Abstract
With many advantages over other therapeutic agents such as monoclonal antibodies, aptamers have recently emerged as a novel and powerful class of ligands with excellent potential for diagnostic and therapeutic applications. Typically generated through Systematic Evolution of Ligands by EXponential enrichment (SELEX), aptamers have been selected against a wide range of targets such as proteins, phospholipids, sugars, nucleic acids, as well as whole cells. DNA/RNA aptamers are single-stranded DNA/RNA oligonucleotides (with a molecular weight of 5–40 kDa) that can fold into well-defined 3D structures and bind to their target molecules with high affinity and specificity. A number of strategies have been adopted to synthesize aptamers with enhanced in vitro/in vivo stability, aiming at potential therapeutic/diagnostic applications in the clinic. In cardiovascular diseases, aptamers can be developed into therapeutic agents as anti-thrombotics, anti-coagulants, among others. This review focuses on aptamers that were selected against various molecular targets involved in cardiovascular diseases: von Willebrand factor (vWF), thrombin, factor IX, phospholamban, P-selectin, platelet-derived growth factor, integrin αvβ3, CXCL10, vasopressin, among others. With continued effort in the development of aptamer-based therapeutics, aptamers will find their niches in cardiovascular diseases and significantly impact clinical patient management.
Keywords: Aptamers, cardiovascular diseases, von Willebrand factor (vWF), thrombin, factor IX, DNA, RNA, peptide aptamer
INTRODUCTION
Aptamers, typically generated through Systematic Evolution of Ligands by EXponential enrichment (SELEX) [1,2], have quickly emerged as a novel and powerful class of ligands with excellent potential for diagnostic and therapeutic applications [3,4]. To date, aptamers have been selected against a wide range of targets such as proteins (e.g. cytokines, proteases, kinases, cell-surface receptors, and cell-adhesion molecules), phospholipids, sugars, nucleic acids, as well as whole cells [3,5–7]. In general, aptamers can be classified into two major categories: DNA/RNA aptamers and peptide aptamers. DNA/RNA aptamers are single-stranded DNA/RNA oligonucleotides (with a molecular weight of 5–40 kDa) that can fold into well-defined 3D structures and bind to their target molecules with high affinity and specificity. Since wild-type RNA and DNA molecules can be easily degraded by nucleases, various strategies have been adopted to synthesize aptamers with enhanced in vitro/in vivo stability, such as the use of chemically modified oligonucleotides [8–10], unnatural internucleotide linkages [11], polyethylene glycol (PEG) conjugation [12], Spiegelmers (where the sugars are enantiomers of wild-type nucleic acid sugars) [13,14], among others [3].
A peptide aptamer is consisted of a variable peptide loop attached at both ends to a protein scaffold [15,16]. This double structural constraint significantly increases the binding affinity of peptide aptamers to levels comparable to those of antibodies. The variable loop is typically composed of ten to twenty amino acids, while the scaffold may be any protein with good solubility and compacity properties. The first report on peptide aptamers appeared in 1996, where peptide aptamers recognizing different epitopes on the cyclin-dependent kinase 2 (Cdk2) surface were selected with dissociation constants (Kd) in the nanomolar (nM) range [17]. Comparing with DNA/RNA aptamers, the field of peptide aptamers is still in its infancy, but has gradually emerged as an attractive area for chemists as well as biologists in recent years.
Aptamers possess several advantages over other therapeutic agents such as monoclonal antibodies. First, production of aptamers does not rely on biological systems hence is much easier to scale up with low batch-to-batch variability; Second, aptamers are quite thermally stable and can be denatured and renatured multiple times without significant loss of activity [18]; Third, lack of immunogenicity is another favorable advantage of aptamers over antibodies; Lastly, conjugation chemistry for the attachment of various imaging labels or functional groups to aptamers are orthogonal to nucleic acid chemistry, hence they can be readily introduced during aptamer synthesis. On the other hand, the disadvantages of aptamers include faster excretion than antibodies due to smaller size, potentially weaker binding to targets than antibodies, unpredictable toxicity and other systemic properties, susceptibility to serum degradation when unmodified aptamers are used, and intellectual property-related issues [3].
During the last two decades since aptamers were first selected through SELEX [1,2], Pegaptanib (Pfizer/Eyetech), an aptamer that binds to human vascular endothelial growth factor (VEGF), has been approved by the Food and Drug Administration (FDA) for clinical use in treating age-related macular degeneration (AMD) [19]. In addition, a variety of aptamers against other molecular targets are currently in clinical investigation [3]. For applications in cardiovascular diseases, various aptamers can potentially be translated into diagnostics and/or therapeutics for identification and management of patients with or at risk for cardiovascular disorders. For example, aptamers can be used as anti-thrombotics and anti-coagulants, which have been reviewed elsewhere [20–23]. In this review, we will only focus on aptamers selected against various cardiovascular disease-related molecular targets: von Willebrand factor (vWF), thrombin, factor IX, among many others.
APTAMERS THAT BIND TO VWF
VWF, a blood glycoprotein synthesized by endothelial cells, megakaryocytes, and subendothelial connective tissue, plays a pivotal role in both hemostasis and thrombosis [24]. Platelets could be activated by vWF binding (the A1 domain of vWF binds to the glycoprotein Ib [GPIb] receptor on platelets) and undergo conformational change, which leads to thrombosis formation at sites of cardiovascular injury [25]. Plasma level of vWF, a predictive indicator of the state of endothelial damage, has been proposed as a marker for cardiovascular risk in patients with acute coronary syndrome (ACS) [26]. Furthermore, mounting preclinical/clinical evidence has suggested that vWF is a promising target for developing new therapeutic agents in cardiovascular diseases [27].
