Abstract
Studies of the Hepatitis C virus (HCV) life-cycle rely heavily upon Huh7.5 cells, but the reasons why these cells are exceptionally permissive for HCV replication are not clear. Based on recent clinical observations, we hypothesized that the Hedgehog (Hh) pathway, which has not been previously associated with HCV replication, may be involved in the Huh7.5 phenotype of increased permissiveness. We tested this hypothesis by comparing levels of a variety of Hh related cellular markers in Huh7.5 cells with the parental Huh7 cells, which are far less permissive. Here, we demonstrate that Huh7.5 cells, when compared to Huh7 cells, have substantially decreased expression of epithelial markers, increased levels of mesenchymal markers and markedly upregulated Hh pathway activity: Shh, >100 fold, Gli1, >30 fold, Ptc, 2 fold. In Huh7.5 cells, we found that cyclopamine, a Hh pathway antagonist, reduced HCV RNA levels by 50% compared to vehicle and inactive isomer controls. Moreover, in Huh7 cells, treatment with recombinant Shh ligand and SAG, both Hh pathway agonists, stimulated HCV replication by 2 fold and 4 fold, respectively. These effects were observed with both viral infections and a subgenomic replicon. Finally, we demonstrated that GDC-0449 decreased HCV RNA levels in a dose response manner.
Conclusions
We have identified a relationship between HCV and Hh signaling where upregulated pathway activity during infection promotes an environment conducive to replication. Given that Hh activity is very low in most hepatocytes, these findings may serve to further shift the model of HCV liver infection from modest widespread replication in hepatocytes to one where a subset of cells support high level replication. These findings also introduce Hh pathway inhibitors as potential anti-HCV therapeutics.
Hepatitis C virus (HCV) is an important cause of chronic liver disease, with the severe consequences of hepatocellular carcinoma (HCC) and cirrhosis occurring in some patients(1, 2). When considering determinants of HCV persistence and propagation of infection, little consideration has been given to differences between cells within the liver. Recent studies have demonstrated HCV Core protein localized to discrete foci within HCC sections from patients, and laser captured microdissection samples indicated that HCV genomes exist at unexpectedly low average copy numbers per cell(3, 4). These observations suggest that HCV infection is not widespread throughout the liver, but rather selective or restrained in its target cells.
In vitro studies of HCV rely heavily on the Huh7.5 cell line. This cell line was generated after Huh7 cells selected for harboring an HCV subgenomic replicon were cured of replicating viral RNA with interferon-α(5). The resulting cells were highly permissive for HCV replication when re-transfected with replicon constructs. As they support replication of the JFH1 viral isolate and produce infectious virus in tissue culture, Huh7.5 cells have propelled studies of the HCV life-cycle forward(6). Similar cell lines with increased HCV permissivity, like LH86 cells, have been directly isolated from patient samples, although HCV RNA levels are 1–2 log lower compared to Huh7.5 cells(7). The reasons for Huh7.5 cells being exceptionally permissive for HCV replication were attributed to a defect in RIG-I, a pattern recognition receptor that activates type I interferon expression during viral infection(8). However, recent studies found no RIG-I defects in novel cell lines also generated from Huh7 cells with increased permissiveness to HCV(9, 10). Thus RIG-I alone may not explain this phenomenon.
The Hedgehog (Hh) pathway plays an important role during embryogenesis, normal tissue growth, regeneration after injury and carcinogenesis(11–15). Most hepatocytes in healthy adult livers do not express detectable Hh ligands Sonic hedgehog (Shh) or Indian hedgehog (Ihh), whereas some Hh ligand expression can be demonstrated in ductular-type cells(16). After injury, Shh expression increases, causing Gli family transcription factors to accumulate in Hh-responsive cells as part of the regenerative and fibrotic responses(14, 15). Hh pathway activation has also been observed in some HCC cell lines, although significant heterogeneity exists within actual tumors(16). A vigorous debate exists as to whether liver epithelial cells, such as cholangiocytes and hepatocytes, undergo epithelial-to-mesenchymal transitions (EMT) in injured livers, but some evidence supports this concept and suggests Hh-mediated regulation(17–21). In viral hepatitis patients, recent data suggests EMT may occur in response to infection(22). HCV infection of hepatoma cell lines in vitro alters cell polarity to expose gap junction complex proteins key to viral entry (23). To our knowledge, such studies have not addressed the effects of altered cell polarity on HCV replication, or mechanisms by which viral infection might promote EMT.
We hypothesized that Huh7.5 cells are highly permissive for HCV because they possess a “transitional” phenotype skewed towards mesenchymal characteristics due to increased Hh pathway activity. We subsequently asked whether Hh pathway activation may create an environment conducive to viral replication and whether Hh pathway inhibition would inhibit HCV replication.
Experimental Procedures
Cells, Constructs and Reagents
Huh7 cells, Huh7.5 cells (a gift from C. Rice, Rockefeller University), LH86 cells (a gift from C. Liu, University of Florida) and HepG2 cells were used for these studies. Primary human hepatic stellate cells were isolated and cultured as described (17) and primary human hepatocytes were commercially-prepared (a gift from R. Witek, Invitrogen, Durham, NC). JFH1 cDNA (kindly provided by T. Wakita, National Institute of Infectious Diseases, Tokyo, Japan) was used to generate cell culture HCV, and HCV Con1 replicon (a gift from C. Rice, Rockefeller University) to study HCV replication. Primer sequences for qRT-PCR reactions are listed in Supplemental Table 1. Purchased reagents were: Interferon-α A/D (Sigma-Aldrich, St. Louis, MO), cyclopamine and tomatidine (Toronto Research Chemicals, Toronto, Canada), Recombinant N-terminal mouse Shh (StemCell Technologies, Vancouver, Canada), SAG (Enzo Life Sciences, Plymouth Meeting, PA), GDC-0449 (Selleck Chemicals, Houston TX), and mouse IgG1 isotype control antibody (R&D Systems, Minneapolis, MN). Antibodies to Shh and Gli1 (Santa Cruz Biotechnology, Santa Cruz, CA), α-SMA (Dako, Carpinteria, CA) and α-tubulin (Sigma-Aldrich) were used for immunoblotting and α-HCV Core (C7-50, Abcam) was used for immunofluorescence/immunoblotting.
