Abstract
Cryptococcus gattii and C. neoformans are causal agents of cryptococcosis, which manifests as pneumonia and meningitis. C. gattii has recently received widespread attention due to outbreaks in British Columbia, Canada and the US Pacific Northwest. The biology of this tree-dwelling yeast is relatively unexplored, and there are few clues about how it causes infections in humans and animals. In this review, we summarize recent discoveries about C. gattii genetics and its ecological niche and highlight areas ripe for future exploration. Increased focus on epidemiology, ecological modeling, and host-pathogen interactions is expected to yield a better understanding of this enigmatic yeast, and ultimately lead to better measures for its control.
Two etiologic agents of cryptococcosis
Cryptococcus gattii is found as single or budding yeast with round, oval or cylindrical cells enveloped in a polysaccharide capsule. This pathogen causes cryptococcosis that most commonly includes lung and brain infections in healthy as well as immune-compromised individuals (see Glossary). The far better known etiologic agent of cryptococcosis is C. neoformans, the sibling species of C. gattii. Until recently, these two fungi were classified as varieties of the same species 1, 2. Cryptococcosis due to C. neoformans occurs worldwide and it has received major attention as one of the common complications in HIV+ individuals and in AIDS patients. In addition, C. neoformans cryptococcosis has been called an ‘awakening giant’ among opportunistic infections in patients with solid tumors3. The pathogen C. neoformans comprises two varieties that differ significantly - C. neoformans variety (var.) grubii is global in distribution while C. neoformans var. neoformans is more common in Europe 4, 5. In this review, all references to C. neoformans pertain to C. neoformans var. grubii.
| Glossary | |
|---|---|
| Antioxidant | A molecule or compound that prevents or neutralizes oxidative stress caused by reactive oxygen species. |
| Autochthonous | Indigenous or native in origin; not imported. |
| Bacilliform | Shaped like a Bacillus bacterium, rod-shaped. |
| Clamp connection | A tube-like connection between two segments of a hypha to ensure that differing nuclei are equally distributed during the cell division. |
| Clonal | Derived from mother cell by asexual reproduction. |
| Cryptococcosis | Lung and brain infections caused by fungi Cryptococcus gattii or Cryptococcus neoformans. |
| Ecological niche | The specialized space in the environment where an organism lives and interacts with living and non-living objects in its surroundings. |
| Endemic | Native to or originating from a geographical area. |
| Genotype | The genetic makeup revealed by a biological assay. |
| Hybrids | Offspring resulting from crossing of dissimilar parent cells |
| Immunocompetent | Possessing a functioning immune system that mounts effective host defense to infection. |
| Immunocompromised | Unable to mount effective host defense to infection due to underlying immunodeficiency or immunosuppressive treatment. |
| Mating type | Genetic characteristics that distinguish two sexually compatible cells that appear similar in morphology. |
| Meningoencephalitis | Inflammation of the brain and surrounding membrane. |
| Neutrophils | Abundant white blood cells that play important role in immune defense via their ability to engulf and destroy pathogenic microbes. |
| Opportunist pathogen | A fungus that is only able to cause disease in an immunocompromised host. |
| Outbreak | A sudden increase in the incidence of an infection not expected in an area or at a particular period of time. |
| Pathogenicity | The ability of a fungus to produce disease in a given host. |
| Primary pathogen | A pathogen that is able to cause disease in apparently healthy host. |
| Phagocytosis | The process of engulfment of a pathogenic microbe by immune response cells or phagocytes. |
| Recombination | An assortment of genes or characteristics in offspring that distinguish the offspring from their parents. |
| Serotype | Characteristic surface antigens that permit closely related microbes to be distinguished by means of antibodies. |
| Virulence | The degree of pathogenicity of a fungus in a given host. |
Originally, C. gattii was considered to be restricted in distribution to the tropics and sub-tropics 5. During the past decade, compelling evidence has been accumulated regarding the widespread incidence of C. gattii cryptococcosis in Asia, Africa, Australia, South America and outbreaks in parts of North America, which were not previously suspected of harboring this pathogen 6-13. Newly presented data also suggests the possibility of a ‘smoldering outbreak’ throughout the Australian continent 14. This emerging information and ongoing outbreaks have rekindled interest in C. gattii cryptococcosis. This review provides a timely focus on the notable discoveries about population genetics of C. gattii and its ecological niche and highlights areas such as disease surveillance, fungal ecology, and host-pathogen interactions that are now ripe for further exploration.
Distinction of C. gattii from C. neoformans
The available records indicate that a yeast pathogen, initially named Saccharomyces subcutaneous tumefaciens, but subsequently identified as C. gattii, was documented as far back as 1896 15, 16. Cryptococcus neoformans var. gattii was formally proposed as a new taxonomic entity in 1970 from a clinical case that yielded regular round cells, i.e., R forms, and bacilliform (rod-shaped) cells, i.e., B forms, in culture (Figure 1). The B form phenotype was lost after prolonged culture in the laboratory, but this phenotype could be regained by passage in mice 1, 15, 17. This discovery paralleled an earlier characterization of capsular antigens in C. neoformans that revealed three serotypes: A, B, and C; B and C serotypes were exclusive to what would eventually become C. gattii (Figure 2) 18. Further proof of C. gattii being a separate species from C. neoformans was provided by the discovery of the sexual form of the former, which was named as Filobasidiella bacillispora. The sexual state was observed in the laboratory after mating between compatible strains (MATα and MATa) led to the production of bacilliform basidiospores or sexual spores 19. Further experiments revealed that interbreeding between C. gattii and C. neoformans did not yield spores that have an evidence of genetic recombination2. Furthermore, biochemical differences were found between C. gattii and C. neoformans in creatinine, canavanine, and glycine metabolism, which allowed the development of a diagnostic medium to easily differentiate these two pathogens in the laboratory 20. Thus, the separation of C. gattii from C. neoformans met both the morphological and reproductive basis for the recognition of a new species. Further proof of C. gattii as a new species resulted from many genetic studies that showed that the two siblings species have evolved into distinct entities 37- 80 million years ago 21, 22. These observations satisfied the phylogenetic basis for the formal species status for C. gattii 2. The recent discoveries of rare hybrids of C. gattii and C. neoformans provides additional proof of a taxonomic proximity, but not similarity between these two species 23.