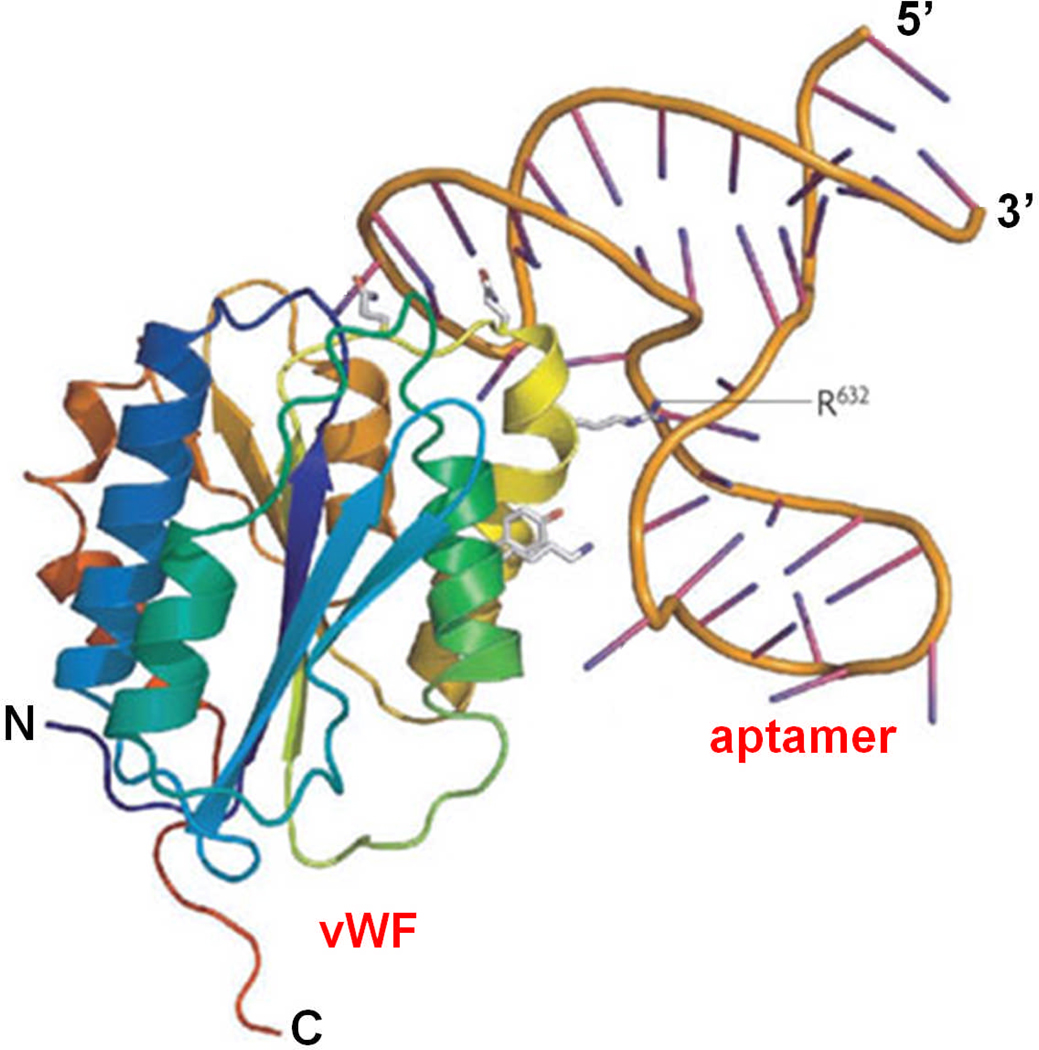
ARC1779 (Archemix Corporation) is an aptamer that binds to the A1 domain of activated vWF with high affinity (Kd of ~2 nM), thereby inhibiting its interaction with the GPIb receptor on platelets (Fig. (1)) [28]. Since vWF is an important mediator in platelet adhesion and aggregation at sites of high shear rates in the arterial side of circulation (e.g. in ACS or ruptured atherosclerotic plaque lesions), ARC1779 can lead to an anti-thrombotic effect without causing significant anti-coagulation. In a Phase I clinical trial with 47 healthy volunteers, ARC1779 exhibited dose- and concentration-dependent inhibition of vWF activity and vWF-dependent platelet function [28].
Fig. (1).
Crystal structure of the all-DNA parent of ARC1779 bound to the A1 domain of von Willebrand factor (vWF). Adapted from [3].
Subsequently, ARC1779 was investigated in acute myocardial infarction (AMI) in an ex vivo dosing study [29]. Since vWF level is increased in the elderly and in the setting of AMI, as reflected by increased shear-dependent platelet function, the goal of this study was to evaluate whether ARC1779 can inhibit ex vivo platelet function. It was found that potent and specific inhibition of vWF activity and vWF-dependent platelet function could be achieved, even in the setting of AMI where vWF activity was increased. In another report, in vitro inhibition of vWF-induced platelet aggregation, as well as in vivo activity of ARC1779 was assessed in a cynomolgus monkey carotid electrical injury thrombosis model [30]. ARC1779 not only inhibited platelet aggregation and reduced adhesion of platelets to collagen-coated matrices, but also inhibited the formation of occlusive thrombi in cynomolgus monkeys. Both of these studies clearly suggested that ARC1779 has therapeutic potential as an anti-vWF agent in vWF-mediated thrombosis such as ACS.
Recently, the effect of ARC1779 on platelet function in patients with coronary artery disease (CAD) on double anti-platelet therapy was investigated [31]. In contrast to abciximab (a monoclonal antibody commonly used for CAD treatment [32]), ARC1779 did not significantly affect platelet aggregation, P-selectin expression, or platelet-leukocyte interaction. With the wide use of anti-platelet drugs in anti-thrombosis surgery, hemorrhage has become one of the major complications [33]. The promising findings in this study suggested that ARC1779 may be a potential replacement for traditional anti-platelet drugs [31]. Besides theses abovementioned reports, ARC1779 has also been evaluated in thrombotic thrombocytopenic purpura (in which ultralarge vWF multimers bind to platelet GPIb and lead to the formation of disseminated fibrin-poor, vWF-rich platelet thrombi) [34–36] and type 2B von Willebrand disease [37].
