatment of acute lymphoblastic leukemia. The six polyketides shown in Table 1 consist of two acyclics, discodermolide (6) and dictyostatin-1 (7), and four macrolides, bryostatin 1 (8), fijianolide B (laulimalide) (9), spongiastatin 1 (10a) ≈ altohyrtin (10b) ≈ cinachryolide A (10c), and aplyronine A (11). Each of these compounds can be considered a relevant target for further modification by synthetic biology approaches, because this technology, to harness polyketide synthesis for heterologous expression, is beginning to mature.38 The two compounds of mixed biogenesis, psymberin (12), and mycothiazole (13) continue to be the subjects of extensive pre-clinical investigation.39–43 The diterpene-glycosides of the pseudopterosin (14) class are of significance because of their potent anti-inflammatory properties.44 A pseudopterosin containing extract45 is employed in the cosmetic cream product marketed as Resilence by Estée Lauder. Two other analogues of 14, the C-10 O-methyl ether and the isomer having the l-fucose at C-9 are being investigated as agents for wound healing.46,47
1.5. The re-supply challenges and the role of analogues— insights from marine micro-organisms
Although the recent experiences at Nereus Pharmaceuticals in developing two marine natural product-inspired anticancer drug candidates do not involve invertebrate products, they provide a benchmark illustration of analogue selection to maximize beneficial properties. Lessons can be learned about effective strategies used by Nereus to provide kilograms of material for anticancer therapeutic development.48 Not unexpectedly, hundreds of analogues were investigated during the early stage work for both marizomib (NPI-0052), and plinabulin (NPI-2358).
The first compound, also known as salinosporamide A and originally isolated from a salt obligate actinomycete, Salinispora tropica, is a proteosome inhibitor currently in Phase I evaluation. The activity differences for salinosporamide A49,50 versus omuralide51 partially illustrate the important role of the chlorethyl group as well as other functional groups that were investigated by the evaluation of analogues obtained by synthesis and fermentation. Ultimately, salinosporamide was chosen for development and was initially supplied through saline fermentation, with the fine-tuned industrial scale 1500 L GMP fermentation achieving a yield of 240 mg/L. More recently, a large-scale fermentation of an engineered strain has been developed that can be grown in deionized water-based media.52
The second compound, plinabulin (NPI-2358), is a unique tubulin inhibitor obtained by total synthesis. It consists of a framework inspired by the structure of (−)-phenylahistin (halimide) isolated from the culture of an Aspergillus sp. derived from a Halimeda lacrimosa (Caribbean alga). Part of the pharmacophore redesign to define this clinical candidate involved the preparation of some 110 analogues resulting in simplifying the side chains attached to the diketopiperazine core.53–55 An update on clinical trials with plinbulin, which has shown a favorable safety profile accompanied by positive therapeutic effects and its use in combination with other anticancer drugs has been published.56
1.6. The re-supply challenges and the role of analogues—a significant milestone based on an entire complex scaffold
The vignettes discussed above illustrate that the difficulties of supply can be overcome once a natural product lead compound has been identified. There is one other classic case to be cited that involves the development of (+)-discodermolide (6), a polyketide tubulin inhibitor, isolated in very low yields (20 mg/kg frozen sponge) from a difficult to collect deepwater sponge, Discodermia dissoluta. This complex heterocycle (C33H55O8N) is chiral-rich (13 asymmetric centers and 3-E/Z double bonds) and has been enantiosynthesized in large amounts, firstly in gram57 and finally in kilogram amounts.58 The transition of discodermolide into a Phase I study (begun in 2004 but discontinued in 2004) was stimulated due to its action against taxol-resistant cells, its enhanced water solubility and the availability of the compound through GMP synthesis. Alternatively, efforts by Kosan Biosciences to develop a cultivation route by biosynthetic engineering techniques were unsuccessful.
1.7. The re-supply challenges and the role of analogues—achievements through synthesis
The view that it is impossible to effectively re-supply a marine invertebrate-derived structure for further development can now be effectively set aside. The nine examples in Table 2, whose structures possess multiple chiral centers, proved ample support for this view. For each, the problem of supply was resolved either by total synthesis or design of a reduced-complexity pharmacophore obtained by synthesis. The naturally occurring polypeptide toxin, comprised of 25 amino acids, marketed as Prialt® (19) is also at the top of the list of significant marine-derived drugs that further illustrate these principles. The drug, produced by synthesis is identical to the snail-derived natural product, but the literature of its development can be tricky to follow because the list of synonyms consists of omega-conotoxin MVIIA, SNX-111, ziconotide, or ziconotide acetate. It is clinically used in the US to manage pain in patients as a continuous infusion, and in the EU as a non-opiod chronic pain management agent. Interestingly, two biopharmaceutical groups market this compound—US: Elan Corporation; UK and Germany: Eisai, in a cooperative agreement with Elan Corporation of Ireland.
Table 2.