Messenger RNA Quantification by Real-Time Reverse-Transcription Polymerase Chain Reaction
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA), reverse-transcribed using Superscript reverse transcriptase (Invitrogen). cDNA samples were used in triplicate for quantitative reverse-transcription polymerase chain reaction (qRT-PCR) using iQ-SYBR Green Supermix (Bio-Rad Laboratories)(17). Targetgene levels are presented as ratio of levels detected in treated cells overlevels detected in control cells, according to the ΔΔCt method(17).
HCV replicon, JFH1 infections and Focus Forming Assay
Huh7.5 cells were grown in supplemented Dulbecco’s modified Eagle medium (DMEM)(5). HCV replicon-harboring Huh7.5 cells were established by transfecting in vitro transcribed Con1 replicon RNA followed by selection with 1 mg/ml G418(24). After selection, cells were propagated in DMEM with 0.75 mg/ml G418. Infectious JFH1 virus was obtained by transfection of Huh7.5 cells with in vitro transcribed RNA and harvesting of cell supernatant as described(6, 25). To generate viral stocks, clarified supernatant was used to infect naïve Huh7.5 cells, supernatants were recovered 7 days post-infection, concentrated using an Amicon 100k device. Virus-containing supernatants were titered by focus-forming assay as previously described (25). Briefly, serial 10-fold dilutions of samples were plated in triplicate on 96-well plates containing subconfluent Huh7.5 cells. After 72 hours of incubation, cells were washed with PBS and fixed with ice-cold methanol. Infected cells were subsequently identified by immunofluorescence using mouseα-Core antibody (C7-50; Abcam) and FFU/ml values were calculated using the mean of 3 separate wells per sample.
Time course infections-protein analysis: Huh7.5 cells (3×105) were seeded onto four T-25 flasks and two of the flasks were infected the following day with HCV at a multiplicity of infection of 0.5–1.0. The virus was removed 16 hrs post infection and replaced with normal growth medium. Uninfected cells were split and grown in culture, for the same time as their respective infected culture counterparts. Cells were harvested and lysed in RIPA buffer (50mM Tris-HCl, 150mM NaCl, 0.5% Sodium Deoxycholate, 1% NP40, 0.1% SDS) on day 2 and day 5 post infection. The cell lysate was collected in 1.5ml microcentrifuge tubes and allowed to sit on ice for 15 minutes prior to centrifugation at 10,000×g. The supernatant was collected as total cell lysate and amount of protein was estimated using Bradford method.
Drug/Antibody Treatment
Drugs were incubated with cells prior to RNA and protein isolation. The following concentrations and incubation times were used: cyclopamine and tomatidine (5μM for 48 hours for initial experiment; 24, 48 and 72 hours for time course), Shh (100ng/ml for 48 hours), SAG (0.3μM for 24 hours), Interferon-α (500 U/ml for 48 hours), GDC-0449 (range 0–25μM for 24 hours) 5E1 (Shh neutralizing antibody, 10 μg/ml, for 48 hours), mouse IgG1 isotype control (10 μg/ml, for 48 hours. If the experiment was done with the JFH1 HCV, drug was added at the time of infection.
LDH release assay
Supernatant media was collected from Huh7.5 cells infected and uninfected cells after 72 hours. LDH assay was performed according to manufacturers protocol (LDH Cytotoxicity Detection Kit, Takara Bio Inc, Japan). Uninfected cells lysed with Triton-X were used as positive control.
Results
Huh7.5 cells are more permissive for JFH1 HCV infection, have decreased epithelial markers, increased mesenchymal markers and markedly higher Hh pathway activity
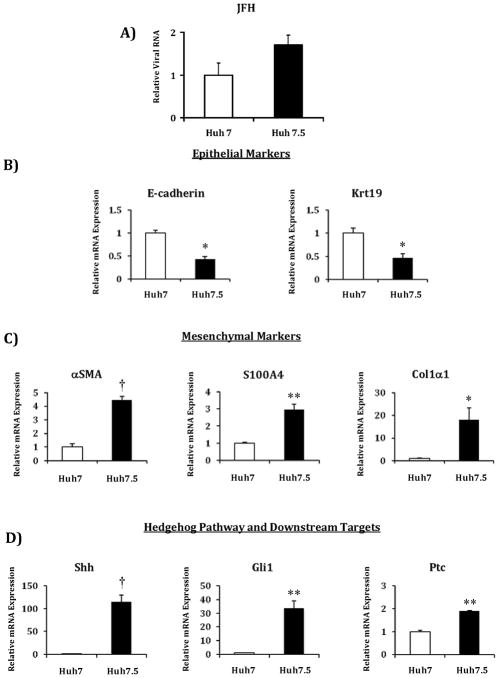
We first compared Huh7.5 cells to Huh7 cells grown under standard conditions and infected with JFH1 HCV to confirm that the cell lines in our lab follow the observation made by other labs. Indeed, we found that Huh7.5 cells were more permissive for HCV infection than Huh7 cells infected under the same conditions (Figure 1a). We next compared uninfected cells grown under standard conditions to assess differences in baseline gene expression. Analysis focused on markers that are differentially expressed primarily in epithelial and mesenchymal cells, as well as Hh pathway markers involved in regulating the “transitional” state. We found that Huh7.5 cells expressed significantly reduced transcript levels of the epithelial markers E-cadherin and Keratin 19 (Krt19) (Figure 1b), and significantly increased transcript levels of mesenchymal markers α–smooth muscle actin (αSMA), collagen, type I, alpha 1 (Col1α1) and S100 calcium binding protein A4 (S100A4) (Figure 1c). We also noted highly significant increases in Hh pathway component transcript levels in Huh7.5 cells compared to Huh7 cells: Shh >150 fold, Gli1 >30 fold and Patched >2 fold (Figure 1d).
Figure 1. Huh7.5 are more permissive than Huh7 cells for HCV infection and RNA expression levels are markedly different for epithelial, mesenchymal and Hh pathway genes.
