Abstract
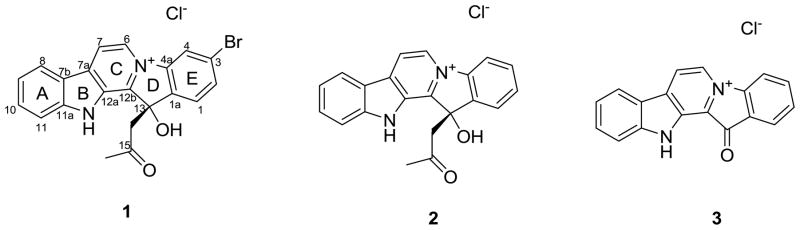
A new fascaplysin analogue, 3-bromohomofascaplysin A (1), along with two known analogues, homofascaplysin A (2) and fascaplysin (3), were isolated from a Fijian Didemnum sp. ascidian. The absolute configurations of 3-bromohomofascaplysin A (1) and homofascaplysin A (2) were determined via experimental and theoretically calculated ECD spectra. The differential activities of 1–3 against different blood-borne life stages of the malaria pathogen Plasmodium falciparum were assessed. Homofascaplysin A (2) displayed an IC50 of 0.55 ± 0.11 nM against ring stage parasites and 105 ± 38 nM against all live parasites. Given the stronger resistance of ring stage parasites against most current antimalarials relative to the other blood stages, homofascaplysin A (2) represents a promising agent for treatment of drug resistant malaria.
Keywords: Fascaplysin analogues, ascidian, antimalarial, absolute configuration, experimental and theoretically calculated ECD
1. Introduction
Fascaplysin (3), originally isolated from a marine sponge Fascaplysinopsis sp.,1 displays a variety of biological activities, such as selective CDK 4 inhibition,2,3 DNA binding,4 and both in vitro and in vivo anti-angiogenic effects.5 We revisited the ascidian, Didemnum sp., that we previously studied6 and discovered a new related compound, 3-bromohomofascaplysin A (1), along with homofascaplysin A (2)7,8 and fascaplysin (3).1 The genus Didemnum is known to contain structurally diverse biologically active compounds such as cyclic peptides with characteristic modified amino acids,9 pyridoacridines,10 β-carbolines,11 lamellarin,12 3,4-substituted maleimides,13 and polysulfide alkaloids.14 Herein we report the isolation and structure elucidation of the new compound 1, the determination of the absolute configuration of 1 and 2 by electronic circular dichroism (ECD) methods, and the differential activities of 1–3 against blood-borne life stages of the malaria pathogen Plasmodium falciparum.
2. Results and Discussion
Compound 1 was isolated as an amorphous pale yellow-brown solid. The positive ESIMS of 1 showed a molecular ion cluster at m/z 407/409 (1:1) M+ diagnostic for a monobrominated molecule. The molecular formula was determined as C21H16N2O2Br by HRESIMS. The 1H NMR spectrum (Table 1) displayed one methyl at δH 1.99, one methylene at δH 4.26/4.16 and nine aromatic protons at δH 9.26, 8.77, 8.54, 8.43, 7.80, 7.82 (2H), 7.83, and 7.49. The 13C NMR spectrum (Table 1) revealed 21 carbons including one carbonyl carbon, 17 aromatic carbons, one oxygenated quaternary carbon, one methylene, and one methyl carbon.
Table 1.