Nine proof-of-concept examples involving comparison of synthetic candidates for potential therapeutic development or in clinical use vs. the parent structures isolated from coral reef invertebrates
| Entry | Synthetic analogue | Natural product frame |
Organism source | Biosynthetic class |
Target | Comment |
|---|---|---|---|---|---|---|
| 1 | GTS-21 (15) (DMBXA) | Anabaseine (16) | Worm Amphiporus angulatus | Alkaloid | Alzheimer and schizophrenia: phase I clinical trial |
Under development by US NCI |
| 2 | Yondelis® (1) (Trabectedin) |
Ecteinascidin 743 (1) (ET-743) |
Ascidian Ecteinascidia turbinata |
Alkaloid | Cancer: phase II clinical trial | Under development by PharmaMar |
| 3 | Zalypsis (17) | Jorumycin (18) | Mollusk Jorunna funebris | Alkaloid | Cancer: phase II clinical trial | Under development by PharmaMar |
| 4 | Prialt® (19) (Ziconotide) | ω-Conotoxin (19) (MVIIA) |
Snail Conus magus | Polypeptide | Analgesic: in clinical use | Elan Pharm. |
| 5 | Soblidotin (20) (TZT- 1027) |
Dolastatin 10 (21) | Mollusk Dolabella auricularia | Polypeptide | Cancer: phase I clinical trial | Possibly marinemicrobe derived |
| 6 | E7974 (22) | Hemiasterlin (23) (Milnamide B) |
Sponge, Hemiasterella minor Auletta cf. constricta |
Tripeptide | Cancer: phase I clinical trial | Under development by Eisai |
| 7 | Irvalec® (24) (Elisidepsin, PM02743) |
Kahalalide F (25) | Mollusk Elysia rufescens; Alga Bryopsis sp. |
Desipeptide | Cancer: phase II clinical trial | Under development by PharmaMar |
| 8 | Halaven® (26) (Eribulin mesylate, E7389) |
Halichondrin B (27) | Sponge Lissodendoryx sp. | Polyketide macrolide |
Cancer: clinical use US | US FDA approved November 2010; Eisai |
| 9 | LAF-389 (28) | Bengamide B (29) | Sponge Jaspis cf. coriacea | Polyketide peptide | Cancer: phase I clinical trail | Novartis; discontinued 2002 |
The remaining compounds shown in Table 2 are synthetics advanced to clinical evaluation which are divided into two categories: (a) scaffolds advanced that were nearly identical to the natural product, and (b) reduced complexity analogues. While most of these compounds have been the subject of previous comprehensive reviews there are some important new perspectives to discuss. Three compounds, Yondelis® (1), Irvalec® (24) and LAF-389 (29) fall into the first category. Irvalec® (24) is an unnatural salt of isokaha-lalide F, a natural product that co-isolated with kahalalide F.59,60 Extensive evaluations of the SAR of 24 and its diastereomers revealed that two of the amino acids in the original structures were misassigned61 which clarified the bioactive scaffold for further clinical evaluation.62 Evaluation of almost 100 bengamide B (29) analogues revealed that only its extremities could be modified giving rise to 28 which was prepared in kilogram quantities. A remarkable illustration of the second circumstance pertains to eribulin mesylate (E7389) 26 marketed as Halavan® (Eisai) that was US FDA approved in November 2010 for treatment of metastatic breast cancer unresponsive to other drug treatments.63,64 The elegant coupled synthesis—cytotoxicity evaluation revealed that the entire western portion of 27 could be stripped away while retaining a positive therapeutic effect. Side-by-side inspection of the remaining examples of Table 1 shows parallel functional outcomes based on pharmacophore manipulation for selected alkaloids: GTS-21 (15) versus anabaseine (16), and Zalypsis® (17) versus jorumycin (18); and for selected polypeptides: soblidotin (20) versus dolastatin 10 (21); and E7974 (22) versus hemiasterlin (23).
1.8. Demystifying the major microbial associants of three invertebrates
It is widely known that many marine invertebrates, especially sponges, host a rich array of associated micro-organisms, some of which are true symbionts.65 Through the use of 16S ribosomal DNA (rDNA) library construction, the phylogeny of complex microbial consortia of marine invertebrates is now beginning to come to light.66 It has been demonstrated that marine sponges, which pump seawater through their tissues at rates up to 1 L/cc tissue/h provide a rich environment to house diverse microbial populations including archaea, eubacteria (including cyanobacteria), microalgae, fungal spores, and protozoa.67 A more detailed phylogenetic analysis is shown in Figure 2.
Figure 2.
Marine sponge associated micro-organisms as potential candidates for producing secondary metabolites. (Adapted from Ref. 76)
The microbial diversity associated with bryozoans and ascidians is somewhat less well-studied in comparison to that of sponges. However, one survey of North Sea bryozoan samples revealed a phylogenetically mixed bacterial population including γ- and α-proteobacteria and Gram-positive bacteria with both low and high GC content,68 all common inhabitants of the marine environment. A phylogenetic analysis of the intracellular bacteria associated with the ascidian Ecteinascidia turbinata indicated the presence of more than 30 bacterial species, with three predominant strains—one of which is Candidatus Endoecteinascidia fru-mentensis, a γ-proteobacterium that appears to be a true symbiont while the other two frequently occurring are Mollicutes, novel ‘Spiroplasma-like’ bacteria.69
1.9. The possible role of micro-organisms in producing secondary metabolites
Marine derived micro-organisms continue to be implicated in the production of certain invertebrate natural products. We agree with the 2007 view expressed by Newman and Craig,70 ‘we wish to draw the attention of readers to the rapidly evolving recognition that a significant number of natural product drugs/leads are actually produced by microbes and/or microbial interactions with the ‘host from whence it was isolated, and therefore we consider that this area of natural product research should be expanded significantly.’ The compilation of Table 3 adds to the record by underscoring 21 compounds fully or putatively illustrating this situation. As a further point, from a conservation viewpoint we also agree with Kingston48 ‘that these macroorganisms need to be preserved and valued as the source of novel lead compounds.’ An important rational is that in the future, as new tools and strategies are created, it may be possible to engage in the effective culture of such biosynthetically rich micro-organisms. An illustration of the potential of this paradigm is the development of manzamine A (2) which putatively can be produced by the culture of Micromonsopora M42.71
Table 3.