A) Comparison of relative HCV RNA levels in Huh7.5 and Huh7 cells after infection with JFH1 HCV. RNA was isolated at 72 hours post-infection. Comparison of relative RNA levels of Huh7.5 cells compared to Huh7 cells for: B) epithelial markers E-cadherin and keratin 19 (Krt19); C) mesenchymal markers α–smooth muscle actin (αSMA), collagen, type I, alpha 1 (Col1α1) and S100 calcium binding protein A4 (S100A4); D) Hedgehog pathway and downstream targets sonic hedgehog (Shh), glioblastoma 1 (Gli1), Patched (Ptc). Results are expressed as relative fold expression with Huh7 expression indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
We next asked if these findings were unique to Huh7.5 cells or also observed in LH86 cells, another cell line permissive for HCV replication(7). We found LH 86 cells had: i) increased transcript levels of mesenchymal markers, significantly higher than Huh7 cells but lower than Huh7.5 cells (Supplemental Figure 1a); ii) markedly increased Shh transcript levels comparable to Huh7.5 cells; iii) significantly higher levels of Gli1 and Ptc transcripts compared to Huh7 cells, but significantly lower than Huh7.5 cells (Supplemental Figure 1b). We confirmed the relevance of observed changes in mRNA levels by analyzing protein expression. αSMA and Shh proteins were undetectable in Huh7 cells, but robustly expressed in both Huh7.5 and LH86 cells (Supplemental Figure 2). To further confirm our results, we profiled HepG2 cells, which are non-permissive for HCV infection, and determined that these cells exhibit markers consistent with an epithelial phenotype and express Shh and Gli1 at levels even lower than parental Huh7 cells and far lower than Huh7.5 cells (data not shown).
Interferon treatment of Huh7 cells causes increased mesenchymal marker transcripts and higher transcripts of upstream and downstream Hh pathway components
We evaluated the possible role of interferon treatment in the genesis of the Huh7.5 cell phenotype by asking if Huh7 cells treated with interferon-α demonstrated similar changes in gene expression. We observed that interferon-α caused the expected changes in Huh7 cells: increased αSMA and Col1α1 transcripts; significantly increased Shh and Gli1 transcripts (Supplemental Figure 3).
Hh pathway inhibition and activation modulate HCV replication
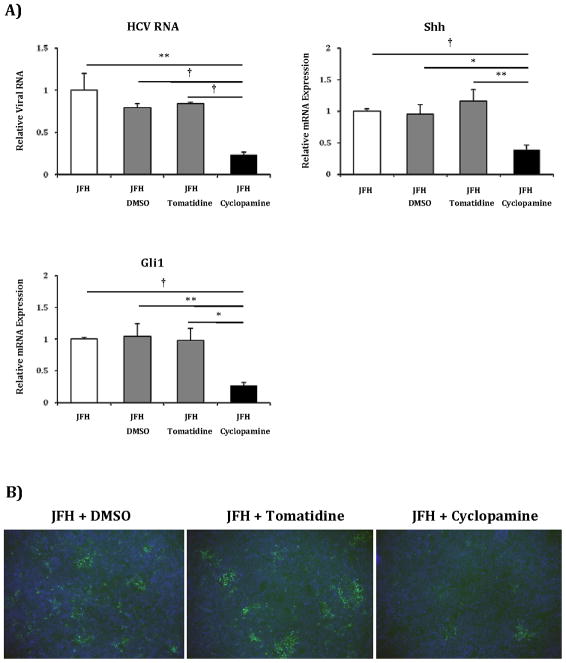
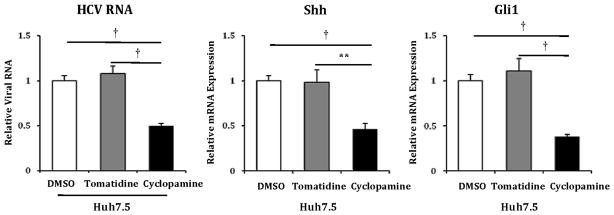
Given the significantly upregulated Hh pathway activity in Huh7.5 cells, we examined if manipulation of the Hh pathway would inhibit or promote HCV viral RNA replication. Huh7.5 cells were infected with JFH1 HCV and treated with cyclopamine (Smoothened antagonist), tomatidine (biologically inactive isomer), or vehicle control (26). Compared to control treatments, cyclopamine reduced cell-associated HCV RNA by 70%, mirroring the observed decreases in Shh and Gli1 expression (Figure 2a). Cyclopamine treatment also noticeably reduced the extent of infection as ascertained by immunofluorescence using antibody to the viral Core protein (Figure 2b). Reductions in HCV Core content correlated strongly with the drop in Shh expression that followed cyclopamine treatment.
Figure 2. Hh pathway blockade with cyclopamine results in reduced HCV RNA and protein levels.
A) Huh7.5 cells grown on 12-well plates were infected with 1500 focus-forming units of JFH1 HCV virus alone, or virus plus vehicle control (DMSO), inactive analog (tomatidine 5μM) or Hh inhibitor (cyclopamine 5μM) and incubated for 72 hours. Relative RNA expression for HCV RNA, Shh and Gli1 were analyzed with virus-alone infected cells indexed at 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
B) Immunofluorescent analysis using α-HCV Core of Huh7.5 cells infected with virus plus DMSO, virus plus tomatidine or virus plus cyclopamine.
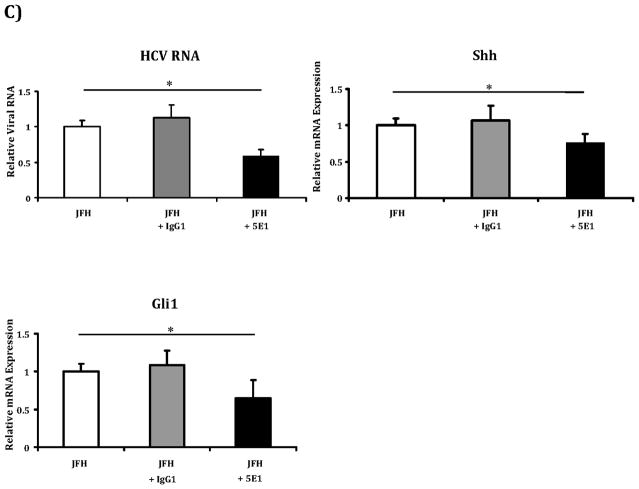
C) Huh7.5 were grown as described above. Cells were infected with virus alone, virus plus antibody control (mouse IgG1, 10 μg/ml) or virus plus a Shh neutralizing antibody (5E1, 10 μg/ml) and incubated for 48 hours. Relative RNA expression for HCV RNA, Shh and Gli1 were analyzed with virus alone infected cells indexed at 1. Key to statistical comparisons: *p<.05.