ARC1172 (Archemix Corporation), a 41-mer DNA aptamer, also binds to the A1 domain of vWF [38]. A derivative of ARC1172, developed to increase intravascular survival, was shown to be able to inhibit carotid artery thrombosis in a cynomolgus macaque model, as well as vWF-dependent platelet aggregation in humans. In another report, a vWF-binding RNA aptamer was demonstrated to inhibit vWF-mediated platelet adhesion and aggregation [39]. In addition, an antidote molecule was designed to reverse the effect of the aptamer and restore platelet function quickly and effectively over a clinically relevant period. Such an aptamer-antidote pair represents a reversible anti-platelet agent, an important step towards safer drugs in the clinic.
APTAMERS THAT BIND TO THROMBIN
Thrombin (also known as coagulation factor II), a serine protease that plays multiple roles in endothelial and smooth muscle cell functions as well as coagulation and hemostasis, can convert soluble fibrinogen into insoluble strands of fibrin thereby producing fibrin clots [40,41]. In addition, thrombin can also promote platelet activation and aggregation. Thrombotic disorders and their common clinical phenotypes of AMI, ischemic stroke, and venous thromboembolism cause substantial health care expenditures, morbidity, and mortality worldwide. Inhibition of thrombin in vivo, such as the use of aptamers [42], is a promising method to prevent and/or treat these diseases [43,44].
The first report of aptamers with thrombin-specific inhibitory activities appeared almost two decades ago [45]. Subsequently, other DNA aptamers were also selected as thrombin inhibitors, which can compete with their exosite binding substrates such as fibrinogen and the platelet thrombin receptor [46]. A short-acting aptamer which binds to thrombin was tested in a canine cardiopulmonary bypass (CPB) model to determine its anti-coagulant efficacy and potential as a substitute for heparin in CPB and other clinical situations [47]. It was found that this aptamer was both safe and effective as an anti-coagulant in the canine CPB model with predictable pharmacokinetics. Combinatorial libraries and protein engineering was also employed to produce a thrombin binding aptamer, which exhibited potent and rapid anti-coagulation effect and was successfully used to replace heparin in a canine CPB model [48].
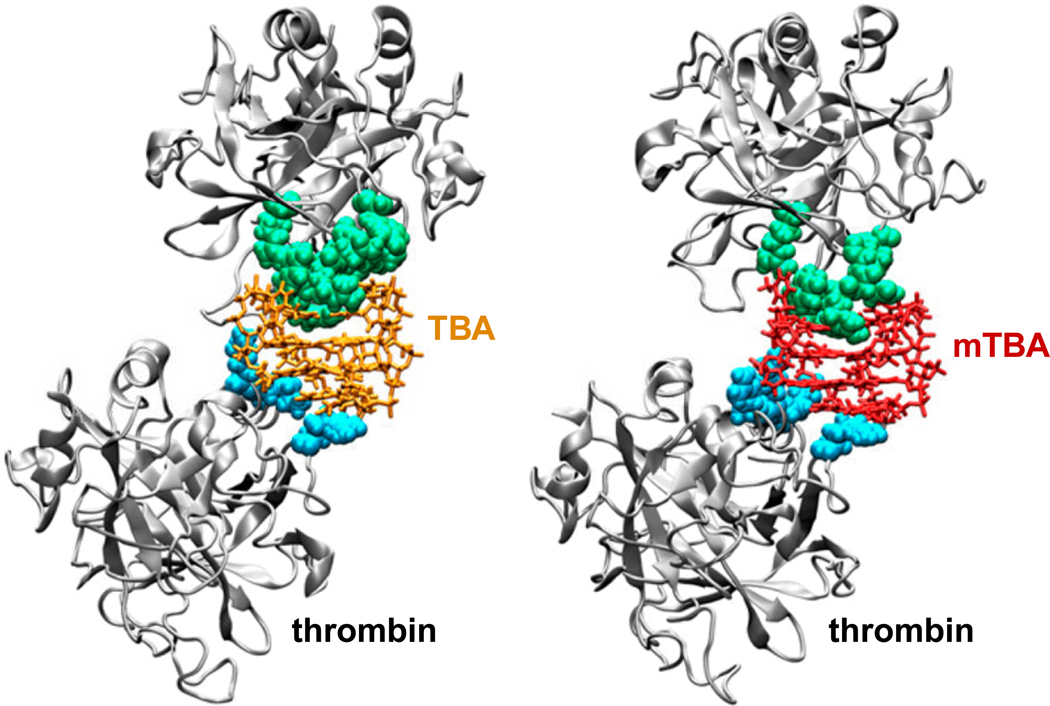
The thrombin binding aptamer (TBA) folds into a well-defined quadruplex structure and binds to its target with good specificity and affinity [49,50]. A recent study showed that a modified TBA (mTBA), which also folds into a quadruplex structure, was more stable and exhibited higher thrombin binding affinity than TBA [51]. Molecular dynamics simulation studies of the aptamers provided in-depth information about the different stability behaviors of the molecules. In addition, structural models of complexes with thrombin molecules provided a detailed view of the interactions occurring between each aptamer and its target (Fig. (2)), suggesting that detailed analysis of the molecular recognition process is fundamental for rational design of aptamers.
Fig. (2).
Structural models of the complex of thrombin binding aptamer (TBA) or modified thrombin binding aptamer (mTBA) with two thrombin molecules. The interacting residues of exosite I (green) and exosite II (cyan) are shown. Adapted from [51].