Figure 1.
C. gattii under the microscope. Some photographs courtesy of Ping Ren and Sunanda S. Rajkumar. (a) Yeast cells are round (R form) or rod-shaped (B form) in fungal culture [NIH444 (ATCC 32609), Serotype B, MATα, VGIIa] grown on yeast-peptone dextrose agar at 30°C for 24-hrs and imaged by differential interference contrast (DIC) microscopy (Bar = 6.25 μm). (b) More details of yeast cells seen with a scanning electron microscope [scanning electron microscopy (SEM), Bar = 2.0 μm]. (c) When two sexually compatible yeast cultures (NIH444 {MATα} X NIH198 {MATa}) are grown under dry conditions and nitrogen limitation, they produce characteristic slender thread-like structure (hypha) with bulb-like head (basidium) bearing sexual spores (basidiospores, thick arrow); also seen is a clamp connection (thin arrow) between two hyphal segments. Some yeast cells (gray arrow) can also be seen in the same field (Bar = 6.25 μm). (d) SEM image of hyphal segment (arrow) with clamp connection (Bar = 2.0 μm). (e) Further SEM images highlighting basidium and basidiospores (Bar = 1.0 μm). (f) Yeast cells make large capsules when they are in tissues as seen here in a DIC image of lung tissue from an infected mouse (Bar = 6.25 μm).
Figure 2.
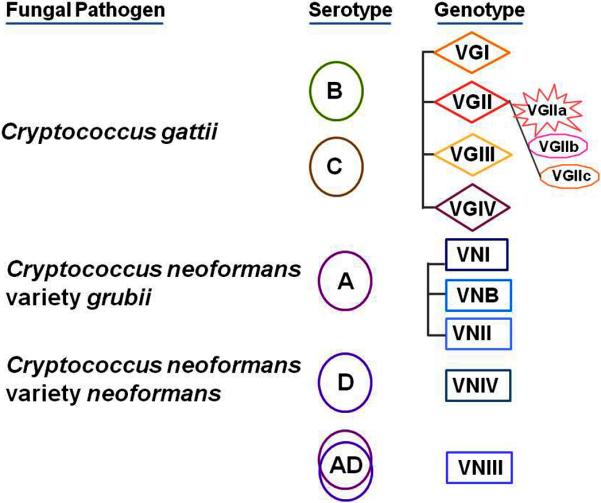
Serotypes and genotypes of C. gattii and C. neoformans. C. gattii strains are either serotype B or C; BC serotype strains have been rarely reported. C. neoformans variety grubii strains are serotype A while C. neoformans variety neoformans strains are serotype D; the taxonomic status of AD serotype strains is not clear. C. gattii and C.neoformans genotypes are based upon multilocus sequence typing of various gene fragments. C. gattii outbreaks in North America are variably linked to VGII sub-genotypes. Details of C. neoformans VNI sub-genotypes are not shown.
Currently, it is believed that C. gattii comprises four distinct genotypes (VGI – VGIV), which are distinct from C. neoformans genotypes (VNI, VNB, VNII and VNIV) as determined by multilocus sequence typing of multiple gene fragments 24-26. There is considerable genetic divergence among C. gattii genotypes, and there is even a proposal to accord strains in these genotypes independent varietal status 24, 26. Additional details are expected to emerge when functional analyses of whole genomes are carried out to determine how various C. gattii genotypes marshal their cellular processes in response to growth in nature and in animals. Indeed, a recent genome comparison of VGI and VGII strains is a good first step in this direction27.
C. gattii cryptococcosis
The pathogen C. gattii is believed to be clinically more virulent than C. neoformans given that C. gattii causes multiple lesions in the lungs and the brains of affected patients whereas C. neoformans does not 28-30. Similar to other ‘primary pathogenic fungi’, C. gattii causes disseminated skin and lung lesions that can potentially be misdiagnosed as malignancies 31-33. Brain infections (meningoencephalitis) caused by C. gattii are reported to respond slowly to standardized therapy, and they require more diagnostic follow-ups than those seen in C. neoformans infections 34-37. Although the available data are limited, C. gattii isolates susceptible to antifungals in the laboratory can nevertheless cause poor outcomes 38-40.
Other preliminary data suggest that the severity of C. gattii infection is due to defective induction of host immune responses, resulting in low levels of the pro-inflammatory cytokines that are crucial for controlling the spread of the fungus in the body 41. One survey in Papua New Guinea, an area highly endemic for C. gattii, has shown that significant numbers of people show measurable antibody responses to C. gattii protein, but yet they did not develop disease 42. Surveys conducted in Vancouver Island, British Columbia, have revealed that a small number of cats and dogs in the sampled population could act as asymptomatic carriers of C. gattii in their nasal cavity 43. These limited observations from humans and animals would support a scenario where some residents and visitors in high-risk areas might be exposed to C. gattii and not develop overt illness, but still suffer from reactivation of the disease later in life 44.