To further characterize the effect of cyclopamine on infected cells, we performed a time course experiment in which we isolated RNA from JFH1 infected Huh7.5 cells treated with cyclopamine and controls at 24, 48 and 72 hours post-infection (Supplemental Figure 4). HCV RNA mirrored reductions in Gli1 RNA beginning at 24 hours and maximizing by 48–72 hours. In order to ascertain whether cyclopamine treatment was associated with changes in cell viability, we performed an analysis of LDH levels in supernatant media under different conditions. LDH levels were comparable between uninfected and infected cells regardless of treatment (Supplemental Figure 5), indicating that reduced HCV replication was not due to potential toxic effects of cyclopamine treatment. Finally, we performed a focus forming units assay to quantify infectious virus from supernatant media at 72 hours from infected cells after cyclopamine and control treatment (Supplemental Figure 6). Cyclopamine treatment led to a one log reduction in focus forming units/ml compared to control treated cells.
To further verify these results obtained with pharmacologic inhibition, we also used a neutralizing antibody to Shh (5E1) to inhibit pathway activity in Huh7.5 cells and observed similar reductions in HCV RNA, Shh and Gli1 observed with chemical inhibition (Figure 2c).
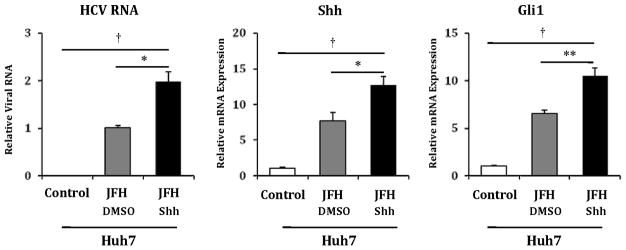
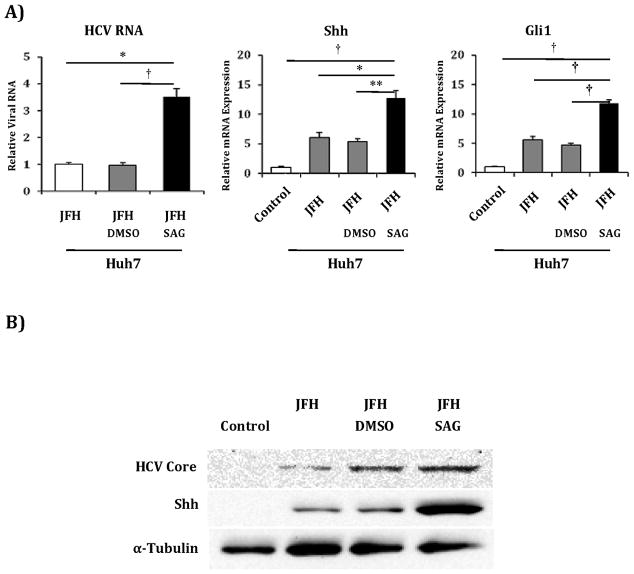
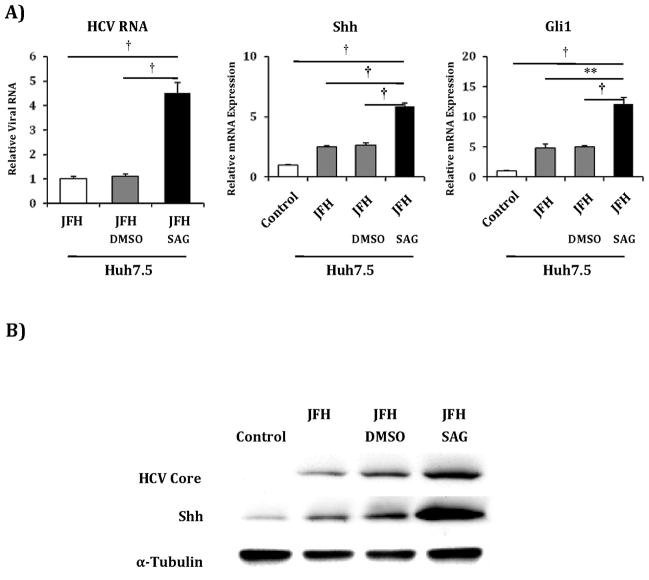
We next examined if recombinant N-terminal fragments of Shh, an agonist of the Hh pathway, would promote HCV viral titers in Huh7 cells. Incubation with exogenous Shh for 48 hours produced increased Shh and Gli1 transcripts, and caused a 2-fold increase in HCV RNA levels (Figure 3). It should be noted that JFH1 infection alone produced increased Shh and Gli1 transcripts and protein, and the increase in Shh expression paralleled the increase in core protein over time post infection (Supplemental Figure 7)(22). We replicated the increase in Huh7 permissiveness results using SAG, a Hh agonist that acts downstream by directly binding to Smoothened. SAG treatment resulted in a 3-fold increase in Hh pathway transcripts, and a corresponding 3-fold increase in HCV RNA levels (Figure 4a). Corresponding increases in protein expression levels were observed (Figure 4b). To confirm that this increase in HCV RNA correlated with functional virus, we used supernatants collected from the above experiment to infect naïve Huh7 cells. We observed foci of HCV Core-staining cells in Huh7 cells infected with supernatants from SAG treated cells but no foci in cells infected with supernatants from untreated or vehicle-treated cells (data not shown). SAG also potentiated HCV RNA accumulation in Huh7.5 cells, with a 3-fold increase in Shh and Gli1 transcripts accompanied by a 4-fold increase in HCV RNA levels (Figure 5a). Corresponding increases in protein expression levels were observed (Figure 5b). We subsequently treated commercially prepared primary human hepatocytes with SAG to assess if they would support HCV replication. Given that mature hepatocytes do not express Hh pathway intermediaries and have little detectable Hh activity, these cells were not SAG responsive and did not support HCV replication (data not shown).