The consensus thrombin aptamer, C15-mer, is a single-strand DNA of 15 nucleotides selected from a large combinatorial library of oligonucleotides [45]. In a recent study, various analogs of the original thrombin-inhibiting sequence, in which some of the thymidylate residues were replaced by 4-thio-deoxyuridylates, were synthesized and thoroughly investigated [52]. It was found that replacement of four thymidylate residues in C15-mer by 4-thio-deoxyuridylates resulted in a new thrombin aptamer with increased anti-coagulant and anti-thrombotic properties.
Taken together, these studies confirmed that thrombin-specific aptamers can be promising anti-thrombotic agents [53]. To the best of our knowledge, two thrombin-specific DNA aptamers have entered clinical investigation. ARCA Biopharma, Inc. is sponsoring a Phase 2 study of NU172 for anti-coagulation in patients undergoing off-pump coronary artery bypass graft surgery (http://clinicaltrials.gov). ARC183 (Archemix Corporation), a direct thrombin inhibitor, has also been tested in a Phase 1 clinical trial for potential use in acute cardiovascular procedures. However, limited information is available in the literature.
APTAMERS THAT BIND TO FACTOR IX
Another attractive target in cardiovascular diseases is factor IX, a serine protease that plays an important role in generating the critical quantity of thrombin necessary in coagulation [54,55]. Factor IX is produced as a zymogen, an inactive precursor. When activated into factor IXa in the presence of Ca2+, membrane phospholipids, and a Factor VIII cofactor, it hydrolyzes one arginine-isoleucine bond in factor X to form factor Xa, which can convert prothrombin into thrombin [56].
In one study, an aptamer-antidote pair targeting factor IXa was investigated in a porcine CPB model, which did not cause adverse effects on thrombin generation, inflammation, and cardiac physiology that are typically associated with the use of heparin or protamine [57]. It was suggested that this anti-coagulant-antidote pair may be useful in patients diagnosed with heparin-induced thrombocytopenia or those who have been sensitized to protamine, particularly patients with insulin-dependent diabetes.
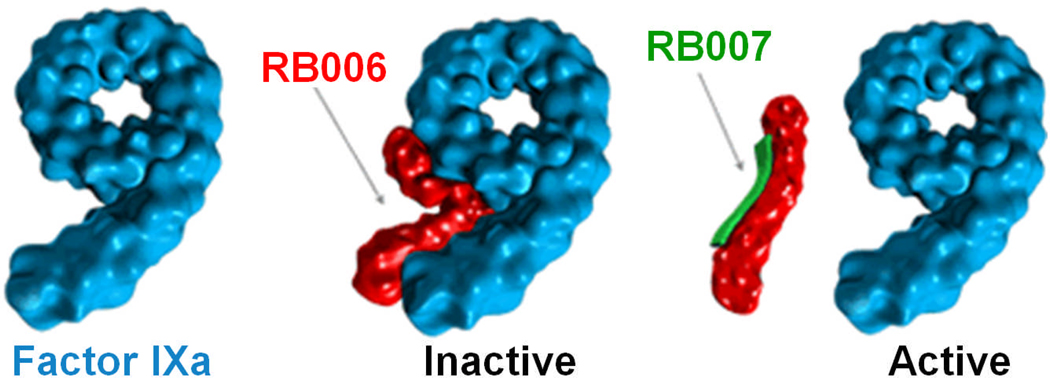
REG1 is an anti-coagulation system (Fig. (3)) that includes a factor IXa-specific aptamer (RB006) and its oligonucleotide antidote (RB007) [58]. RB006 was initially discovered in a 2′-ribo purine/2′-fluoro pyrimidine transcript library using SELEX [9]. The aptamer was then truncated to 34 nucleotides, conjugated to a 40 kDa PEG to reduce the renal clearance rate, and capped at the 3′-terminus with an inverted nucleotide to reduce 3′-exonuclease-mediated degradation. RB006 binds to factor IXa with a Kd of 2.8 nM while RB007 is a fully 2′-O-methyl substituted 17-mer oligonucleotide that is complementary to the 5′-terminal region of RB006. RB007 rapidly disrupts the structure of RB006 and inhibits its anti-coagulation function, thereby serving as an antidote.
Fig. (3).
REG1 is an anti-coagulation system that includes a factor IXa-specific aptamer (RB006) and its oligonucleotide antidote (RB007).
To determine the safety profile and characterize the pharmacodynamic responses of REG1, a subject-blinded, placebo-controlled Phase 1a study was carried out [58]. Eight-five healthy volunteers were randomized to each receive a bolus of drug or placebo followed 3 hours later by a bolus of antidote or placebo. No significant differences were found in median hemoglobin, platelet, creatinine, or liver function parameters. Both the drug (i.e. RB006) and the antidote (i.e. RB007) were well tolerated and no significant bleeding was associated with RB006. Subsequently, multiple repeat-dose safety, intra-individual pharmacodynamic reproducibility, and graded active reversibility of REG1 was also investigated in another study with 39 healthy volunteers [59]. Afterwards, a phase Ib study in patients with stable CAD [60] and a phase IIa trial in patients undergoing elective percutaneous coronary intervention [61] of REG1 both gave satisfactory results, demonstrating clinical safety and predictable pharmacodynamic effects [62]. Detailed pharmacokinetic and pharmacodynamic modeling of REG1 has been performed, with a focus on the level of target inhibition, to translate phase 1 results to phase 2 dose selection [63], which could guide the currently ongoing phase IIb trial in ACS patients undergoing cardiac catheterization [64]. This phase IIb trial is expected to answer important questions about each component of the REG1 anti-coagulation system and define the best strategy to support the development of adequately powered phase III clinical trials. Findings from this trial are eagerly awaited.