Overall evidence suggests that C. gattii cryptococcosis is different from C. neoformans cryptococcosis in its manifestations, and it requires accurate diagnosis and careful management. These characteristics have finally caught the attention of physicians, as documented in recently published clinical reports and in the recent guidelines for the treatment of cryptococcosis 37 Nevertheless, more systematic surveys are urgently warranted in relevant areas to document the extent of exposure, asymptomatic carriage, and sub-clinical disease among residents and visitors. Similarly, the need for clinical trials to examine if C. gattii cryptococcosis requires a more specific therapy and management cannot be overemphasized.
Epidemiology of C. gattii cryptococcosis
The epidemiological profile of C. gattii cryptococcosis has undergone significant change in the past decade, with the recognition that this disease afflicts the immune-competent as well as immune-compromised patients (including AIDS patients) in areas of the world beyond the tropics and subtropics 6, 8-11, 45. Many published reports describe autochthonous cases of human and animal infections from unsuspected areas, thereby providing evidence for the global distribution of C. gattii disease. On the other extreme, travel-related C. gattii infections have been reported from residents of areas that are considered free of this pathogen 34, 44. Overall, epidemiology of this disease is not easily understood because the routine differentiation of C. gattii infections from those caused by C. neoformans is not a standard practice in many parts of the world. This stems from the fact that the cryptococcosis caused by the two pathogens is not pathognomonic, and that laboratory reagents such as serotyping kits and the specialized culture media, needed to distinguish the two are not easy to commercially obtain. Additionally, many of the genotyping strategies described for the differentiation of two pathogens are only suitable for specialized laboratories. Given these limitations, our understanding of the occurrence and prevalence of C. gattii cryptococcosis is expected to remain biased and underestimated unless species differentiation can be integrated into the routine workflow of clinical laboratories.
Recent outbreaks of C. gattii
An ongoing outbreak of C. gattii cryptococcosis in Vancouver Island, British Columbia, Canada has greatly expanded our understanding of C. gattii cryptococcosis10. This outbreak originated in 1999, suggesting that the fungus has either recently spread or reappeared in a hospitable environment. Vancouver Island has a distinct climate from the rest of Canada (in the temperate climate zone) similar to other C. gattii affected areas such as Australia, Spain, South Africa and Southeast Asia, all of which have dry summer subtropical or Mediterranean climates 46, 47. Douglas fir and western hemlock trees in Vancouver Island have a high colonization of C. gattii similar to the fungal association with other tree species worldwide 47, 48. Moreover, in addition to humans, a number of animals have either been infected or asymptomatically carry the fungus, which suggests that C. gattii has a wide host range 43, 49, 50. Prior to 2010, there were 218 cases of C. gattii cryptococcosis identified in British Columbia, Canada with a rather high human case fatality rate of nearly 9%, which led to a sustained public health effort to increase the awareness among healthcare professionals and the public30.
True to an often repeated maxim ‘infectious diseases know no geographical boundaries’, the C. gattii outbreak has also been recognized in the neighboring Pacific Northwest area of the United States with 52 patients reported between 2004 and 2009 51, 52. Unlike Vancouver Island, areas of California and Washington State were already known to harbor C. gattii and it is not clear if the recent outbreak represents a spread of the outbreak strain from Vancouver Island or a re-emergence of local strains 5, 8, 53. The last point is critical considering that a strain very similar to the Vancouver Island major genotype was isolated from the sputum of a patient from Seattle, WA, in 1970 19. Additional details of the C. gattii outbreak in the US are being vigorously investigated considering the high case fatalities (20%) documented so far 51.
There have been no other recent outbreaks reported from parts of the world with documented C. gattii cryptocococcosis although a recent study raises the possibility of a widespread Australian outbreak 14. Similarly, high incidence of endemic C. gattii cryptococcosis has been reported from children in Eastern Amazon areas of Brazil13. It would be prudent to plan surveillance studies in relevant geographical areas by means of seroepidemiological and natural surveys to catch early signs of the resurgence of this pathogen.
Ecology of C. gattii
Many years after its first recovery from clinical specimens, C. gattii was isolated from the environment for the first time by sampling of Eucalyptus camaldulensis trees 54. This tree species was initially thought to be the exclusive natural habitat of C. gattii both in its native Australia and around the world given that the tree has been exported extensively. However, after a decade and a half, it became clear that trees beside E. camaldulensis also harbor C. gattii 55. A key finding was that the decaying hollows in trunks and branches of a number of tree species are the preferred habitat of the fungus 56. Tree and tree hollows represent a specialized niche for basidiomycetous yeasts of the genera Cryptococcus, and presumably, they offer selective advantages in competition vis-à-vis other microbes 57. This association with trees has held true worldwide, including the most recent major outbreak in North America that encompasses the Douglas fir-Western Hemlock biogeoclimatic zones 47, 48. To date, more than 50 tree species have been reported as harboring the fungus 48. The fungus has also been found in the air, the soil, and bodies of water in heavily infested areas 58. An equally important corollary is that the fungus has not been recovered with any regularity from pigeon guano, which is a well-known ecological niche for C. neoformans; C. gattii has been shown to be unable to complete mating on this substrate 59.