Examples of invertebrate-derived versus micro-organism products with identical or parallel structures
| Entry | Sponge metabolite | Sponge source | Micro-organism metabolite | Micro-organism source |
|---|---|---|---|---|
| 1 | Andrimid (30) | Hyatella sp. | Andrimid (30) | Vibrio sp.b |
| 2 | Arenastatin (31) | Dysidea arenaria | Arenastatin (31)(Cryptophycin-24) Cryptophycin-1 (32) |
Nostoc sp.d |
| 3 | Bengamide E (33) | Jaspis cf. coriacea | Bengamide E (33), E′ (34) | Myxococcus virescensb |
| 4 | Discodermide (35) | Discodermia dissoluta | Alteramide Ae (36) Ikarugamycin (37) |
Alteromonas sp.a,e Streptomyces sp.a |
| 5 | Herbacic acid (38) | Dysidea herbacea | Barbamide (39) | Lyngbya majusculad |
| 6 | Jasplakinolide (40) |
Jaspis splendens Auletta sp. Hemiasterella minor |
Chondramide D (41) | Chondromyces crocatusb |
| 7 | Latrunculin A (42) |
Cacospongia mycofijiensis Negombata magnifica |
Epothilone B (43) | Sorangium cellulsoumb |
| 8 | Makaluvamine A (44) | Zyzzya cf. marsalis | Makaluvamine A (44) | Didymium bahiensec |
| 9 | Manzamine A (2) | Haliclona sp. | Manzamine A (2) | Micromonospora M42a |
| 10 | Mimosamycin (45) | Petrosia sp. | Mimosamycin (45) | Streptomyces lavendulaea |
| 11 | Motuporin (46) (Nodularin-V) | Theonella swinhoei | Nodularin (47) |
Microcystis aerugionsad Planktothrix agardhiid |
| 12 | Mycalamide A (48) | Mycale hentscheli | Pederin (49) | Undescribed endosymbiont of Paederus (beetle) |
| 13 | Psymberin (12) (Irciniastatin A) |
Psammocinia sp. Ircinia sp. |
Unknown | Pseudomonas sp.a |
| 14 | Okadaic acid (50) |
Halichonbdria okadai H. melandocia |
Okadiac acid (50) | Procentrum limai,d P. concavum,d Dinophysis sppd |
| 15 | Renieramycin E (51) | Reniera sp. | Saframycin Mx1 (52) Saframycin A (53) |
Myxococus xanthusb Pseudomonas fluorescensb |
| 16 | Salicylihalamide A (54) | Haliclona sp. | Apicularen A (55) | Chondromyces sp.b |
| 17 | Sphinxolide (56) | Neosiphonia superstes | Rhizopodin (57) | Myxococus stipitusb |
| 18 | Swinholide A (58) | Theonella swinhoei | Swinholide A (58) | Symploca sp.d |
| Tunicate metabolite | Tunicate source | Micro-organism metabolite | Micro-organism source | |
| 19 | Iejimalide A (59) | Eudistoma cf. rigida | Archazolid B (60) | Archangium gephyra,b Cystobacter sp.b |
| 20 | Namenamicin (61) |
Polysyncraton lithostrotum Didemnum proliferum |
Esperamicin A1 (62) | Actinomadura verrucosospora a |
| 21 | Patellamide A (63) | Lissoclinum patella | Patellamide A (63) | Prochloron didemnid |
| Bryozoan metabolite | Bryozoan source | Micro-organism metabolite | Micro-organism source | |
| 22 | Bryostatin 1 (8) | Bugula neritina | Bryostatin 1f (8) | Candidatus Endobugula sertulaa |
Gram-positive bacterium.
Gram-negative bacterium.
Slime mold.
Cyanobacterium.
Obtained from culturing a bacterium isolated from the sponge Halichondria okadai.
Recent studies have shown B. neritina appears to have the genetic machinery to biosynthesize bryostatins.
A growing collection of circumstantial evidence suggests that there are varied biosynthetic categories of marine natural products assembled through the action of sponge–micro-organism interactions. The 18 examples shown in Table 3 are presented to explore this point since they are all based on parallel or identical structures isolated from sponges versus those from micro-organisms. We also note that somewhat similar information has appeared in a recent encyclopedic, but uncritical review examining natural products from sponge symbionts.72 Although sponges feed on micro-organisms, many of them host phylogenetically diverse populations of microbes.73 These micro-organisms are located mainly extracellularly in the sponge mesohyl, sometimes contributing as much as 40–60% of the sponge’s biomass.74 Such sponges are therefore termed high-microbial-abundance (HMA) sponges, in contrast to low-microbial-abundance (LMA) sponges.75 Recent discoveries by marine ecologists have shown that sponges engage in ‘inter-generational transmission’ of microbes.76 Culture-independent methods are now recognized as being powerful tools to identify sponge symbiotic marine bacterial communities and this will be discussed later in this review. It is important to note that comparative genetic studies on the basis of either the 16S rDNA77 of sponges reveal a considerable degree of coevolution between host and some symbionts and that there is little species overlap between planktonic bacteria and those within sponge tissues.78 It is also anticipated that as clone-free sequencing, such as pyrosequencing,79 continues to evolve and become affordable this will allow new opportunities. In particular this may facilitate investigations into the microbial diversity and population dynamics of host-associated microorganisms, which could offer a huge reservoir of new and useful information to examine differences in population structure.