NMR data for compound 1 (1H 500 MHz, 13C 125 MHz, δ ppm) in methanol-d4
| position | δC | δH mult. (J in Hz) | 1H-1H COSY | HMBC |
|---|---|---|---|---|
| 1 | 127.5 CH | 7.80 m | H-2 | C-3, C-4a, C-13 |
| 1a | 138.0 C | |||
| 2 | 135.0 CH | 7.82 m | C-1a, C-4 | |
| 3 | 125.8 C | |||
| 4 | 119.2 CH | 8.54 d (1.2) | H-2 | C-4a, C-1a, C-2, C-3 |
| 4a | 144.1 C | |||
| 6 | 125.3 CH | 9.26 d (6.8) | H-7 | C-12b, C-4a, C-7a, C-7 |
| 7 | 118.7 CH | 8.77 d (6.8) | C-12a, C-7b, C-6, C-12b | |
| 7a | 137.5 C | |||
| 7b | 121.9 C | |||
| 8 | 124.8 CH | 8.43 d (8.4) | H-9, H-10 | C-11a, C-10, C-7b |
| 9 | 124.0 CH | 7.49 dd (8.4, 7.6) | H-10 | C-10, C-7b, C-11 |
| 10 | 134.6 CH | 7.83 m | H-11 | C-11a, C-8, C-9 |
| 11 | 114.5 CH | 7.82 m | C-11a, C-9 | |
| 11a | 147.5 C | |||
| 12a | 132.4 C | |||
| 12b | 145.0 C | |||
| 13 | 79.6 C | |||
| 14 | 51.6 CH2 | 4.16 d (18.4) | C-15, C-12b, C-1a, C-13 | |
| 15 | 206.7 C | 4.26 d (18.4) | ||
| 16 | 30.4 CH3 | 1.99 s | C-15, C-14, C-13 |
The 1H NMR data were similar to those reported for homofascaplysin A (2),7 the major difference being the loss of an aromatic proton signal in 1 compared to 2. Comparison of the molecular formulae of 1 and 2 showed that 1 was the bromo-analogue of 2. Inspection of the 1H NMR spectrum of 1 revealed the presence of one meta-coupled aromatic proton (δH 8.54, J = 1.2 Hz), indicating the Br was positioned at either C-2, C-3, C-9 or C-10. Comparison of the 13C NMR data of 1 with 2 indicated significant shifts of the E-ring carbons, suggesting attachment of the Br atom at C-2 or C-3. The ROESY experiment showed a correlation between H-6 and the meta-coupled proton allowing assignment of this signal to H-4 and hence attachment of the Br atom at C-3. HMBC data provided unambiguous evidence confirming the structural assignment.
The identity of the known compounds homofascaplysin A (2) and fascaplysin (3) were readily confirmed by comparison of their spectroscopic properties with literature data.1,7,8
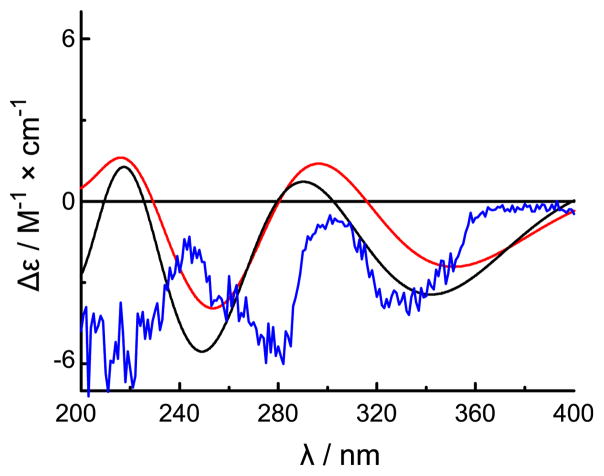
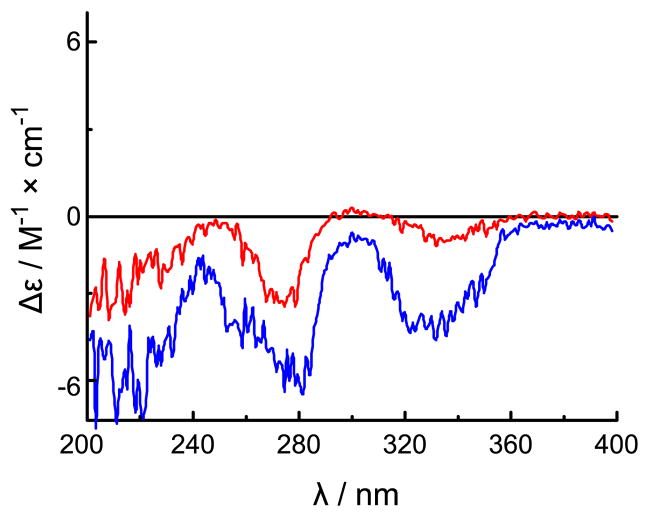
The absolute configuration of 2 was assigned by comparison of the experimental ECD spectrum with simulated spectra calculated by time-dependent density functional theory (TDDFT).15 The model was arbitrarily assigned a configuration of 13S. The potential energy surface was scanned in the gas phase at the AM1 level by rotating about the C-14-C-13 and C-15-C-14 bonds (Figure S-1). Of the six conformers that were located, only three (2a-2c) were relocated at the B3LYP/6-31G** level in the gas phase with conformational distributions of 20.5%, 0.4% and 79.1%, respectively, as calculated from electronic and zero-point energies (Figure S-2 and Table S-1). The distributions for 2a–2c shifted to 41.8%, 2.2% and 56.0%, respectively, at the B3LYP-SCRF/6-31G**//B3LYP/6-31G** level in MeOH with COSMO model, indicating