Experimental studies provide further proof of a C. gattii ecological niche in trees: the fungal cells can survive and proliferate on inoculated almond seedlings (Terminalia catappa) and on plant derived-substrates, such as leaves and wood 60, 61. C. gattii can complete its sexual cycle on experimentally inoculated Arabidopsis thaliana plants (Thale cress or mouse ear cress), and the fungus makes special structures (extracellular fibrils) that are hypothesized to be important for plant colonization and animal virulence 62, 63.
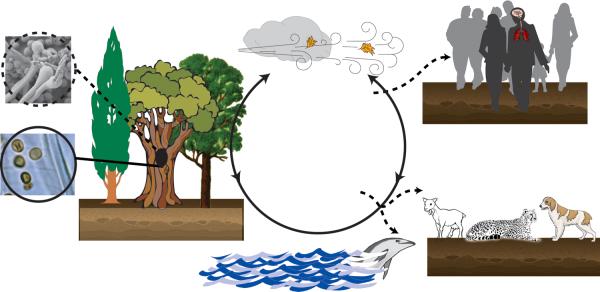
The use of sophisticated modeling tools has revealed that drier coastal areas at an elevation of approximately 100 meters, January temperatures of 2.8°C, and the predominance of Douglas fir (Pseudotsuga menziesii) and western hemlock (Tsuga heterophylla) trees were characteristics of a C. gattii ecological niche in outbreak areas of British Columbia 47. Future studies should focus on how C. gattii colonizes trees and what role this colonization plays in the fitness of the fungus for mammalian virulence. This ecological knowledge will be valuable in clarifying certain epidemiological aspects of C. gattii cryptococcosis such as the likely natural reservoirs of this pathogen and the role that outdoor occupations or recreational activities might play in exposure to C. gattii (Figure 3).
Figure 3.
Suggested natural cycle of Cryptococcus gattii and events leading to cryptococcosis. The available information indicates the pathogen can cycle through plants, soil, air and water without any intermediate live hosts (solid line with arrows). Yeast cells of either mating type (MATα or MATa) have been repeatedly isolated from trees especially hollows (solid circle); it is not yet known if cells of C. gattii cells complete their sexual cycle in nature as has been shown in the laboratory (dotted circle). Humans and animals (domestic and wildlife) coming in contact with fungal propagules (yeast cells or sexual spores) mostly remain asymptomatic (dotted lines with arrows), but a small number of infected humans and animals develop serious infections of the lungs and brain. These drawings are not to exact scale. Graphic artist: Andrew Bentley.
C. gattii virulence mechanisms
We know a great deal about the different structural and regulatory elements of C. neoformans that help to make it a successful pathogen. Capsular polysaccharide, melanin pigment, thermal tolerance, cellular signaling pathways, strategies to parasitize host phagocytes and mechanisms to breach the blood-brain barrier are all critical for C. neoformans virulence 36. It is safe to extrapolate that these attributes are equally critical for virulence in C. gattii due to the close proximity of these sibling species. However, distinctions in these traits or some yet unrecognized virulence factors must exist to explain unique features of cryptococcosis caused by C. gattii.
Published studies have indicated that some C. gattii virulence attributes subtly differ from those seen in C. neoformans. The clues to these distinctions resulted from the finding that C. gattii is able to arrest migration of neutrophils under laboratory conditions and in infected animals; such an effect on neutrophils would explain why healthy people could fall victim to this infection 64. Further investigations in animal models revealed that C. gattii metabolites especially acetoin and dihydroxyacetone, elicit less pro-inflammatory responses than do C. neoformans metabolites thereby promoting fungal survival and multiplication in the infected host 65, 66. The prominent antioxidant superoxide dismutase (SOD1p) in C. gattii protects the functional integrity of a number of virulence factors. Further experiments showed that SOD1p is required for infection in an animal model and for protection from killing by human phagocytes; some of these cellular functions differ remarkably from a homologous protein in C. neoformans 67, 68 (Table 1). The mitochondrial superoxide dismutase (SOD2p) was equally critical for growth at 37°C and virulence (inhalation model) for both C. gattii and C.neoformans 69, 70. Trehalose is another important antioxidant and stress protectant in fungi. Two genes encoding the key biosynthetic enzyme trehalose-6- phosphate (T6P) synthase (TPS1p, and TPS2p), were found to be critical for thermotolerance, virulence and the integrity of various virulence factors such as the capsule and melanin of C. gattii. The homologous proteins in C. neoformans were essential for thermotolerance, but not for expression of capsule and melanin 71, 72. Functional divergence was again seen in transcription factor Ste12α, which regulated mating, melanin, virulence and ecological fitness in C. gattii, but only mating and capsule size in C. neoformans 61, 73. Thus, there is a closer link between some important cellular processes and virulence in C. gattii, which is not the case for similar processes in C. neoformans.
Table 1.
The evolution of cell functions in similar proteins as revealed by gene knockout mutants of C. gattii (Cg) and C. neoformans (Cn)a
| Target gene | Cellular function | Fungus | Fungal features examinedb | Ref. | ||||