APTAMERS THAT BIND TO OTHER TARGETS
Besides vWF, thrombin, and factor IX(a), aptamers that specifically bind to various other targets can potentially be used for therapeutic applications in cardiovascular diseases as well. Mesenchymal stem cells (MSCs) represent a promising cell source for cellular cardiomyoplasty [65,66]. Aptamer-based selection of MSCs was employed to provide “ready to transplant” cells directly after isolation [67]. Tracking of newly isolated and freshly transplanted MSCs with magnetic resonance imaging was carried out in a porcine heart perfusion model, after aptamer-based isolation and appropriate labeling of the cells. This study demonstrated that isolating MSCs by MSC-specific aptamers linked to magnetic particles is feasible and effective, which combines specific separation and labeling into a “one stop shop” strategy.
In another study, aptamers that can capture CD31-positive endothelial cells were used for coating of intracoronary stents to improve endothelialization and vascular healing [68]. However, after implanted in farm-raised swine, stents coated with endothelial cell-capturing aptamers showed similar late loss and angiographic restenosis rates as uncoated stents. No advantage of aptamer-coating on neointimal proliferation of intracoronary stents was observed. A number of studies have also reported aptamers that can bind to other cardiovascular disease-related targets such as phospholamban (a sarcoplasmic reticulum membrane protein) [69], P-selectin [70,71], platelet-derived growth factor [72], integrin αvβ3 [73], advanced glycation end products [74], CXCL10 [75], vasopressin [76], among many others [3].
CONCLUSIONS
A variety of aptamers that can bind to molecular targets relevant to cardiovascular diseases have been selected and optimized, many of which have already entered clinical trials (Table 1). The major limitations of existing therapies/therapeutic agents include target non-selectivity, variable onset and offset of pharmacodynamic effects, a narrow efficacy-safety profile, and the absence of a safe and reliable platform for either accurate titration (based on existing patient-specific, disease-specific, and clinical conditions) or active reversibility. From anti-platelet to intracoronary stent coating, aptamers possess enormous potential for biomedical applications in treating cardiovascular diseases. Perhaps aptamers can be considered as “chemical antibodies”, in the sense that they offer high specificity and affinity in a relatively small, chemically synthesized molecule free from cell culture-derived contaminants, yet they do not elicit immunogenic responses in vivo.
Table 1.
Aptamers that have entered clinical investigation in cardiovascular diseases.
| Aptamer | Target | Company | Development Stage |
|---|---|---|---|
| ARC1779 | vWF | Archemix | Phase I, II |
| ARC1172 | vWF | Archemix | Phase I |
| ARC183 | Thrombin | Archemix | Phase I |
| NU172 | Thrombin | ARCA Biopharma, Inc. | Phase I, II |
| REG1 (RB006/RB007) | Factor IXa | Regado Biosciences | Phase I, IIa, IIb |
The development of more efficient selection methods and easier strategies for aptamer conjugation are needed for potential clinical translation. To date, researchers have identified high affinity aptamers that bind to a broad array of targets. Since aptamers can be selected against most protein targets, the possible therapeutic applications with aptamers or aptamer-based agents range far and wide. One disadvantage of aptamers is the hard-to-predict and poorly understood pharmacokinetics, toxicity, and other properties upon systemic delivery. Therefore, although aptamers are unlikely to elicit immunogenic responses, the safety profile of each individual aptamer (conjugate) must be examined carefully. Off-target effects due to inhibition/activation of other proteins with similar structural conformations as the target protein need to be investigated empirically since they will likely differ for each aptamer. With more and more academic groups and commercial entities getting involved in this area of research after the earliest intellectual property on aptamers expires [77], the development of aptamer-based therapeutics will speed up and aptamers will find their own niches in treating cardiovascular diseases and significantly impact clinical patient management.
ACKNOWLEDGEMENTS
The authors acknowledge financial support from the University of Wisconsin Carbone Cancer Center, NCRR 1UL1RR025011, a DOD BCRP Postdoctoral Fellowship, and a DOD PCRP IDEA Award.
REFERENCES
- 1.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gold L, Janjic N, Jarvis T, Schneider D, Walker JJ, Wilcox SK, Zichi D. Aptamers and the RNA World, Past and Present. Cold Spring Harb. Perspect. Biol. 2010 doi: 10.1101/cshperspect.a003582. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiel KW, Giangrande PH. Therapeutic applications of DNA and RNA aptamers. Oligonucleotides. 2009;19:209–222. doi: 10.1089/oli.2009.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X, Tan W. Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach. Acc. Chem. Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer G. The chemical biology of aptamers. Angew. Chem. Int. Ed. Engl. 2009;48:2672–2689. doi: 10.1002/anie.200804643. [DOI] [PubMed] [Google Scholar]