We further note that the putative role of bacteria in producing the various compounds observed from invertebrates is also richly illustrated in the compilation of Table 3. These entries are divided into four categories according to the possible micro-organism source ranging from: (a) Gram-positive bacterium, (b) Gram-negative bacterium, (c) cyanobacterium, and (d) slime mold. Significantly, there are ten compounds in Table 3 which have been isolated from both an invertebrate and an associated micro-organism pair as follows: Gram-positive—manzamine A (2), mimosamycin (45) bryostatin 1 (8); Gram-negative—andrimid (30), bengamide E (33); Cyanos—arenastatin (31), okadaic acid, (50), swinholide A (58), and patellamide A (63); and Slime Mold —makaluvamine A (44). These parallels are powerful and the reader is urged to examine the original publications/patents describing these findings.
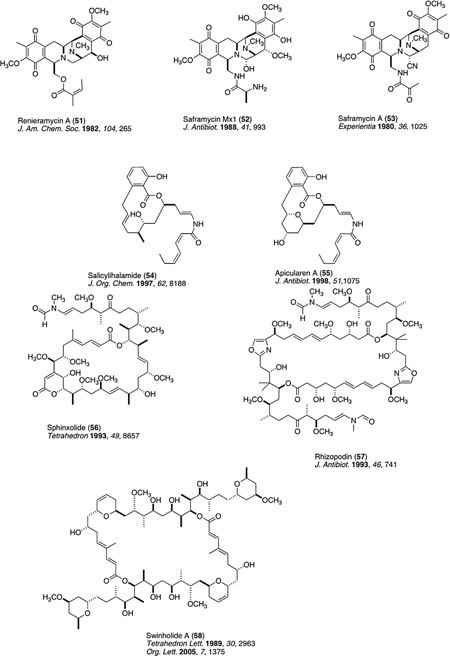
There are parallel constitutions that exist for an additional set of 22 structures shown in Table 3. These are intriguing because parallel biosynthetic networks may be turned on in disparate organisms to create similar chirality and/or functionality located in very specific regions within the carbon framework of the compounds isolated from a corresponding set of invertebrate— micro-organisms. The easiest parallel to visualize are those involving: (i) herbacic acid (38)—barbamide (39); (ii) jasplakinolide (40)—chondramide D (41); (iii) motuporin (46)—nodularin (47); (iv) mycalamide (48)/psymberin (12)—pederin (49); (v) renieramycin E (51)—saframycin Mx1 (52)/saframycin A (53); (vi) salicylihalamide A (54)—apicularen A (55); and (vii) sphinoxolide A (56)—rhizopodin (57). Finally, there are more distant parallels in an additional set of structures. These are as follows: (viii) latrunculin A (42)—epothilone B (43); (ix) iejimalide A (59)—archazolid B (60); and (x) nemenamicin (61)—esperamicin A1 (62).
1.10. Some important next steps
In our view, a clear pattern is emerging intimating that microbial symbionts are important producers of marine derived-natural products. The obvious challenge is to transform this understanding into workable technology that can reproducibly yield secondary metabolites of therapeutic importance. Work on cultured bacteria has also contributed to our understanding of the sponge-microbe association. So far, however, attempts to cultivate bacterial producers have been mostly limited to α- and γ-proteobacteria80,81 and actinobacteria,82 although strains in other bacterial phyla, such as Planctomycetes, Verrucomicrobia, and δ-Proteobacteria, have been cultured from sponges as well. However, none of these studies were able to link the chemistry of the sponge to the cultures of the symbiont, with the exception of manzamine A, mentioned earlier obtained from the culture of Micromonospora M42. The other two exceptions include two terrestrial bacteria found to produce similar or identical metabolites isolated from sponges and these include Myxococcus virescens, Chondromyces crocatus, and Didymium bahiense. In summary, none of the other micro-organisms listed in Table 3 can be either cultured or cultured in such a way to produce either the sponge metabolites or its close analogues thereof. This problem has been recognized for several years,83,84 but in collaborative projects involving microbial ecology, molecular genetics, genomics, and natural products chemistry no breakthroughs of general utility have been published to date.
1.11. The emerging use of sponge-derived compounds as chemical probes
A number of suppliers of fine chemicals have recently recognized the value of employing marine sponge-derived natural products as tools for biological research. The rationale is that the complexity and diversity of natural products enables them to selectively target macromolecules of biological importance. A recent review summarizes a number of examples based on terrestrial natural products while also providing an important historical per-spective.85 The importance of using small molecules as chemical probes and their value as seeds for further investigations are now firmly appreciated.
The fourteen molecules summarized in Table 4 can all be considered as possessing ‘privileged scaffolds.’ All of these compounds are especially useful because knowledge is in hand describing their molecular targets. There has been widespread speculation that they may have evolved to tightly interact with large biomolecules. An important early discovery in this category was an account appearing in 199686 showing that synthetic samples of trapoxin, a fungal metabolite, target Histone Deacetylases (HDACs). Actually, preceding these classic findings by more than a decade were those pertaining to describing the molecular targets of the sponge metabolite latrunculin A (42).
Table 4.