|---|---|---|---|---|---|---|---|---|
| 37°C growth | Capsule | Melanin | Mating | Virulence | ||||
| SOD1 | Cytoplasmic antioxidant |
Cg
Cn |
↔
↔ |
↓
↔ |
↓
↔ |
↔ ND |
↓
↓ |
67, 68 |
| SOD2 | Mitochondrial antioxidant |
Cg
Cn |
↓
↓ |
↔ ND |
↔ ND |
↔ ND |
X X |
69, 70 |
| TPS1 | Trehalose biosynthesis pathway |
Cg
Cn |
↓
↓ |
↓
↔ |
↓
↔ |
↓ ND |
↓ X |
71, 72 |
| TPS2 | Trehalose biosynthesis pathway |
Cg
Cn |
X ↓ |
↓
↔ |
↓
↔ |
↔ ND |
↓
↔ |
71, 72 |
| PKA1 | Regulation of cellular pathways |
Cg
Cn |
ND ND |
↓ X |
↔ X |
↔ X |
ND X |
74, 75 |
| PKA2 | Regulation of cellular pathways |
Cg
Cn |
ND ND |
↓
↔ |
↓
↔ |
↑
↔ |
ND ND |
74, 75 |
| STE12α | Regulation of mating |
Cg
Cn |
ND ↔ |
↔
↓ |
↓
↔ |
↓
↓ |
↓
↔ |
61, 73 |
C. neoformans refers to C. neoformans var. grubii.
Abbreviations: ND, not done; ↔, no change; ↓, reduced; ↑, enhanced; X, absent.
An interesting evolutionary switch in functions has been documented for C. gattii and C. neoformans Gα protein cAMP-protein kinase A (Pka1p and Pka2p). Pka1p controls mating, capsule, melanin and virulence in C. neoformans, but only influences capsule production in C. gattii. Pka2p regulates mating, capsule and melanin in C. gattii while the corresponding protein has no such role in C. neoformans 74, 75. These studies have elegantly connected C. gattii – C. neoformans speciation to ‘rapid re-wiring’ of a key cellular signaling pathway75.
Although functional differences among four C. gattii genotypes are not yet fully known, a good beginning was recently made when a group of highly virulent strains was found to differ from other C. gattii strains in their ability to parasitize phagocytes more effectively via increased activity of their mitochondria 76. Most recently, C. gattii was found to make extracellular fibrils on plant surfaces that impart an increased ability to evade killing by human neutrophils in laboratory tests63. This mechanism could possibly allow the fungus to more readily colonize lungs in infected individuals by avoiding initial attack by phagocytes. The emerging theme from molecular pathogenesis is that the two sibling pathogens have evolved to use similar genes for different purposes tailored to different environments and pathogenic modes. Less progress has been made to find de novo virulence traits in C. gattii; this could be either because such factors do not exist, or, more likely, is due to the lack of incisive experiments attempted so far. In either case, basic experimental tools are now available to carry out detailed host-pathogen analyses of C. gattii cryptococcosis to discover the reason for its unique virulence.
Hypervirulent clone(s) in North America?
C. gattii genotypes have been co-opted as tools for tracking down strains from recent outbreaks and for the determination of the ancestry of virulent strains. A rare C. gattii genotype (VGII) was found among clinical and environmental strains collected in connection with the outbreak of C. gattii cryotococcosis on the Vancouver Island 77, 78. It was further suggested that C. gattii strains from Vancouver Island comprised two sub-genotypes (VGIIa and VGIIb)77, 78. The C. gattii VGIIa strains were abundant in the outbreak, tested hypervirulent in mice and presented genetic evidence of spread from Australia to Canada by means of clonal expansion 78. A more recent analysis along similar lines suggested that a C. gattii outbreak in the Pacific Northwest included the Vancouver Island outbreak major sub-genotype VGIIa, as well as a new hypervirulent sub-genotype VGIIc, which possibly emerged by recombination events in the VGIIa strains 11. Thus, there is strong analytical evidence to suggest that reassortment of genetic information either by asexual or sexual processes could lead to the emergence and rapid expansion of hypervirulent C. gattii strains in a new locale.
The insightful discoveries on the emergence of hypervirulent clones cannot be fully reconciled with other published data. The type strain of Vancouver Island outbreak R265, is very similar in genotype and virulence attributes of a clinical strain NIH444, which was reported from Seattle, WA, in 1970 19, 79. Furthermore, the hypervirulence trait is shared by many VGIIa strains collected from clinical and environmental sources in different parts of the world 80. Paradoxically, the hypervirulence trait documented by mouse experiments seems to have no bearing on the course of human disease as both VGIIa and VGIIb strains cause similar morbidity and mortality among patients in the Vancouver Island outbreak 30. Clearly, much more remains to be done to elucidate if the C. gattii genotypes are uniform predictors of hypervirulence and emergence of outbreak strains, and if genotypes have important bearing on the course of cryptococcosis.
Concluding remarks
With the recent outbreaks in North America, C. gattii has finally caught the attention of healthcare professionals in its own right. While it might take awhile for reagents and diagnostic procedures to catch up with the clinical need for rapid and routine differentiation of C. neoformans from C. gattii, this is an important challenge for clinical laboratories to address. In addition, the new guidelines for C. gattii disease will hopefully mean that clinicians can better treat cryptococcosis and that they will look for clusters of C. gattii cases, which suggest a common origin. A parallel awareness in veterinary medicine would ensure prompt diagnosis and treatment of domestic animals. The recent rise in the incidence of infections in North America and Australia promises that biomedical researchers will devote greater resources to investigations of the host-pathogen factors that underlie C. gattii disease (Box 1). Such efforts would benefit greatly by the expected rapid expansion of knowledge on the genomics of this pathogen.
Box 1. Outstanding questions.
In some parts of the world, C. gattii cryptococcosis has been reported, but there have been no isolations of the fungus; or, the converse is true. What causes these disconnect between ecology and epidemiology?
Do forested or semi-forested areas pose higher risks for C. gattii infection?
Are some trees species particularly prone to C. gattii colonization? Do interactions with plants make C. gattii more pathogenic?