Figure 3. Huh7 cells treated with a Hh pathway agonist demonstrate increased permissiveness for HCV replication.
Using identical conditions to those described in figure 2A, Huh7 cells were mock infected (control), infected with JFH1 HCV plus vehicle (JFH+DMSO) and infected with JFH1 HCV and then treated with N-terminal fragment Shh ligand. After 72 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with mock-infected expression indexed to 1, except for HCV RNA sample, in which case JFH1+DMSO was indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Figure 4. Huh7 cells treated with SAG, a Hh pathway agonist with a different target, also demonstrate increased permissiveness for HCV replication.
A) Huh7 cells were mock infected (control), infected with JFH1 HCV alone, JFH1 HCV plus vehicle (JFH+DMSO) and JFH1 HCV plus SAG 0.3μM. After 72 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with mock-infected expression indexed to 1, except for HCV RNA sample, in which case JFH1 HCV alone was indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
B) Protein lysates were created from the above described experiment. Antibodies to HCV Core, Shh and α-tubulin were used for analysis.
Figure 5. SAG treatment of Huh7.5 cells further increases their permissivity for HCV replication.
Huh7.5 cells were mock infected (control), infected with JFH1 HCV alone, JFH1 HCV plus vehicle (JFH+DMSO) and JFH1 HCV plus SAG 0.3μM. After 72 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with mock infected expression indexed to 1, except for HCV RNA sample, in which case JFH1 HCV alone was indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
B) Protein lysates were created from the above described experiment. Antibodies to HCV Core, Shh and α-tubulin were used for analysis.
Hh pathway activity modulates replication of a sub-genomic HCV replicon
We next attempted to determine if upregulated Hh pathway activity promotes HCV RNA replication versus some other event such as assembly or entry. We treated Huh7.5 cells harboring the Con1 HCV subgenomic replicon with cyclopamine, tomatidine or vehicle. Cyclopamine-treated cells exhibited a 50% reduction in HCV RNA compared to cells treated with either tomatidine or vehicle, corresponding to the reduction in Shh and Gli1 transcript levels (Figure 6).
Figure 6. Blockade of Hh pathway with Cyclopamine in Huh7.5 cells reduces replication of HCV subgenomic replicon.
Huh7.5 cells harboring HCV Con1 subgenomic replicon RNA were treated with vehicle control (DMSO), inactive analog (tomatidine 5μM) or Hh pathway antagonist (cyclopamine 5μM). After 48 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with vehicle control sample (DMSO) indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
We also performed an experiment to assess the effect of Hh pathway on HCV cell entry. Huh7.5 cells were infected after 24 hours incubation with cyclopamine, tomatidine or vehicle plus or minus the presence of antibody to CD81. HCV RNA levels at 4 hours post-infection were equivalent in infected cells in the presence of absence of cyclopamine (data not shown). Antibody to CD81 inhibited association of HCV with target cells under all conditions. This suggests that the Hh pathway promotes HCV replication, at least partially through enhancing HCV RNA synthesis and/or translation, and does not alter viral attachment.
GDC-0449 has anti-HCV activity
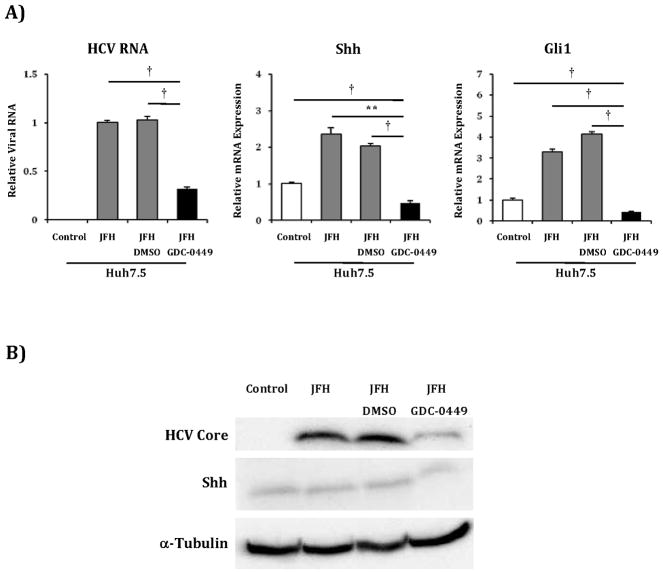
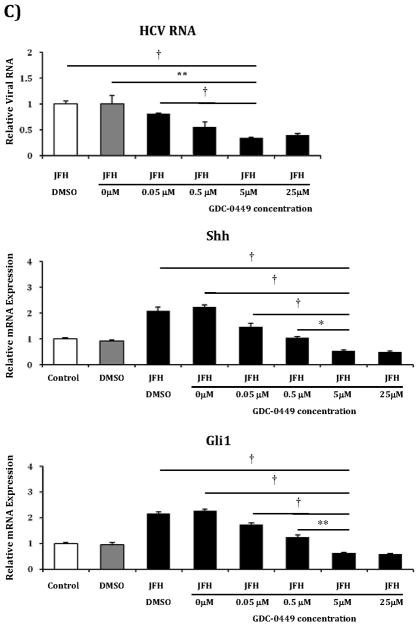
While cyclopamine treatment was able to reduce HCV viral titers, this agent has well described toxicity, and thus, no potential as a future anti-HCV pharmaceutical. In contrast, GDC-0449 is a Smoothened antagonist currently in phase 1and 2 clinical studies as a chemotherapeutic agent for various malignancies with an excellent safety profile and thus much greater potential as an HCV treatment(27–30). Therefore, we tested GDC-0449 in Huh7.5 cells infected with the JFH1 virus. GDC-0449 at 5μM concentration inhibited HCV RNA by >50% when compared with untreated or vehicle-treated cells (Figure 7a). Reductions in HCV RNA mirrored the decreases in Shh and Gli1 transcripts, and similar reductions in protein levels were observed (Figure 7b). We subsequently determined that GDC-0449 resulted in Hh pathway and HCV RNA inhibition in a dose response fashion beginning at concentrations as low as 0.05μM with a plateau at 5μM (Figure 7c). Based on this curve, the IC50 is estimated to be 0.16 μM.