A selected summary of sponge-derived commercially available products widely used as cell biology molecular probes or bioassay positive controls
| Entry | Sponge metabolite | Biosyn. class | Biological target | Commercial source | CAS # |
|---|---|---|---|---|---|
| 1 | Jasplakinolide (40) | PK-NRP | Actin | Invitrogen | 102396-24-7 |
| 2 | Latrunculin A (42) | PK-NRP | Actin | Invitrogen | 76343-93-6 |
| 3 | Swinholide A (58) | PK | Actin | Enzo Life Sciences | 95927-67-6 |
| 4 | Fascaplysin (64) | ALK | CDK4, DNA-intercol. | Sigma Aldrich | 114719-57-2 |
| 5 | Ilimaquinone (65) | PK | Golgi | Calbiochem | 71678-03-0 |
| 6 | Psammaplin A | NRP | HDAC, DNA MT | Sigma Aldrich | 110659-91-1 |
| 7 | Fijianolide B (9) (Laulimalide) |
PK | Microtubules | Pac Mar Bioactives | 115268-43-4 |
| 8 | Mycothiazole (13) | PK-NRP | Mitochondrial complex I | Pac Mar Bioactives | 114582-75-1 |
| 9 | Aaptamine (66) | ALK | Monoaminooxidase | Enzo Life Sciences | 85547-22-4 |
| 10 | Heteronemin (67) | TERP | NFκB | Sigma Aldrich | 62008-04-2 |
| 11 | Okadaic Acid (52) | PK | Protein phosphatase | Santa Cruz Biotech | 78111-17-8 |
| 12 | Mycalamide A (49) | PK | Protein synthesis | Pac Mar Bioactives | 115185-92-7 |
| 13 | Pateamine (68) | PK | Translation initiation | Pac Mar Bioactives | 139220-18-1 |
| 14 | Manzamine A (2) | ALK | GSK-3B | Santa Cruz Biotech | 104196-68-1 |
ALK = alkaloid, NRP = nonribosomal peptide, PK = polyketide, TERP = terpene.
A somewhat convoluted pathway was indeed associated with identifying the molecular target of latrunculin A (42). It began with an observation, in the early 1970s, that squeezings from the Red Sea sponge, Negombata magnifica and the Indo-Pacific sponge Caco-spongia mycofijiensis (see Table 3) were ichthyotoxic.87 Subsequently, the active constituent was identified as latrunculin A, and its molecular structure was described in 1983.88 Simultaneously, results from in vitro experiments were also published revealing 42 rapidly disrupts mammalian actin microfilament organization.89 Additional investigations by many other labs have revealed a host of other biological effects for 42 such as modulating cell shape, disrupting of meiosis fertilization, effecting early cell development;90 disrupting protein kinase C signaling,91 1:1 binding of 42 with monomeric actin, and rapid blocking of actin polymerization either in vitro or in cells.92
There were two additional key developments which occurred that propelled latrunculin A (42) into the limelight as a chemical probe. First, in 1997, the ability of latrunculin A to cause complete disruption of the yeast actin cytoskeleton in less than five minutes was disclosed.93 Second, during that same year, latrunculin A became commercially available. To date, there have been more than a thousand publications (more than 95 in 2010 alone) describing the use of latrunculin A as a research tool. The plot of Figure 3 shows the steady upward trend in its use in cell biology or in experimental therapeutics studies. Similar intense interest continues to be shown in three other compounds, jasplakinolide (40), fijianolide B (9), and okadaic acid (50), as they were subjects of more than 219 papers published in 2010.
Figure 3.
Histogram of peer-review publications on the latrunculin class isolated from marine sponges covering 1983–2010.
In summary, we believe that the marine natural products shown in Table 4 extend the general conclusion in the Carlson review. He wrote ‘… natural products have seen great success as therapeutic agents. However, this vast pool of compounds holds much promise beyond the development of future drugs. These features also make them ideal tools for the study of biological systems.’
1.12. Future prospects
Enormous structural diversity is represented in all of the marine-invertebrate derived bioactive molecules discussed in this review. It is evident that future investigations on unexplored taxa will have the potential to greatly expand the entries in all of the graphics presented in this review. There is optimism for the future because the international marine bioorganic community clearly recognizes that invertebrates must be harvested and studied in an environmentally sustainable manner. Up-and-coming efforts ranging from aquaculture of robust and chemically important invertebrates to expanding current inventories of biodiversity for future study must be further encouraged. These efforts require a broad array of expertise and must involve a collaborative approach.
Fortunately, US funding mechanisms have greatly stimulated past research discussed in this review. Examples of especially important programs began with the NCI marine organisms collection contracts begun in 1980s which created a vast National repository of invertebrate derived compounds and extracts from specimens rigorously identified by taxonomic experts. Another important model has been the Academic-Industry alliances created through NIH National Cooperative Natural Product Drug Discovery Group programs (unfortunately discontinued in 2010). A very different collaborative network has been created through the multi-agency supported International Cooperative Biodiversity Group programs which have emphasized drug discovery alongside biodiversity conservation and stimulating ‘host country’ scientific infrastructure and their economies. It is vital that these initiatives be continued. They continue to serve as an integral tool to promote biodiversity assessment and mining to uncover novel bioactive molecules with potential applications in human and fundamental studies.
There are other important issues deserving of comment. First, almost no progress has been made in the last decade in culturing potentially chemically prolific micro-organism associants of sponges. This area is ripe for collaborative teams to engage in future pioneering efforts employing marine microbiology, natural history insights, and knowledge of biosynthetic pathways (see Table 3). Second, the insights from culture-independent data shown in Figure 2 could also provide stimulus for future innovative culture efforts. Third, the new generation of relatively small biophar-maceutic organizations, such as Eisai, PharmaMar and Nereus, has been nimble and innovative in setting aside past dogma that has tended to derail the development of clinical candidates based on marine natural products. We look forward to the next generation of marine invertebrate-derived biosynthetic products and believe many will be derived from focused investigation of invertebrate associants.