Do C. gattii genotypes other than VGII have hypervirulent strains? Are hypervirulent strains clonal or undergo asexual or sexual assortment of characters?
Are current genotyping methods sufficient or are more refined typing methods are needed based on whole genome analyses?
How common are asymptomatic infections, especially in people and animals living in areas of high incidence, and what triggers re-activation of the infection?
Do neutrophils fight or flee in response to C. gattii infection? Are some phagocytes more important than others?
Are C. gattii cells evolving in response to antifungals and are instances of clinical or laboratory resistance not currently recognized?
What are the intricate details of the capsular material and its organization in C. gattii?
Acknowledgments
We are grateful for the contributions of the following individuals: Andrew J. Hamilton, Birgit Stein (neé Rodeghier), Brian L. Wickes, the late Charles Lowry, Ping Ren, Srinivas D. Narasipura, Soumitra K. Saha, Jinjiang Fan, Madhu Dyavaiah, Deborah J. Springer, Manoj Iyer, Guan Zhu, Melissa J. Behr, and William A. Samsonoff. We also thank an anonymous reviewer for many suggestions for the improvement of this manuscript. Investigations in our laboratories were supported with funds from the National Institutes of Health (AI-41968, AI-48462, AI-05887701 and AI-053732), Pfizer Inc., and the Clinical Laboratory Reference Systems of the Wadsworth Center. The authors apologize to those whose studies could not be included as the citation of published literature is selective due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vanbreuseghem R, Takashio M. An atypical strain of Cryptococcus neoformans (San Felice) Vuillemin 1894. II. Cryptococcus neoformans var. gattii var. nov. Ann Soc Belg Med Trop. 1970;50:695–702. [PubMed] [Google Scholar]
- 2.Kwon-Chung KJ, et al. Proposal to conserve the name Cryptococcus gattii against C. hondurianus and C. bacillisporus (Basidiomycota, Hymenomycetes, Tremellomycetidae). Taxon. 2002;51:804–806. [Google Scholar]
- 3.Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; 1998. [Google Scholar]
- 4.Franzot SP, et al. Cryptococcus neoformans var. grubii: separate varietal status for Cryptococcus neoformans serotype A isolates. J. Clin. Microbiol. 1999;37:838–840. doi: 10.1128/jcm.37.3.838-840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon-Chung KJ, Bennett JE. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am. J. Epidemiol. 1984;120:123–130. doi: 10.1093/oxfordjournals.aje.a113861. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, et al. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerg Infect Dis. 2008;14:755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morgan J, et al. Cryptococcus gattii infection: characteristics and epidemiology of cases identified in a South African province with high HIV seroprevalence, 2002-2004. Clin Infect Dis. 2006;43:1077–1080. doi: 10.1086/507897. [DOI] [PubMed] [Google Scholar]
- 8.Chaturvedi S, et al. Cryptococcus gattii in AIDS Patients, Southern California. Emerg Infect Dis. 2005;11:1686–1692. doi: 10.3201/eid1111.040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Litvintseva AP, et al. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- 10.Stephen C, et al. Multispecies outbreak of cryptococcosis on southern Vancouver Island, British Columbia. Can. Vet. J. 2002;43:792–794. [PMC free article] [PubMed] [Google Scholar]
- 11.Byrnes EJ, 3rd, et al. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS pathogens. 2010;6:e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer W, et al. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9:189–195. doi: 10.3201/eid0902.020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos WRA, et al. Primary endemic Cryptococcosis gattii by molecular type VGII in the state of Pará, Brazil. Memórias do Instituto Oswaldo Cruz. 2008;103:813–818. doi: 10.1590/s0074-02762008000800012. [DOI] [PubMed] [Google Scholar]
- 14.Carriconde F, et al. Clonality and α-a Recombination in the Australian Cryptococcus gattii VGII Population - An Emerging Outbreak in Australia. PLoS One. 2011;6:e16936. doi: 10.1371/journal.pone.0016936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtis F. Contribution ‘a l'’etude de la saccharomycose humaine. Annales de l'Institut Pasteur. 1896;10:449–468. [Google Scholar]
- 16.Barnett JA. A history of research on yeasts 14:1 medical yeasts part 2, Cryptococcus neoformans. Yeast. 2010;27:875–904. doi: 10.1002/yea.1786. [DOI] [PubMed] [Google Scholar]
- 17.Gatti F, Eeckels R. An atypical strain of Cryptococcus neoformans (San Felice) Vuillemin 1894. I. Description of the disease and of the strain. Ann Soc Belges Med Trop Parasitol Mycol. 1970;50:689–693. [PubMed] [Google Scholar]
- 18.Evans EE. The Antigenic composition of Cryptococcus neoformans: I. A serologic classification by means of the capsular and agglutination reactions. J Immunol. 1950;64:423–430. [PubMed] [Google Scholar]
- 19.Kwon-Chung KJ. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- 20.Kwon-Chung KJ, et al. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, et al. Multiple gene genealogies reveal recent dispersion and hybridization in the human pathogenic fungus Cryptococcus neoformans. Mol Ecol. 2000;9:1471–1481. doi: 10.1046/j.1365-294x.2000.01021.x. [DOI] [PubMed] [Google Scholar]