that solvent effects are significant for this cationic system. The prominent differences between the optimized geometries are the presence of (C-15)O···H-O(C-13) hydrogen bonding in 2a and both (C-15)O···H-O(C-13) and (C15)O···H-N-12 hydrogen bonding in 2c (Figure S-2 and Table S-3). ECD spectra of individual conformers have been calculated at the B3LYP/6-31G** level in the gas phase and at the B3LYP-SCRF/6-31G**//B3LYP/6-31G** level in MeOH with the COSMO model (Figure S-3). The weighted average ECD spectra of conformers 2a–2c in both the gas phase and MeOH are depicted in Figure 1 alongside the experimental ECD spectrum for 2. Experimentally observed negative Cotton effects at 334 and 275 nm correlate well with predicted electronic transitions at 345 and 272 nm for conformer 2a and at 345 nm and 262 nm for conformer 2c, respectively, calculated in MeOH at the B3LYP-SCRF/6-31G**//B3LYP/6-31G** level (Figure S-3). Thus, an absolute configuration of 13S was unambiguously determined for 2. The absolute configuration of 1 was also assigned as 13S based on the similarity of its CD spectrum to that of 2 (Figure 2). Such assignment was additionally supported by the similar specific rotations of compounds 1 and 2 (−9.0° and −9.38°,7 respectively).
Figure 1.
Weighted ECD spectra of compound 2 in the gas phase at the B3LYP/6-31G** level (red) and in MeOH at the B3LYP-SCRF/6-31G**//B3LYP/6-31G** level with the COSMO Model in MeOH (black) and its experimental ECD spectrum in MeOH (blue).
Figure 2.
The experimental CD spectra of compounds 1 (red) and 2 (blue) in MeOH.
A previous study had shown that homofascaplysin A (2) and fascaplysin (3) each exhibited activity with IC50 values of 38 nM and 160 nM,8 respectively, against the chloroquine- and pyrimethamine-resistant Plasmodium falciparum strain K1. In order to further probe the potential for use of fascaplysins against resistant P. falciparum, the blood-borne life stage-specific activities16 of compounds 1–3 against the highly resistant strain W2-Mef were determined. The results are shown in Table 2. Compounds 1 and 2 displayed overall IC50 values comparable to the previously reported values against the strain K1.16 Moreover, each of 1–3 exhibited greater potency against the ring stage parasites compared to the trophozoite and schizont stages. Of the three compounds, 1 was the least potent and 3 the most potent overall. However, 2 exhibited the strongest selectivity for ring stage parasites with a stage-specific IC50 value of 0.55 nM, representing a 450-fold and 170-fold selectivity for ring stage over trophozoite and schizont stages, respectively. Since ring-stage parasites are typically two-fold less sensitive to many first line antimalarials such as chloroquine,16 the fascaplysins and particularly 2 represent promising lead compounds for the treatment of resistant malaria.
Table 2.