Figure 7. GDC-0449, a pre-clinical Hh pathway inhibitor, inhibits replication of JFH1 HCV in a dose response manner.
A) Huh7.5 cells were mock infected (control), infected with JFH1 HCV alone, JFH1 HCV plus vehicle (JFH+DMSO) and JFH1 HCV plus GDC-0449 5μM concentration. After 72 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with mock infected expression indexed to 1, except for HCV RNA sample, in which case JFH1 HCV alone was indexed to 1.
B) Protein lysates were created from the above described experiment. Antibodies to HCV Core, Shh and α-tubulin were used for analysis.
C) The above experiment was replicated with varying concentrations of GDC-0449 to assess dose response of anti-HCV activity. Concentrations used were: 0 μM, 0.05 μM, 0.5 μM, 5 μM and 25 μM. After 72 hours, relative RNA expression was analyzed for HCV RNA, Shh and Gli1. Results are expressed as relative fold expression with mock infected expression indexed to 1, except for HCV RNA sample, in which case JFH1 HCV alone was indexed to 1.
Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Discussion
We have demonstrated a significant association between HCV infection and Hh pathway activity in liver-derived cells. Cells with dramatically increased Hh pathway activity such as Huh7.5 and LH86 cells support higher levels of HCV replication compared to Huh7 cells with virtually undetectable Hh pathway activity. Pharmacological and antibody treatment to either antagonize or promote Hh signaling in liver cells at different points in the pathway had the same direct effects on HCV replication. Included in the current study was GDC-0449, a Hh pathway antagonist already in pre-clinical development for other indications, which we have shown can inhibit HCV replication.
The observation that HCV replication is closely associated with Hh pathway activity has not been previously appreciated. Certainly it has never been explored as an explanation for why Huh7.5 cells are exceptionally permissive for HCV replication. Hh pathway components were previously identified in a functional genomic primary screen, but were not identified in the secondary screen(31). This may be due to the broad changes in the intracellular environment induced by Hh pathway activation that may not be detected in single gene siRNA knockdowns. It may be that the intracellular changes that occur upon Hh pathway activation and its possible association with EMT are part of the same continuum when considering an environment conducive to HCV infection. The concept of a continuum is supported by the mixed phenotype we detected in LH86 cells compared to Huh7.5 cells and how that correlates with the original observation that LH86 cells were less permissive than Huh7.5 cells(7). One of the key remaining questions is at what level does Hh pathway activation exerts its effects on HCV replication. While we used multiple agonists and antagonists, these agents all act close to the “top” of the pathway. We suspect the changes may occur further downstream and are likely to involve other pathways known to be associated with Hh in promoting an environment conducive for HCV replication.
Our results may be further cause to re-evaluate the model of HCV infection in the liver. Liang et al demonstrated in their analysis of explanted livers from HCV patients with HCC that Core protein was detected in a minority of cells despite this advanced-stage of disease(3). Perhaps Huh7.5 cells are representative of a minor population of liver cells that retain an active Hh pathway (18). HCV may preferentially infect and replicate in this minority population of cells so hospitable to efficient replication. Infection of these cells combined with a possible contribution of the resulting interferon release may induce further Hh pathway activation including increased Shh ligand production. Moreover, increased Shh ligand may serve as a paracrine factor that allows neighboring cells to become more easily infected by promoting a “transitional” phenotype, exposing the gap junction complexes to facilitate viral entry and/or altering the intracellular environment.
The association between HCV and the Hh pathway will not alone settle the vigorous debate regarding the occurrence of EMT within the liver(17, 19–21, 32). The results we present should be examined when considering the cellular environment that is most conducive for replication. Given that HCV has evolved to infect humans over a long period of time, like other viruses, it has developed patterns of infection that most benefit its propagation and/or persistence. Recent data suggests that de-differentiated cells within hepatoma cell lines had increased Hh activity and demonstrated increased proliferation and invasion in a mouse xenograft model(33). Given the increased proliferation of these de-differentiated cells with increased Hh activity, one could hypothesize that HCV prefers this type of cell as a target because proliferation allows persistence of chronic infection.
The results of our studies have certain limitations that must be acknowledged. All of these observations have been made in vitro, and should be interpreted with caution when considering in vivo pathogenesis. Our in vitro evidence linking Hh signaling and viral infection are supported by observations that have been made about Hh pathway activity in liver samples from patients with chronic HCV(22). Another pertinent caveat is that most of our data relied upon real time PCR analysis to quantify RNA expression. We have included selected analyses of protein levels and infectious virus levels to support these findings and all have correlated with the RNA results. Finally, our findings do not indicate whether the association between HCV replication and Hh pathway activity is direct or indirect. Further exploration of changes in the microenvironment of Hh-responsive cells will be required to determine the specific proteins that mediate changes in cellular content of HCV RNA and the contribution of viral entry and replication in supporting this microenvironment.
In conclusion, we have discovered a significant association between HCV replication and Hh pathway activity that has not been previously described. The implications of this work are that subpopulations of hepatocytes may support disproportionately high levels of HCV replication and Hh-mediated effects on HCV replication may represent an important new therapeutic target or reveal other intracellular targets against HCV.
Supplementary Material
Comparison of relative RNA levels of LH 86 cells compared to Huh7.5 and Huh7 cells for: A) mesenchymal markers α–smooth muscle actin (α SMA) and Collagen 1α1 (Col1α1); B) Hedgehog pathway and downstream targets sonic hedgehog (Shh), glioblastoma 1 (Gli1), Patched (Ptc). Results are expressed as relative fold expression with Huh7 expression indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Protein lysates were created from primary human hepatic stellate cells (HSC, + control), Huh7.5 cells, Huh7 cells and LH86 cells. Antibodies to αSMA, Shh and α-tubulin were used for analysis.