- 22.Sharpton T, et al. Mechanisms of intron gain and loss in Cryptococcus. Genome biology. 2008;9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bovers M, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 24.Meyer W, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Med Mycol. 2009;47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bovers M, et al. Six monophyletic lineages identified within Cryptococcus neoformans and Cryptococcus gattii by multi-locus sequence typing. Fungal Genet Biol. 2008;45:400–421. doi: 10.1016/j.fgb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Ngamskulrungroj P, et al. Genetic diversity of the Cryptococcus species complex suggests that Cryptococcus gattii deserves to have varieties. PLoS One. 2009;4:e5862. doi: 10.1371/journal.pone.0005862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D'Souza CA, et al. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. MBio. 2011;2:e00342–00310. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitchell DH, et al. Cryptococcal disease of the CNS in immunocompetent hosts: Influence of cryptococcal variety on clinical manifestations and outcome. Clin Infect Dis. 1995;20:611–616. doi: 10.1093/clinids/20.3.611. [DOI] [PubMed] [Google Scholar]
- 29.Chen S, et al. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Australasian Cryptococcal Study Group. Clin Infect Dis. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 30.Galanis E, Macdougall L. Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999-2007. Emerg Infect Dis. 2010;16:251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dora JM, et al. Cutaneous cryptococccosis due to Cryptococcus gattii in immunocompetent hosts: case report and review. Mycopathologia. 2006;161:235–238. doi: 10.1007/s11046-006-0277-5. [DOI] [PubMed] [Google Scholar]
- 32.Dewar GJ, Kelly JK. Cryptococcus gattii: an emerging cause of pulmonary nodules. Can Respir J. 2008;15:153–157. doi: 10.1155/2008/392350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oliveira FM, et al. Cryptococcus gattii fungemia: report of a case with lung and brain lesions mimicking radiological features of malignancy. Rev Inst Med Trop Sao Paulo. 2007;49:263–265. doi: 10.1590/s0036-46652007000400014. [DOI] [PubMed] [Google Scholar]
- 34.Tsunemi T, et al. Immunohistochemical diagnosis of Cryptococcus neoformans var. gattii infection in chronic meningoencephalitis: the first case in Japan. Intern Med. 2001;40:1241–1244. doi: 10.2169/internalmedicine.40.1241. [DOI] [PubMed] [Google Scholar]
- 35.Speed B, Dunt D. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin Infect Dis. 1995;21:28–34. doi: 10.1093/clinids/21.1.28. discussion 35-26. [DOI] [PubMed] [Google Scholar]
- 36.Heitman J, et al., editors. Cryptococcus: From Human Pathogen to Model Yeast. ASM Press; 2010. [Google Scholar]
- 37.Perfect, John R, et al. Clinical practice guidelines for the management of Cryptococcal disease: 2010 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peachey PR, et al. The association between cryptococcal variety and immunocompetent and immunocompromised hosts. Pharmacotherapy. 1998;18:255–264. [PubMed] [Google Scholar]
- 39.Soares BM, et al. Cerebral infection caused by Cryptococcus gattii: a case report and antifungal susceptibility testing. Rev Iberoam Micol. 2008;25:242–245. [PubMed] [Google Scholar]
- 40.Mendes FE, et al. Correlation of the in vitro antifungal drug susceptibility with the in vivo activity of fluconazole in a murine model of cerebral infection caused by Cryptococcus gattii. Eur J Clin Microbiol Infect Dis. 2010;29:1525–1532. doi: 10.1007/s10096-010-1034-8. [DOI] [PubMed] [Google Scholar]
- 41.Brouwer AE, et al. Immune dysfunction in HIV-seronegative, Cryptococcus gattii meningitis. J Infect. 2007;54:e165–168. doi: 10.1016/j.jinf.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Seaton RA, et al. Exposure to Cryptococcus neoformans var. gattii--a seroepidemiological study. Trans Roy Soc Trop Med Hyg. 1996;90:508–512. doi: 10.1016/s0035-9203(96)90297-7. [DOI] [PubMed] [Google Scholar]
- 43.Duncan C, et al. Sub-clinical infection and asymptomatic carriage of Cryptococcus gattii in dogs and cats during an outbreak of cryptococcosis. Med Mycol. 2005;43:511–516. doi: 10.1080/13693780500036019. [DOI] [PubMed] [Google Scholar]
- 44.Hagen F, et al. Activated dormant Cryptococcus gattii infection in a Dutch tourist who visited Vancouver Island (Canada): a molecular epidemiological approach. Med Mycol. 2010;48:528–531. doi: 10.3109/13693780903300319. [DOI] [PubMed] [Google Scholar]
- 45.Tintelnot K, et al. Follow-up of epidemiological data of cryptococcosis in Austria, Germany and Switzerland with special focus on the characterization of clinical isolates. Mycoses. 2004;47:455–464. doi: 10.1111/j.1439-0507.2004.01072.x. [DOI] [PubMed] [Google Scholar]
- 46.Peel MC, et al. Updated world map of the Koppen-Geiger climate classification. Hydrol. Earth Syst. Sci. Discuss. 2007;4:439–473. [Google Scholar]
- 47.Mak S, et al. Ecological niche modeling of Cryptococcus gattii in British Columbia, Canada. Environ Health Perspect. 2010;118:653–658. doi: 10.1289/ehp.0901448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerg Infect Dis. 2010;16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan C, et al. Cryptococcus gattii in wildlife of Vancouver Island, British Columbia, Canada. J Wildl Dis. 2006;42:175–178. doi: 10.7589/0090-3558-42.1.175. [DOI] [PubMed] [Google Scholar]
- 50.Duncan C, et al. Clinical characteristics and predictors of mortality for Cryptococcus gattii infection in dogs and cats of southwestern British Columbia. Can Vet J. 2006;47:993–998. [PMC free article] [PubMed] [Google Scholar]
- 51.Emergence of Cryptococcus gattii-- Pacific Northwest, 2004-2010. MMWR Morb Mortal Wkly Rep. 2010;59:865–868. [PubMed] [Google Scholar]