Life stage-specific IC50 valuesa of 1–3 against Plasmodium falciparum strain W2-Mef
| Cmpd. | All live parasites | Rings | Trophozoites | Schizonts |
|---|---|---|---|---|
| 1 | 805 ± 50 | 574 ± 27 | 1189 ± 34 | 765 ± 81 |
| 2 | 105 ± 38 | 0.55 ± 0.11 | 252.18 ± 0.08 | 94 ± 90 |
| 3 | 48.2 ± 3.3 | 7.82 ± 0.13 | 401 ± 65 | 65.2 ± 9.8 |
| chloroquine | 149 ± 17 | 174 ± 32 | 162 ± 12 | 80 ± 2.8 |
| artemisinin | 6.245 ± 0.07 | 5.92 ± 0.17 | 6.46 ± 0.20 | 5.91 ± 0.27 |
IC50 values given in nM.
The fascaplysins bear strong structural similarities to the cryptolepines, another indole-containing family of antimalarial compounds.17 Like fascaplysin, cryptolepine has DNA intercalating activity.18 Cryptolepine has additionally been shown to inhibit formation of hemozoin in P. falciparum,17 though synthetic studies with other cryptolepine analogues have suggested that other mechanisms are responsible for their activity.19 It is interesting to note that bromination of cryptolepines generally increased potency19 whereas for the fascaplysins, 1 is considerably less potent than 2. In the earlier study describing antimalarial activities for 2 and 3,8 cytotoxicity against skeletal rat muscle myoblast cells (L6) showed the same trends as antimalarial activity with 2 (1.1 μg/mL) being ~2-fold more potent than 3 (2.5 μg/mL), though the MIC values were at concentrations 50–100-fold higher than those needed for activity against P. falciparum. Interestingly, CDKs have been implicated as a promising target in P. falciparum20 and kinase inhibitors have exhibited modest selective inhibition of ring stage parasites.16 Thus, further elucidation of the relative contributions of the CDK 4 inhibitory, DNA intercalating, and hemozoin inhibitory activities to the remarkable selectivity of fascaplysin for ring stage over trophozoite and schizont stages holds promise for discovering effective treatments of malaria.
3. Experimental Section
3.1 General Experimental Procedures
Optical rotations were measured on a Jasco DIP-370 polarimeter. UV spectra were acquired in spectroscopy grade MeOH using a Hewlett-Packard 8452A diode array spectrophotometer. IR spectra were recorded on a JASCO FT/IR-420 spectrophotometer. NMR data of 1 were collected using a Varian INOVA 500 (1H 500 MHz, 13C 125 MHz) NMR spectrometer with a 3 mm Nalorac MDBG probe with a z-axis gradient and utilized residual solvent signals for referencing (δH 3.30, δC 49.00 for methanol-d4). NMR data for 2 and 3 were obtained using a Varian Mercury 400 (1H 400 MHz, 13C 100 MHz) instrument equipped with a 5 mm Nalorac probe and utilized residual solvent signals for referencing as for compound 1. High-resolution mass spectra (HRMS) were obtained using a Micromass Q-ToF micro. Analytical and semipreparative HPLC were accomplished utilizing a Beckman System Gold 126 solvent module equipped with a 168 PDA detector. All reagents were purchased and used without additional purification.
3.2 Biological Material
The Didemnum sp. ascidian was collected by SCUBA from Pratt Reef, Fiji Islands.6
3.3 Extraction and Isolation
This study started with material partially purified during the isolation of bengacarboline and fascaplysin.6 A red fraction obtained from prior LH-20 column chromatography was subjected to HPLC on a Phenomenex Luna C18 column (250 × 10 mm) employing a gradient of 80:20 to 50:50 H2O-MeCN (0.1% TFA) at 4 mL/min over 20 min to yield compound 1 (2.0 mg, tR= 18.7 min), 2 (8.0 mg, tR= 15.0 min), and 3 (11.0 mg, tR= 13.4 min).