- 8.Lin Y, Nieuwlandt D, Magallanez A, Feistner B, Jayasena SD. High-affinity and specific recognition of human thyroid stimulating hormone (hTSH) by in vitro-selected 2'-amino-modified RNA. Nucleic Acids Res. 1996;24:3407–3414. doi: 10.1093/nar/24.17.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D, Sullenger BA. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 10.Pagratis NC, Bell C, Chang YF, Jennings S, Fitzwater T, Jellinek D, Dang C. Potent 2'-amino-, and 2'-fluoro-2'-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 1997;15:68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 11.King DJ, Safar JG, Legname G, Prusiner SB. Thioaptamer interactions with prion proteins: sequence-specific and non-specific binding sites. J. Mol. Biol. 2007;369:1001–1014. doi: 10.1016/j.jmb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Takafuji Y, Jo JI, Tabata Y. Simple PEG Modification of DNA Aptamer Based on Copper Ion Coordination for Tumor Targeting. J. Biomater. Sci. Polym. Ed. 2010 doi: 10.1163/092050610X501713. Epub. [DOI] [PubMed] [Google Scholar]
- 13.Eulberg D, Klussmann S. Spiegelmers: biostable aptamers. Chembiochem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 14.Vater A, Klussmann S. Toward third-generation aptamers: Spiegelmers and their therapeutic prospects. Curr. Opin. Drug Discov. Devel. 2003;6:253–261. [PubMed] [Google Scholar]
- 15.Borghouts C, Kunz C, Groner B. Peptide aptamers: recent developments for cancer therapy. Expert Opin. Biol. Ther. 2005;5:783–797. doi: 10.1517/14712598.5.6.783. [DOI] [PubMed] [Google Scholar]
- 16.Hoppe-Seyler F, Crnkovic-Mertens I, Tomai E, Butz K. Peptide aptamers: specific inhibitors of protein function. Curr. Mol. Med. 2004;4:529–538. doi: 10.2174/1566524043360519. [DOI] [PubMed] [Google Scholar]
- 17.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 18.Liss M, Petersen B, Wolf H, Prohaska E. An aptamer-based quartz crystal protein biosensor. Anal. Chem. 2002;74:4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 19.Ng EW, Shima DT, Calias P, Cunningham ET, Jr, Guyer DR, Adamis AP. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 20.Keefe AD, Schaub RG. Aptamers as candidate therapeutics for cardiovascular indications. Curr. Opin. Pharmacol. 2008;8:147–152. doi: 10.1016/j.coph.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 21.Kuliczkowski W, Floyd J, Malinin A, Serebruany V. Aptamers: the emerging class of future anticoagulation for vascular disease. Expert Rev. Cardiovasc. Ther. 2010;8:503–507. doi: 10.1586/erc.09.182. [DOI] [PubMed] [Google Scholar]
- 22.White RR, Sullenger BA, Rusconi CP. Developing aptamers into therapeutics. J. Clin. Invest. 2000;106:929–934. doi: 10.1172/JCI11325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker RC, Oney S, Becker KC, Rusconi CP, Sullenger B. Nucleic acid aptamers and their complimentary antidotes. Entering an era of antithrombotic pharmacobiologic therapy. Hamostaseologie. 2007;27:378–382. [PubMed] [Google Scholar]
- 24.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 25.Moake JL, Chow TW. Increased von Willebrand factor (vWf) binding to platelets associated with impaired vWf breakdown in thrombotic thrombocytopenic purpura. J. Clin. Apher. 1998;13:126–132. doi: 10.1002/(sici)1098-1101(1998)13:3<126::aid-jca6>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 26.Lip GY, Blann A. von Willebrand factor: a marker of endothelial dysfunction in vascular disorders? Cardiovasc. Res. 1997;34:255–265. doi: 10.1016/s0008-6363(97)00039-4. [DOI] [PubMed] [Google Scholar]
- 27.Paulinska P, Spiel A, Jilma B. Role of von Willebrand factor in vascular disease. Hamostaseologie. 2009;29:32–38. [PubMed] [Google Scholar]
- 28.Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, Horvath CJ, Merlino PG, Marsh HN, Healy JM, Boufakhreddine S, Holohan TV, Schaub RG. First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation. 2007;116:2678–2686. doi: 10.1161/CIRCULATIONAHA.107.724864. [DOI] [PubMed] [Google Scholar]
- 29.Spiel AO, Mayr FB, Ladani N, Wagner PG, Schaub RG, Gilbert JC, Jilma B. The aptamer ARC1779 is a potent and specific inhibitor of von Willebrand Factor mediated ex vivo platelet function in acute myocardial infarction. Platelets. 2009;20:334–340. doi: 10.1080/09537100903085927. [DOI] [PubMed] [Google Scholar]
- 30.Diener JL, Daniel Lagasse HA, Duerschmied D, Merhi Y, Tanguay JF, Hutabarat R, Gilbert J, Wagner DD, Schaub R. Inhibition of von Willebrand factor-mediated platelet activation and thrombosis by the anti-von Willebrand factor A1-domain aptamer ARC1779. J. Thromb. Haemost. 2009;7:1155–1162. doi: 10.1111/j.1538-7836.2009.03459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arzamendi D, Dandachli F, Theoret JF, Ducrocq G, Chan M, Mourad W, Gilbert JG, Schaub RG, Tanguay JF, Merhi Y. An Anti-von Willebrand Factor Aptamer Reduces Platelet Adhesion Among Patients Receiving Aspirin and Clopidogrel in an Ex Vivo Shear-Induced Arterial Thrombosis. Clin. Appl. Thromb. Hemost. 2010 doi: 10.1177/1076029610384114. Epub. [DOI] [PubMed] [Google Scholar]