- 52.MacDougall L, et al. Spread of Cryptococcus gattii in British Columbia, Canada, and detection in the Pacific Northwest, USA. Emerg Infect Dis. 2007;13:42–50. doi: 10.3201/eid1301.060827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bennett JE, et al. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am. J. Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 54.Ellis DH, Pfeiffer TJ. Natural habitat of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1990;28:1642–1644. doi: 10.1128/jcm.28.7.1642-1644.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sorrell TC, et al. Natural environmental sources of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1996;34:1261–1263. doi: 10.1128/jcm.34.5.1261-1263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lazera MS, et al. Cryptococcus neoformans var. gattii--evidence for a natural habitat related to decaying wood in a pottery tree hollow. Med Mycol. 1998;36:119–122. [PubMed] [Google Scholar]
- 57.Boddy L, et al. Ecology of saprotrophic basidiomycetes. Elsevier Academic Press; 2008. [Google Scholar]
- 58.Kidd SE, et al. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl. Environ. Microbiol. 2007;73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen K, et al. Cryptococcus neoformans mates on pigeon guano: implications for the realized ecological niche and globalization. Eukaryot Cell. 2007;6:949–959. doi: 10.1128/EC.00097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huerfano S, et al. Experimental infection of almond trees seedlings (Terminalia catappa) with an environmental isolate of Cryptococcus neoformans var. gattii, serotype C. Rev Iberoam Micol. 2001;18:131–132. [PubMed] [Google Scholar]
- 61.Ren P, et al. Transcription factor STE12alpha has distinct roles in morphogenesis, virulence, and ecological fitness of the primary pathogenic yeast Cryptococcus gattii. Eukaryot Cell. 2006;5:1065–1080. doi: 10.1128/EC.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xue C, et al. The human fungal pathogen Cryptococcus can complete its sexual cycle during a pathogenic association with plants. Cell host & microbe. 2007;1:263–273. doi: 10.1016/j.chom.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 63.Springer DJ, et al. Extracellular fibrils of pathogenic yeast Cryptococcus gattii are important in ecological niche colonization and mammalian virulence. PLoS One. 2010;5:e10978. doi: 10.1371/journal.pone.0010978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dong ZM, Murphy JW. Effects of the two varieties of Cryptococcus neoformans cells and culture filtrate antigens on neutrophil locomotion. Infect Immun. 1995;63:2632–2644. doi: 10.1128/iai.63.7.2632-2644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wright L, et al. Metabolites released by Cryptococcus neoformans var. neoformans and var. gattii differentially affect human neutrophil function. Microbes Infect. 2002;4:1427–1438. doi: 10.1016/s1286-4579(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 66.Cheng PY, et al. Cryptococcus gattii isolates from the British Columbia cryptococcosis outbreak induce less protective inflammation in a murine model of infection than Cryptococcus neoformans. Infect Immun. 2009;77:4284–4294. doi: 10.1128/IAI.00628-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Narasipura SD, et al. Characterization of Cu,Zn superoxide dismutase (SOD1) gene knock-out mutant of Cryptococcus neoformans var. gattii: role in biology and virulence. Molecular microbiology. 2003;47:1681–1694. doi: 10.1046/j.1365-2958.2003.03393.x. [DOI] [PubMed] [Google Scholar]
- 68.Cox GM, et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect. Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Giles SS, et al. Cryptococcus neoformans mitochondrial superoxide dismutase: an essential link between antioxidant function and high-temperature growth. Eukaryot. Cell. 2005;4:46–54. doi: 10.1128/EC.4.1.46-54.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Narasipura SD, et al. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 2005;55:1782–1800. doi: 10.1111/j.1365-2958.2005.04503.x. [DOI] [PubMed] [Google Scholar]
- 71.Ngamskulrungroj P, et al. The trehalose synthesis pathway is an integral part of the virulence composite for Cryptococcus gattii. Infection and Immunity. 2009;77:4584. doi: 10.1128/IAI.00565-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petzold EW, et al. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun. 2006;74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yue C, et al. The STE12alpha homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.D'Souza CA, et al. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol. Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hicks JK, Heitman J. Divergence of protein kinase A catalytic subunits in Cryptococcus neoformans and Cryptococcus gattii illustrates evolutionary reconfiguration of a signaling cascade. Eukaryot Cell. 2007;6:413–420. doi: 10.1128/EC.00213-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma H, et al. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc Natl Acad Sci U S A. 2009;106:12980–12985. doi: 10.1073/pnas.0902963106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kidd SE, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fraser JA, et al. Same-sex mating and the origin of the Vancouver Island Cryptococcus gattii outbreak. Nature. 2005;437:1360–1364. doi: 10.1038/nature04220. [DOI] [PubMed] [Google Scholar]
- 79.Chaturvedi S, et al. Selection of optimal host strain for molecular pathogenesis studies on Cryptococcus gattii. Mycopathologia. 2005;160:207–215. doi: 10.1007/s11046-005-0162-7. [DOI] [PubMed] [Google Scholar]
- 80.Ngamskulrungroj P, et al. Global VGIIa isolates are of comparable virulence to the major fatal Cryptococcus gattii Vancouver Island outbreak genotype. Clinical Microbiology and Infection. 2011;17:251–258. doi: 10.1111/j.1469-0691.2010.03222.x. [DOI] [PubMed] [Google Scholar]