3.3.1 3-Bromohomofascaplysin A (1)
amorphous pale yellow-brown solid; [α]20D −9 (c 0.1, MeOH); UV (MeOH) λmax (log ε) 404 (3.36), 336 (3.75), 270 (3.85), 232 (3.80), 212 (3.90) nm; IR (film) νmax 3102, 2928, 2853, 1714, 1651, 1623, 1521, 1471, 1434, 1336, 1205, 1068, 1029, 952, 841, 802, 779, 751, 724, 645, 622,597,518 cm−1; 1H and 13C NMR Table 1; HRESIMS m/z 409.0358 [M+H]+ (calcd for C21H16N2O281Br, 409.0375; Δ −4 ppm).
3.4 Computational Chemistry
All calculations were performed using the Gaussian03 program. The potential energy surface of compound 2 in the gas phase was scanned at the AM1 level by rotating about the C-14-C-13 and C-15-C-14 bonds. Six conformers were located, of which three were relocated by full optimization at the B3LYP/6-31G** level in the gas phase. The geometries of the ground states were then used to calculate the ECD spectra by using TDDFT at the B3LYP/6-31G** level in the gas phase and at the B3LYP-SCRF/6-31G**//B3LYP/6-31G** level in MeOH with the COSMO model. The calculated excitation energies ΔEi (in nm) and rotatory strength (Ri) were used to compute simulated ECD curves by using the Gaussian function:
where σ is the width of the band at height 1/e and i represents an index over all transitions. In the current work, a value of σ = 0.25 eV and rotatory strength in the dipole length form (Rlen) were used. The detailed calculation data are shown in the Supporting Information.
3.5 Detection of Life Stage-Specific Inhibition of Plasmodium falciparum
The flow cytometric analysis of malaria parasite growth was carried out as previously described.16 In brief, asynchronous cultures of the chloroquine- and mefloquine-resistant P. falciparum strainW2-Mef (MRA-615 deposited to ATCC/MR4 by A. F. Cowman) were maintained at a 5% hematocrit in RPMI-1640 based malaria culture media. To determine the IC50 value, parasites were exposed to test drugs which were diluted in ½ log10 steps from 10 μM down to 0.33 nM. Cultures were then grown in triplicate for 48 hr. Cells were then stained with Hoescht 33342 (DNA), thiazole orange (RNA), DiIC1-5 (membrane potential of live cells), and propidium iodide (membrane integrity) for 40 min. The combination of the DNA and RNA levels allowed for the determination of parasite erythrocytic life cycle stages (rings, trophozoites, or schizonts).21 Calculation of IC50 values were carried out as described previously16 by comparing the averaged number of parasites found in each concentration of drug across replicates and divided by the average number of corresponding parasites in control wells grown in the absence of any drug. To calculate IC50 values, a non-linear sigmoid dose-response curve for variable slope was fitted to the data using GraphPad Prism version 5.00 for Windows (San Diego, CA).
Supplementary Material
Scheme.
Acknowledgments
We thank the Mississippi Center for Supercomputing Research (MCSR) for computational facilities. Funding for the Varian INOVA 500 MHz NMR spectrometer was provided through NIH grant RR06262. Flow cytometry based analysis of parasite growth was provided for by the NIH AI079388, the International Society for Advancement of Cytometry Scholars Program, and the CWRU Vision Fund (B.T.G.) in conjunction with the CWRU Division of Infectious Diseases and HIV Medicine, and Metrohealth Hospital (D.R.D). This work was supported by NIH grant CA36622 (C.M.I.) and the USDA Agricultural Research Service Specific Cooperative Agreement No. 58-6408-2-0009 (NCNPR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Roll DM, Ireland CM, Lu HS, Clardy J. J Org Chem. 1988;53:3276–3278. [Google Scholar]