- 32.Gabriel HM, Oliveira EI. Role of abciximab in the treatment of coronary artery disease. Expert Opin. Biol. Ther. 2006;6:935–942. doi: 10.1517/14712598.6.9.935. [DOI] [PubMed] [Google Scholar]
- 33.Chassot PG, Delabays A, Ravussin P, Spahn DR. Antiplatelet drugs and intraoperative hemorrhage. Rev. Med. Suisse. 2006;2:2684–2687. [PubMed] [Google Scholar]
- 34.Jilma-Stohlawetz P, Gorczyca ME, Jilma B, Siller-Matula J, Gilbert JC, Knobl P. Inhibition of von Willebrand factor by ARC1779 in patients with acute thrombotic thrombocytopenic purpura. Thromb. Haemost. 2011;105:545–552. doi: 10.1160/TH10-08-0520. [DOI] [PubMed] [Google Scholar]
- 35.Knobl P, Jilma B, Gilbert JC, Hutabarat RM, Wagner PG, Jilma-Stohlawetz P. Anti-von Willebrand factor aptamer ARC1779 for refractory thrombotic thrombocytopenic purpura. Transfusion. 2009;49:2181–2185. doi: 10.1111/j.1537-2995.2009.02232.x. [DOI] [PubMed] [Google Scholar]
- 36.Mayr FB, Knobl P, Jilma B, Siller-Matula JM, Wagner PG, Schaub RG, Gilbert JC, Jilma-Stohlawetz P. The aptamer ARC1779 blocks von Willebrand factor-dependent platelet function in patients with thrombotic thrombocytopenic purpura ex vivo. Transfusion. 2010;50:1079–1087. doi: 10.1111/j.1537-2995.2009.02554.x. [DOI] [PubMed] [Google Scholar]
- 37.Jilma B, Paulinska P, Jilma-Stohlawetz P, Gilbert JC, Hutabarat R, Knobl P. A randomised pilot trial of the anti-von Willebrand factor aptamer ARC1779 in patients with type 2b von Willebrand disease. Thromb. Haemost. 2010;104:563–570. doi: 10.1160/TH10-01-0027. [DOI] [PubMed] [Google Scholar]
- 38.Huang RH, Fremont DH, Diener JL, Schaub RG, Sadler JE. A structural explanation for the antithrombotic activity of ARC1172, a DNA aptamer that binds von Willebrand factor domain A1. Structure. 2009;17:1476–1484. doi: 10.1016/j.str.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oney S, Nimjee SM, Layzer J, Que-Gewirth N, Ginsburg D, Becker RC, Arepally G, Sullenger BA. Antidote-controlled platelet inhibition targeting von Willebrand factor with aptamers. Oligonucleotides. 2007;17:265–274. doi: 10.1089/oli.2007.0089. [DOI] [PubMed] [Google Scholar]
- 40.Lancellotti S, De Cristofaro R. Nucleotide-derived thrombin inhibitors: a new tool for an old issue. Cardiovasc. Hematol. Agents Med. Chem. 2009;7:19–28. doi: 10.2174/187152509787047658. [DOI] [PubMed] [Google Scholar]
- 41.Huntington JA. Molecular recognition mechanisms of thrombin. J. Thromb. Haemost. 2005;3:1861–1872. doi: 10.1111/j.1538-7836.2005.01363.x. [DOI] [PubMed] [Google Scholar]
- 42.Becker RC, Rusconi C, Sullenger B. Nucleic acid aptamers in therapeutic anticoagulation. Technology, development and clinical application. Thromb. Haemost. 2005;93:1014–1020. doi: 10.1160/TH04-12-0790. [DOI] [PubMed] [Google Scholar]
- 43.Cirino G, Severino B. Thrombin receptors and their antagonists: an update on the patent literature. Expert Opin. Ther. Pat. 2010;20:875–884. doi: 10.1517/13543776.2010.487864. [DOI] [PubMed] [Google Scholar]
- 44.Leger AJ, Covic L, Kuliopulos A. Protease-activated receptors in cardiovascular diseases. Circulation. 2006;114:1070–1077. doi: 10.1161/CIRCULATIONAHA.105.574830. [DOI] [PubMed] [Google Scholar]
- 45.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 46.Paborsky LR, McCurdy SN, Griffin LC, Toole JJ, Leung LL. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993;268:20808–20811. [PubMed] [Google Scholar]
- 47.DeAnda A, Jr, Coutre SE, Moon MR, Vial CM, Griffin LC, Law VS, Komeda M, Leung LL, Miller DC. Pilot study of the efficacy of a thrombin inhibitor for use during cardiopulmonary bypass. Ann. Thorac. Surg. 1994;58:344–350. doi: 10.1016/0003-4975(94)92206-3. [DOI] [PubMed] [Google Scholar]
- 48.Leung LL. Application of combinatorial libraries and protein engineering to the discovery of novel anti-thrombotic drugs. Thromb. Haemost. 1995;74:373–376. [PubMed] [Google Scholar]
- 49.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang KY, McCurdy S, Shea RG, Swaminathan S, Bolton PH. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 51.Pagano B, Martino L, Randazzo A, Giancola C. Stability and binding properties of a modified thrombin binding aptamer. Biophys. J. 2008;94:562–569. doi: 10.1529/biophysj.107.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mendelboum Raviv S, Horvath A, Aradi J, Bagoly Z, Fazakas F, Batta Z, Muszbek L, Harsfalvi J. 4-thio-deoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J. Thromb. Haemost. 2008;6:1764–1771. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 53.Bini A, Minunni M, Tombelli S, Centi S, Mascini M. Analytical performances of aptamer-based sensing for thrombin detection. Anal. Chem. 2007;79:3016–3019. doi: 10.1021/ac070096g. [DOI] [PubMed] [Google Scholar]
- 54.Lowe GD. Factor IX and thrombosis. Br. J. Haematol. 2001;115:507–513. doi: 10.1046/j.1365-2141.2001.03186.x. [DOI] [PubMed] [Google Scholar]
- 55.Smith SB, Gailani D. Update on the physiology and pathology of factor IX activation by factor XIa. Expert Rev. Hematol. 2008;1:87–98. doi: 10.1586/17474086.1.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt AE, Bajaj SP. Structure-function relationships in factor IX and factor IXa. Trends Cardiovasc. Med. 2003;13:39–45. doi: 10.1016/s1050-1738(02)00210-4. [DOI] [PubMed] [Google Scholar]