Acknowledgments
The authors thank the NIH Project Research Grant R01 CA047135 (P.C. lab) and the US Civilian Research and Development Foundation IDBI-21003-JA-08 for the support received in P.C. and R.G.L. labs. O.K.R. thanks the Directorate General of Higher Education, Indonesian Ministry of National Education through the Program Academic Recharging (PAR)-C No. 1696/D4.4/2010 for the research fellowship received.
References
- 1.Ann. N.Y. Acad. Sci. 1960;90:617. doi: 10.1111/j.1749-6632.1960.tb26409.x. [DOI] [PubMed] [Google Scholar]
- 2.Ruggieri GD. Science. 1976;194:491. doi: 10.1126/science.9691. [DOI] [PubMed] [Google Scholar]
- 3.Rinehart K, Gloer JB, Cook JC. J. Am. Chem. Soc. 1981;103:1857. [Google Scholar]
- 4.Newman DJ, Cragg GM. J. Nat. Prod. 2004;67:1216. doi: 10.1021/np040031y. [DOI] [PubMed] [Google Scholar]
- 5.National Research Council. Marine Biotechnology in the Twenty-first Century: Problems, Promise, and Products. Washington, DC: National Academy Press; 2002. [PubMed] [Google Scholar]
- 6.Pidwirny M, Ocean JE, Duffy . In: Encyclopedia of Earth, Environmental Information Coalition. Cleveland J, editor. Washington, D.C: National Council for Science and the Environment; 2010. (Topic Ed.) [Google Scholar]
- 7. [accessed April 28, 2011]; http://www.coml.org/
- 8.Hooper JNA. In: Systema Porifera: A guide to the classification of sponges. van Soest RWM, editor. New York: Plenum; 2002. [Google Scholar]
- 9.Sverdrup HU, Johnson MW, Fleming RH. The Oceans, Their Physics, Chemistry, and General Biology. New York: Prentice-Hall; 1942. [Google Scholar]
- 10. [accessed April 28, 2011]; dmnp.chemnetbase.com.
- 11.Donia MS, Fricke WF, Ravel J, Schmidt EW. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017897. Published Online 2011 March 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donia MS, Ravel J, Schmidt EW. Nat. Chem. Biol. 2008;4:341. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendola D. Biomol. Eng. 2003;20:441. doi: 10.1016/s1389-0344(03)00075-3. [DOI] [PubMed] [Google Scholar]
- 14.Sigel MM, Wellham LL, Lichter W, Dudeck LE, Gargus J, Lucas AH. In: Food-Drugs from the Sea Proceedings. Younghen HW Jr, editor. Washington, DC: Marine Technology Society; 1969. pp. 281–294. [Google Scholar]
- 15.Rinehart KL, Holt TG, Fregeau NL, Stroh JG, Keifer PA, Sun F, Li LH, Martin DG. J. Org. Chem. 1990;55:4512. [Google Scholar]
- 16.Wright AE, Forleo DA, Gunawardana GP, Gunasekera SP, Koehn FE, McConnell OJ. J. Org. Chem. 1990;55:4508. [Google Scholar]
- 17.Dumdei EJ, Flowers AE, Garson MJ, Moore CJ. Compd. Biochem. Physiol. A. 1997;118:1385. [Google Scholar]
- 18.Dove SG, Lovell C, Fine M, Deckenback J, Hoegh-Guldberg O, Iglesias-Prieto R, Anthony KRN. Plant Cell Environ. 2008;31:1523. doi: 10.1111/j.1365-3040.2008.01852.x. [DOI] [PubMed] [Google Scholar]
- 19.Dove SG, Takabayashi M, Hoegh-Guldberg O. Biol. Bull. 1995;189:288. doi: 10.2307/1542146. [DOI] [PubMed] [Google Scholar]
- 20.McClintock JB, Amsler CD, Baker BJ. Integ. Comp. Biol. 2010;50:967. doi: 10.1093/icb/icq035. [DOI] [PubMed] [Google Scholar]
- 21.Mayer AMS, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. Trends Pharmacol. Sci. 2010;31:255. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Salomon CE, Magarvey NA, Sherman DH. Nat. Prod. Rep. 2004;21:105. doi: 10.1039/b301384g. [DOI] [PubMed] [Google Scholar]
- 23.Sashidhara KV, White KN, Crews P. J. Nat. Prod. 2009;72:588. doi: 10.1021/np800817y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lipkus AH, Yuan Q, Lucas KA, Funk SA, Bartelt WF, III, Schenck RJ, Trippe AJ. J. Org. Chem. 2008;73:4443. doi: 10.1021/jo8001276. [DOI] [PubMed] [Google Scholar]
- 25.Fenical W. Oceanography. 2006;19(2):110. [Google Scholar]
- 26.Molinski TF, Dalisay DS, Lievens SL, Saludes JP. Nat. Rev. Drug Disc. 2009;8:69. doi: 10.1038/nrd2487. [DOI] [PubMed] [Google Scholar]
- 27.Hughes CC, Fenical W. Chem. Eur. J. 2010;16:12512. doi: 10.1002/chem.201001279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curr. Opin. Biotech. 2010;21(Special issue):711. [Google Scholar]
- 29.Tsuji N. JP 59225189. Japanese Patent. 1985
- 30.Cuevas C, Francesch A. Nat. Prod. Rep. 2009;26:322. doi: 10.1039/b808331m. [DOI] [PubMed] [Google Scholar]
- 31.Cuevas C, Pérez M, Martín MJ, Chicharro JL, Fernández-Rivas C, Flores M, Francesch A, Gallego P, Zarzuelo M, de la Calle F, García J, Polanco C, Rodríguez I, Manzanares I. Org. Lett. 2000;2:2545. doi: 10.1021/ol0062502. [DOI] [PubMed] [Google Scholar]