- 2.Soni R, Muller L, Furet P, Schoepfer J, Stephan C, Zunstein-Mecher S, Fretz H, Chaudhuri B. Biochem Biophys Res Commum. 2000;275:877–884. doi: 10.1006/bbrc.2000.3349. [DOI] [PubMed] [Google Scholar]
- 3.Hörmann A, Chaudhuri B, Fretz H. Bioorg Med Chem. 2001;9:917–921. doi: 10.1016/s0968-0896(00)00313-8. [DOI] [PubMed] [Google Scholar]
- 4.Kinder FR, Bair KW, Bontempo J, Crews P, Czutchta AM, Nemzek R, Thale Z, Vattay A, Versace RW, Weltchek S, Wood A, Zabludoff SD, Phillips PE. Proc Am Assoc Cancer Res. 2000;41:600–601. [Google Scholar]
- 5.Lin J, Yan XJ, Chen HM. Cancer Chemother Pharmacol. 2007;59:439–445. doi: 10.1007/s00280-006-0282-x. [DOI] [PubMed] [Google Scholar]
- 6.Foderaro TA, Barrows LR, Lassota P, Ireland CM. J Org Chem. 1997;62:6064–6065. [Google Scholar]
- 7.Jimènez C, Quiñoà E, Adamczeski M, Hunter LM, Crews P. J Org Chem. 1991;56:3403–3410. [Google Scholar]
- 8.Kirsch G, König GM, Wright AD, Kaminsky R. J Nat Prod. 2000;63:825–829. doi: 10.1021/np990555b. [DOI] [PubMed] [Google Scholar]
- 9.(a) Ireland C, Scheuer PJ. J Am Chem Soc. 1980;102:5688–5691. [Google Scholar]; (b) Roy RS, Gehring AM, Milne JC, Belshaw PJ, Walsh CT. Nat Prod Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 10.Molinski TF. Chem Rev. 1993;93:1825–1838. [Google Scholar]
- 11.Schumacher RW, Davidson BS. Tetrahedron. 1995;51:10125–10130. [Google Scholar]
- 12.Krishnaiah P, Reddy VLN, Venkataramana G, Ravinder K, Srinivasulu M, Raju TV, Ravikumar K, Chandrasekar D, Ramakrishna S, Venkateswarlu Y. J Nat Prod. 2004;67:1168–1171. doi: 10.1021/np030503t. [DOI] [PubMed] [Google Scholar]
- 13.Vervoort HC, Pawlik JR, Fenical W. Mar Ecol Prog Ser. 1998;164:221–228. [Google Scholar]
- 14.Davis RA, Sandoval IT, Concepcion GP, da Rocha RM, Ireland CM. Tetrahedron. 2003;59:2855–2859. [Google Scholar]
- 15.Ding Y, Li XC, Ferreira D. J Org Chem. 2007;72:9010–9017. doi: 10.1021/jo071134z. [DOI] [PubMed] [Google Scholar]
- 16.Grimberg BT, Jaworska MM, Hough LB, Zimmerman PA, Phillips JG. Bioorg Med Chem Lett. 2009;19:5452–5457. doi: 10.1016/j.bmcl.2009.07.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wright CW, Addae-Kyereme J, Breen AG, Brown JE, Cox MF, Croft SL, Gökçek Y, Kendrick H, Phillips RM, Pollet PL. J Med Chem. 2001;44:3187–3194. doi: 10.1021/jm010929+. [DOI] [PubMed] [Google Scholar]
- 18.Bonjean K, De Pauw-Gillet MC, Defresne MP, Colson P, Houssier C, Dassonneville L, Bailly C, Greimers R, Wright C, Quetin-Leclercq J, Tits M, Angenot L. Biochemistry. 1998;37:5136–5146. doi: 10.1021/bi972927q. [DOI] [PubMed] [Google Scholar]
- 19.Onyeibor O, Croft SL, Dodson HI, Feiz-Haddad M, Kendrick H, Millington NJ, Parapini S, Phillips RM, Seville S, Shnyder SD, Taramelli D, Wright CW. J Med Chem. 2005;48:2701–2709. doi: 10.1021/jm040893w. [DOI] [PubMed] [Google Scholar]
- 20.Geyer JA, Prigge ST, Waters NC. Biochim Biophys Acta. 2005;1754:160–170. doi: 10.1016/j.bbapap.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 21.Grimberg BT, Erickson JJ, Sramkoski RM, Jacobberger JW, Zimmerman PA. Cytometry A. 2008;73:546–554. doi: 10.1002/cyto.a.20541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.