Huh7 cells were incubated with vehicle control and interferon-α 500 U/ml. After 72 hours, relative RNA expression was analyzed for A) mesenchymal markers α–smooth muscle actin (α-SMA) and Collagen 1α1 (Col1α1); B) Hedgehog pathway and downstream targets sonic hedgehog (Shh) and glioblastoma 1 (Gli1). Results are expressed as relative fold expression with vehicle control sample indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Huh7.5 cells grown on 6-well plates were infected with 1500 focus-forming units of JFH1 HCV virus plus vehicle control (D), inactive analog (tomatidine (T) 5 μM,) or Hh inhibitor (cyclopamine (C) 5 μM,) RNA was isolated at 24 hours, 48 hours and 72 hours post-infection (p.i.). Relative RNA expression for HCV RNA and Gli1 were analyzed with virus-alone infected cells indexed at 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Huh7.5 cells grown on 6-well plates were mock infected (control), infected with 1500 focus-forming units of JFH1 HCV virus alone (JFH), or virus plus vehicle control (DMSO), inactive analog (tomatidine 5 μM) or Hh inhibitor (cyclopamine 5 μM) and incubated for 72 hours. Supernatant media was collected in triplicate samples and analyzed using commercial LDH release assay. Uninfected cells lysed with Triton-X were used as positive control. Results expressed as absorbance units.
Huh7.5 cells grown on 6-well plates were mock infected, infected with 1500 focus-forming units of JFH1 HCV virus plus vehicle control (DMSO), inactive analog (tomatidine 5 μM) or Hh inhibitor (cyclopamine 5 μM) and incubated for 72 hours. Supernatant media was collected as used to infect naïve Huh7.5 cells as described. Results expressed at log focus forming units/ml.
Huh7.5 cells were mock infected (−) or infected with JFH11 HCV alone (+). Cells were incubated and lysates collected at 2 and 5 days post infection. Protein lysates were analyzed by western blotting. Antibodies to HCV Core, Shh and α-tubulin were used for analysis.
Acknowledgments
Financial Support: NIH: SC: K08DK080980; SB: K01DK082613; RJ: K08DK076598, R03DK084120
The authors would like to thank Thiago Pereira, Rafal Witek and all members of the Diehl lab for their support of this work. We would also like to thank Michael Cohen-Wolkowiez for assistance with IC50 estimation, Carlos Goller for assistance with the LDH release assays and Stan Lemon, Patrick Seed, Raphael Valdivia, Bryan Cullen and Robert Mook for their constructive comments on this project and manuscript.
Contributor Information
Steve S. Choi, Email: steve.choi@duke.edu.
Shelton Bradrick, Email: sbrad@duke.edu.
Guan Qiang, Email: Qiang.guan@duke.edu.
Anahita Mostafavi, Email: anahita.mostafavi@duke.edu.
Gaurav Chaturvedi, Email: gchaturvedi@kumc.edu.
Steven A. Weinman, Email: sweinman@kumc.edu.
Anna Mae Diehl, Email: diehl004@mc.duke.edu.
Ravi Jhaveri, Email: ravi.jhaveri@duke.edu.
References
- 1.Seeff LB, Hollinger FB, Alter HJ, Wright EC, Cain CM, Buskell ZJ, Ishak KG, et al. Long-term mortality and morbidity of transfusion-associated non-A, non-B, and type C hepatitis: A National Heart, Lung, and Blood Institute collaborative study. Hepatology. 2001;33:455–463. doi: 10.1053/jhep.2001.21905. [DOI] [PubMed] [Google Scholar]
- 2.Tremolada F, Casarin C, Alberti A, Drago C, Tagger A, Ribero ML, Realdi G. Long-term follow-up of non-A, non-B (type C) post-transfusion hepatitis. J Hepatol. 1992;16:273–281. doi: 10.1016/s0168-8278(05)80657-9. [DOI] [PubMed] [Google Scholar]
- 3.Liang Y, Shilagard T, Xiao SY, Snyder N, Lau D, Cicalese L, Weiss H, et al. Visualizing hepatitis C virus infections in human liver by two-photon microscopy. Gastroenterology. 2009;137:1448–1458. doi: 10.1053/j.gastro.2009.07.050. [DOI] [PubMed] [Google Scholar]
- 4.Stiffler JD, Nguyen M, Sohn JA, Liu C, Kaplan D, Seeger C. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H, Dong H, Eksioglu E, Hemming A, Cao M, Crawford JM, Nelson DR, et al. Hepatitis C virus triggers apoptosis of a newly developed hepatoma cell line through antiviral defense system. Gastroenterology. 2007;133:1649–1659. doi: 10.1053/j.gastro.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 8.Sumpter R, Jr, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, et al. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binder M, Kochs G, Bartenschlager R, Lohmann V. Hepatitis C virus escape from the interferon regulatory factor 3 pathway by a passive and active evasion strategy. Hepatology. 2007;46:1365–1374. doi: 10.1002/hep.21829. [DOI] [PubMed] [Google Scholar]
- 10.Feigelstock DA, Mihalik KB, Kaplan G, Feinstone SM. Increased susceptibility of Huh7 cells to HCV replication does not require mutations in RIG-I. Virol J. 2010;7:44. doi: 10.1186/1743-422X-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekker SC, Ungar AR, Greenstein P, von Kessler DP, Porter JA, Moon RT, Beachy PA. Patterning activities of vertebrate hedgehog proteins in the developing eye and brain. Curr Biol. 1995;5:944–955. doi: 10.1016/s0960-9822(95)00185-0. [DOI] [PubMed] [Google Scholar]
- 12.Krauss S, Concordet JP, Ingham PW. A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell. 1993;75:1431–1444. doi: 10.1016/0092-8674(93)90628-4. [DOI] [PubMed] [Google Scholar]
- 13.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 14.Fleig SV, Choi SS, Yang L, Jung Y, Omenetti A, VanDongen HM, Huang J, et al. Hepatic accumulation of Hedgehog-reactive progenitors increases with severity of fatty liver damage in mice. Lab Invest. 2007;87:1227–1239. doi: 10.1038/labinvest.3700689. [DOI] [PubMed] [Google Scholar]
- 15.Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, Zubiaga AM, et al. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology. 51:1712–1723. doi: 10.1002/hep.23525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, Ludlow JW, et al. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27:748–757. doi: 10.1093/carcin/bgi292. [DOI] [PubMed] [Google Scholar]