- 32.Carrol J, Crews P. Macromarines: A Selective Account of the Potential of Marine Sponges* Molluscs* Soft Corals and Tunicates as a Source of Therapeutically Important Molecular Structures. Chapter 6. In: Buss AD, Butler MS, editors. Natural Product Chemistry for Drug Discovery. Cambridge: Royal Society of Chemistry; 2010. p. 174. [Google Scholar]
- 33.Winkler JD, Axten JM. J. Am. Chem. Soc. 1998;120:6425. doi: 10.1021/ja981303k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toma T, Kita Y, Fukuyama T. J. Am. Chem. Soc. 2010;132:10233. doi: 10.1021/ja103721s. [DOI] [PubMed] [Google Scholar]
- 35.Coffey DS, McDonald AI, Overman LE, Rabinowitz MH, Renhowe PA. J. Am. Chem. Soc. 2000;122:4893. [Google Scholar]
- 36.Ahaidar A, Fernández D, Danelón G, Cuevas C, Manzanares I, Albericio F, Joule JA, Álvarez M. J. Org. Chem. 2003;68:10020. doi: 10.1021/jo035332b. [DOI] [PubMed] [Google Scholar]
- 37.Anderson RJ, Hill JB, Morris JC. J. Org. Chem. 2005;70:6204. doi: 10.1021/jo050523v. [DOI] [PubMed] [Google Scholar]
- 38.Fujii I. Nat. Prod. Rep. 2009;26:155. doi: 10.1039/b817092b. [DOI] [PubMed] [Google Scholar]
- 39.Cichewicz RH, Valeriote FA, Crews P. Org. Lett. 2004;6:1951. doi: 10.1021/ol049503q. [DOI] [PubMed] [Google Scholar]
- 40.Robinson SJ, Tenney K, Yee DF, Martinez L, Media JE, Valeriote FA, van Soest RWM, Crews P. J. Nat. Prod. 2007;70:1002. doi: 10.1021/np070171i. [DOI] [PubMed] [Google Scholar]
- 41.Crews P, Kakou Y, Quinoa E. J. Am. Chem. Soc. 1988;110:4365. [Google Scholar]
- 42.Sonnenschein RN, Johnson TA, Tenney K, Valeriote FA, Crews P. J. Nat. Prod. 2006;69:145. doi: 10.1021/np0503597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morgan JB, Mahdi F, Liu Y, Coothankandaswamy V, Jekabsons MB, Johnson TA, Sashidhara KV, Crews P, Nagle DG, Zhou Y-D. Bioorg. Med. Chem. 2010;18:5988. doi: 10.1016/j.bmc.2010.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Look SA, Fenical W, Jacobs RS, Clardy J. Proc. Natl. Acad. Sci. 1986;83:6238. doi: 10.1073/pnas.83.17.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerr RG. 6046041. US Patent. 2000
- 46.Hoarau C, Day D, Moya C, Wu G, Hackim A, Jacobs RS, Little RD. Tetrahedron Lett. 2008;49:4604. [Google Scholar]
- 47.Haimes HB, Jimenez PA. 5597808. US Patent. 1997
- 48.Kingston DG. J. Nat. Prod. 2011;74:496. doi: 10.1021/np100550t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feeling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem., Int. Ed. 2003;42:355. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]
- 50.Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Bioorg. Med. Chem. 2009;17:2175. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corey EJ, Li WD. Chem. Pharm. Bull. 1999;47:1. doi: 10.1248/cpb.47.1. [DOI] [PubMed] [Google Scholar]
- 52.Macherla VR, Mitchell SS, Manam RR, Reed KA, Chao T-H, Nicholson B, Deyanat-Yazdi G, Mai B, Jensen PR, Fenical WF, Neuteboom STC, Lam KS, Palladino MA, Potts BCM. J. Med. Chem. 2005;48:3684. doi: 10.1021/jm048995+. [DOI] [PubMed] [Google Scholar]
- 53.Sumikura M, Yamazaki Y, Yoshida T, Mori Y, Yasui H, Kiso Y, Neuteboom S, Potts B, Lloyd GK, Hayashi Y. Pept. Sci. 2010;46:315. [Google Scholar]
- 54.Hayashi Y, Nicholson B, Tanaka K, Oda A, Lloyd GK, Akamatsu M, Palladino MA, Kiso Y. Pept. Sci. 2005;41:405. [Google Scholar]
- 55.Palladino M, Lloyd GK, Hayashi Y. WO 2007035841, A1 20070329. PCT Int. Appl. 2007
- 56.Mita MM, Spear MA, Yee LK, Mita AC, Heath EI, Papadopoulos KP, Federico KC, Reich SD, Romero O, Malburg L, Pilat M, Lloyd GK, Neuteboom STC, Cropp G, Ashton E, LoRusso PM. Clin. Cancer Res. 2010;16:5892. doi: 10.1158/1078-0432.CCR-10-1096. [DOI] [PubMed] [Google Scholar]
- 57.Smith AB, III, Beauchamp TJ, LaMarche MJ, Kaufman MD, Qiu Y, Arimoto H, Jones DR, Kobayashi K. J. Am. Chem. Soc. 2000;122:8654. [Google Scholar]
- 58.Mickel SJ. Pure Appl. Chem. 2007;79:685. [Google Scholar]
- 59.Gao J, Caballero-George C, Wang B, Rao KV, Shilabin AG, Hamann MT. J. Nat. Prod. 2009;72:2172. doi: 10.1021/np900287e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamann MT, Scheuer PJ. J. Am. Chem. Soc. 1993;115:5825. [Google Scholar]
- 61.Bonnard I, Manzanares I, Rinehart KL. J. Nat Prod. 2003;66:1466. doi: 10.1021/np030334c. [DOI] [PubMed] [Google Scholar]
- 62.Ling Y-H, Aracil M, Jimeno J, Perez-Soler R, Zou Y. Eur. J. Cancer. 2009;45:1855. doi: 10.1016/j.ejca.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Landers P, Wall Street J. [accessed April 28, 2011]; http://online.wsj.com/article/SB10001424052748704111504576059772498413328.html.
- 64. [accessed April 28, 2011]; http://www.eisai.com/news/news201064.html.
- 65.Hochmuth T, Niederkrüger H, Gernert C, Siegl A, Taudien S, Platzer M, Crews P, Hentschel U, Piel J. ChemBioChem. 2010;11:2572. doi: 10.1002/cbic.201000510. [DOI] [PubMed] [Google Scholar]
- 66.Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, Horn M, Wagner M. Environ. Microbiol. 2010;12:2070. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Osinga R, Armstrong E, Burgess JG, Hoffmann F, Reitner J, Schumann-Kindel G. Hydrobiologia. 2001;461:55. [Google Scholar]
- 68.Pukall R, Kramer I, Rohde M, Stackebrandt E. Syst. Appl. Microbiol. 2001;24:623. doi: 10.1078/0723-2020-00073. [DOI] [PubMed] [Google Scholar]
- 69.Moss C, Green DH, Pérez B, Velasco A, Henríquez R, McKenzie JD. Mar. Biol. 2003;143:99. [Google Scholar]
- 70.Newman DJ, Cragg GM. J. Nat. Prod. 2007;70:461. doi: 10.1021/np068054v. [DOI] [PubMed] [Google Scholar]
- 71.Peraud O. Ph.D. Thesis. College Park: University of Maryland; 2006. Isolation and characterization of a sponge-associated actinomycete that produces manzamines, Dissertations & Theses: A&I Publication No. AAT 3241474. [Google Scholar]
- 72.Thomas TRA, Kavlekar DP, LokaBharathi PA. Mar. Drugs. 2010;8:1417. doi: 10.3390/md8041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U. Appl. Environ. Microbiol. 2008;74:7694. doi: 10.1128/AEM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, Moore BS. Appl. Environ. Microbiol. 2002;68:4431. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Horn M, Hacker J. Microbial Diversity of Marine Sponges. In: Müller WEG, editor. Molecular Marine Biology of Sponges. Heidelberg, Germany: Springer; 2003. p. 59. [DOI] [PubMed] [Google Scholar]
- 76.Taylor MW, Radax R, Steger D, Wagner M. Microbiol. Mol. Biol. Rev. 2007;71:295. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thacker RW, Starnes S. Mar. Biol. 2003;142:643. [Google Scholar]
- 78.Schmitt S, Weisz JB, Lindquist N, Hentschel U. Appl. Environ. Microbiol. 2007;73:2067. doi: 10.1128/AEM.01944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huber JA, Welch DBM, Morrison HG, Huse SM, Neal PR, Butterfield DA, Sogin ML. Science. 2007;318:97. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- 80.Hill RA. Annu. Rep. Prog. Chem. 2006;102:123. [Google Scholar]
- 81.Scheuermayer M, Pimentel-Elardo SM, Fieseler L, Grozdanov L, Hentschel U. Micro-organisms of Sponges: Phylogenetic Diversity and Biotechnological Potential. In: Proksch P, Müller WEG, editors. Frontiers in Marine Biotechnology. London: Horizon Scientific Press; 2006. p. 289. [Google Scholar]
- 82.Abdelmohsen UR, Pimentel-Elardo SM, Hanora A, Radwan M, Abou-El-Ela SH, Ahmed S, Hentschel U. Mar. Drugs. 2010;8:399. doi: 10.3390/md8030399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hildebrand M, Waggoner LE, Liu H, Sudek S, Allen S, Anderson C, Sherman DH, Haygood M. Chem. Biol. 2004;11:1543. doi: 10.1016/j.chembiol.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 84.Piel J. Curr. Med. Chem. 2006;13:39. [PubMed] [Google Scholar]
- 85.Carlson EE. ACS Chem. Biol. 2010;5:639. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taunton J, Collins JL, Schreiber SL. J. Am. Chem. Soc. 1996;118:10412. [Google Scholar]
- 87.Neeman I, Fishelson L, Kashman Y. Mar. Biol. 1975;30:293. [Google Scholar]
- 88.Groweiss A, Shmueli U, Kashman Y. J. Org. Chem. 1983;48:3512. [Google Scholar]
- 89.Spector I, Shochet NR, Kashman Y, Groweiss A. Science. 1983;219:493. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 90.Schatten G, Schatten H, Spector I, Cline C, Paweletz N, Simerly C, Petzelt C. Exp. Cell. Res. 1986;166:191. doi: 10.1016/0014-4827(86)90519-7. [DOI] [PubMed] [Google Scholar]
- 91.Niggli V, Djafarzadeh S, Keller HU. Exp. Cell Res. 1999;250:558. doi: 10.1006/excr.1999.4548. [DOI] [PubMed] [Google Scholar]
- 92.Morton WM, Ayscough KR, McLaughlin PJ. Nat. Cell Biol. 2000;2:376. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 93.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin D. J. Cell Biol. 1997;137:399. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]