- 17.Choi SS, Omenetti A, Witek RP, Moylan CA, Syn WK, Jung Y, Yang L, et al. Hedgehog pathway activation and epithelial-to-mesenchymal transitions during myofibroblastic transformation of rat hepatic cells in culture and cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2009;297:G1093–1106. doi: 10.1152/ajpgi.00292.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sicklick JK, Li YX, Melhem A, Schmelzer E, Zdanowicz M, Huang J, Caballero M, et al. Hedgehog signaling maintains resident hepatic progenitors throughout life. Am J Physiol Gastrointest Liver Physiol. 2006;290:G859–870. doi: 10.1152/ajpgi.00456.2005. [DOI] [PubMed] [Google Scholar]
- 19.Syn WK, Jung Y, Omenetti A, Abdelmalek M, Guy CD, Yang L, Wang J, et al. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137:1478–1488. e1478. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taura K, Miura K, Iwaisako K, Osterreicher CH, Kodama Y, Penz-Osterreicher M, Brenner DA. Hepatocytes do not undergo epithelial-mesenchymal transition in liver fibrosis in mice. Hepatology. 51:1027–1036. doi: 10.1002/hep.23368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeisberg M, Yang C, Martino M, Duncan MB, Rieder F, Tanjore H, Kalluri R. Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J Biol Chem. 2007;282:23337–23347. doi: 10.1074/jbc.M700194200. [DOI] [PubMed] [Google Scholar]
- 22.de Almeida Pereira T, Witek RP, Syn WK, Choi SS, Bradrick S, Karaca GF, Agboola KM, et al. Viral factors induce Hedgehog pathway activation in humans with viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab Invest. 2010 doi: 10.1038/labinvest.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mee CJ, Farquhar MJ, Harris HJ, Hu K, Ramma W, Ahmed A, Maurel P, et al. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology. 2010;138:1134–1142. doi: 10.1053/j.gastro.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 25.Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. Cell culture and infection system for hepatitis C virus. Nat Protoc. 2006;1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- 26.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robarge KD, Brunton SA, Castanedo GM, Cui Y, Dina MS, Goldsmith R, Gould SE, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19:5576–5581. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 28.Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong H, Chen JZ, Chou B, Halladay JS, Kenny JR, La H, Marsters JC, et al. Preclinical assessment of the absorption, distribution, metabolism and excretion of GDC-0449 (2-chloro-N-(4-chloro-3-(pyridin-2-yl)phenyl)-4-(methylsulfonyl)benzamide), an orally bioavailable systemic Hedgehog signalling pathway inhibitor. Xenobiotica. 2009 doi: 10.3109/00498250903180289. [DOI] [PubMed] [Google Scholar]
- 30.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu AS, Diaz R, Hui JJ, Yanger K, Zong Y, Alpini G, Stanger BZ, et al. Lineage tracing demonstrates no evidence of cholangiocyte epithelial-to-mesenchymal transition in murine models of hepatic fibrosis. Hepatology. 2011;53:1685–1695. doi: 10.1002/hep.24206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X, Lingala S, Khoobyari S, Nolta J, Zern MA, Wu J. Epithelial Mesenchymal Transition and Hedgehog Signaling Activation are Associated with Chemoresistance and Invasion of Hepatoma Subpopulations. J Hepatol. 2011 doi: 10.1016/j.jhep.2010.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of relative RNA levels of LH 86 cells compared to Huh7.5 and Huh7 cells for: A) mesenchymal markers α–smooth muscle actin (α SMA) and Collagen 1α1 (Col1α1); B) Hedgehog pathway and downstream targets sonic hedgehog (Shh), glioblastoma 1 (Gli1), Patched (Ptc). Results are expressed as relative fold expression with Huh7 expression indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Protein lysates were created from primary human hepatic stellate cells (HSC, + control), Huh7.5 cells, Huh7 cells and LH86 cells. Antibodies to αSMA, Shh and α-tubulin were used for analysis.
Huh7 cells were incubated with vehicle control and interferon-α 500 U/ml. After 72 hours, relative RNA expression was analyzed for A) mesenchymal markers α–smooth muscle actin (α-SMA) and Collagen 1α1 (Col1α1); B) Hedgehog pathway and downstream targets sonic hedgehog (Shh) and glioblastoma 1 (Gli1). Results are expressed as relative fold expression with vehicle control sample indexed to 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Huh7.5 cells grown on 6-well plates were infected with 1500 focus-forming units of JFH1 HCV virus plus vehicle control (D), inactive analog (tomatidine (T) 5 μM,) or Hh inhibitor (cyclopamine (C) 5 μM,) RNA was isolated at 24 hours, 48 hours and 72 hours post-infection (p.i.). Relative RNA expression for HCV RNA and Gli1 were analyzed with virus-alone infected cells indexed at 1. Key to statistical comparisons: *p<.05, ** p<.01, † p<.005.
Huh7.5 cells grown on 6-well plates were mock infected (control), infected with 1500 focus-forming units of JFH1 HCV virus alone (JFH), or virus plus vehicle control (DMSO), inactive analog (tomatidine 5 μM) or Hh inhibitor (cyclopamine 5 μM) and incubated for 72 hours. Supernatant media was collected in triplicate samples and analyzed using commercial LDH release assay. Uninfected cells lysed with Triton-X were used as positive control. Results expressed as absorbance units.
Huh7.5 cells grown on 6-well plates were mock infected, infected with 1500 focus-forming units of JFH1 HCV virus plus vehicle control (DMSO), inactive analog (tomatidine 5 μM) or Hh inhibitor (cyclopamine 5 μM) and incubated for 72 hours. Supernatant media was collected as used to infect naïve Huh7.5 cells as described. Results expressed at log focus forming units/ml.
Huh7.5 cells were mock infected (−) or infected with JFH11 HCV alone (+). Cells were incubated and lysates collected at 2 and 5 days post infection. Protein lysates were analyzed by western blotting. Antibodies to HCV Core, Shh and α-tubulin were used for analysis.