- 57.Nimjee SM, Keys JR, Pitoc GA, Quick G, Rusconi CP, Sullenger BA. A novel antidote-controlled anticoagulant reduces thrombin generation and inflammation and improves cardiac function in cardiopulmonary bypass surgery. Mol. Ther. 2006;14:408–415. doi: 10.1016/j.ymthe.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 58.Dyke CK, Steinhubl SR, Kleiman NS, Cannon RO, Aberle LG, Lin M, Myles SK, Melloni C, Harrington RA, Alexander JH, Becker RC, Rusconi CP. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: a phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. [DOI] [PubMed] [Google Scholar]
- 59.Chan MY, Rusconi CP, Alexander JH, Tonkens RM, Harrington RA, Becker RC. A randomized, repeat-dose, pharmacodynamic and safety study of an antidote-controlled factor IXa inhibitor. J. Thromb. Haemost. 2008;6:789–796. doi: 10.1111/j.1538-7836.2008.02932.x. [DOI] [PubMed] [Google Scholar]
- 60.Chan MY, Cohen MG, Dyke CK, Myles SK, Aberle LG, Lin M, Walder J, Steinhubl SR, Gilchrist IC, Kleiman NS, Vorchheimer DA, Chronos N, Melloni C, Alexander JH, Harrington RA, Tonkens RM, Becker RC, Rusconi CP. Phase 1b randomized study of antidote-controlled modulation of factor IXa activity in patients with stable coronary artery disease. Circulation. 2008;117:2865–2874. doi: 10.1161/CIRCULATIONAHA.107.745687. [DOI] [PubMed] [Google Scholar]
- 61.Cohen MG, Purdy DA, Rossi JS, Grinfeld LR, Myles SK, Aberle LH, Greenbaum AB, Fry E, Chan MY, Tonkens RM, Zelenkofske S, Alexander JH, Harrington RA, Rusconi CP, Becker RC. First clinical application of an actively reversible direct factor IXa inhibitor as an anticoagulation strategy in patients undergoing percutaneous coronary intervention. Circulation. 2010;122:614–622. doi: 10.1161/CIRCULATIONAHA.109.927756. [DOI] [PubMed] [Google Scholar]
- 62.Becker RC, Chan MY. REG-1, a regimen comprising RB-006, a Factor IXa antagonist, and its oligonucleotide active control agent RB-007 for the potential treatment of arterial thrombosis. Curr. Opin. Mol. Ther. 2009;11:707–715. [PubMed] [Google Scholar]
- 63.Povsic TJ, Cohen MG, Chan MY, Zelenkofske SL, Wargin WA, Harrington RA, Alexander JH, Rusconi CP, Becker RC. Dose Selection for a Direct and Selective Factor IXa Inhibitor and its Complementary Reversal Agent: Translating Pharmacokinetic and Pharmacodynamic Properties of the REG1 System to Clinical Trial Design. J. Thromb. Thrombolysis. 2011 doi: 10.1007/s11239-011-0588-3. Epub. [DOI] [PubMed] [Google Scholar]
- 64.Povsic TJ, Cohen MG, Mehran R, Buller CE, Bode C, Cornel JH, Kasprzak JD, Montalescot G, Joseph D, Wargin WA, Rusconi CP, Zelenkofske SL, Becke RC, Alexander JH. A randomized, partially blinded, multicenter, active-controlled, dose-ranging study assessing the safety, efficacy, and pharmacodynamics of the REG1 anticoagulation system in patients with acute coronary syndromes: Design and rationale of the RADAR Phase IIb trial. Am. Heart J. 2011;161:261–268. doi: 10.1016/j.ahj.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 65.Paul D, Samuel SM, Maulik N. Mesenchymal stem cell: present challenges and prospective cellular cardiomyoplasty approaches for myocardial regeneration. Antioxid. Redox. Signal. 2009;11:1841–1855. doi: 10.1089/ars.2009.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pittenger MF, Martin BJ. Mesenchymal stem cells and their potential as cardiac therapeutics. Circ. Res. 2004;95:9–20. doi: 10.1161/01.RES.0000135902.99383.6f. [DOI] [PubMed] [Google Scholar]
- 67.Schafer R, Wiskirchen J, Guo K, Neumann B, Kehlbach R, Pintaske J, Voth V, Walker T, Scheule AM, Greiner TO, Hermanutz-Klein U, Claussen CD, Northoff H, Ziemer G, Wendel HP. Aptamer-based isolation and subsequent imaging of mesenchymal stem cells in ischemic myocard by magnetic resonance imaging. Rofo. 2007;179:1009–1015. doi: 10.1055/s-2007-963409. [DOI] [PubMed] [Google Scholar]
- 68.Strahm Y, Flueckiger A, Billinger M, Meier P, Mettler D, Weisser S, Schaffner T, Hess O. Endothelial-cell-binding aptamer for coating of intracoronary stents. J. Invasive. Cardiol. 2010;22:481–487. [PubMed] [Google Scholar]
- 69.Tanaka Y, Honda T, Matsuura K, Kimura Y, Inui M. In vitro selection and characterization of DNA aptamers specific for phospholamban. J. Pharmacol. Exp. Ther. 2009;329:57–63. doi: 10.1124/jpet.108.149526. [DOI] [PubMed] [Google Scholar]
- 70.Gutsaeva DR, Parkerson JB, Yerigenahally SD, Kurz JC, Schaub RG, Ikuta T, Head CA. Inhibition of cell adhesion by anti-P-selectin aptamer: a new potential therapeutic agent for sickle cell disease. Blood. 2011;117:727–735. doi: 10.1182/blood-2010-05-285718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jenison RD, Jennings SD, Walker DW, Bargatze RF, Parma D. Oligonucleotide inhibitors of P-selectin-dependent neutrophil-platelet adhesion. Antisense Nucleic Acid Drug Dev. 1998;8:265–279. doi: 10.1089/oli.1.1998.8.265. [DOI] [PubMed] [Google Scholar]
- 72.Leppanen O, Janjic N, Carlsson MA, Pietras K, Levin M, Vargeese C, Green LS, Bergqvist D, Ostman A, Heldin CH. Intimal hyperplasia recurs after removal of PDGF-AB and -BB inhibition in the rat carotid artery injury model. Arterioscler. Thromb. Vasc. Biol. 2000;20:E89–E95. doi: 10.1161/01.atv.20.11.e89. [DOI] [PubMed] [Google Scholar]
- 73.Mi J, Zhang X, Giangrande PH, McNamara JO, 2nd, Nimjee SM, Sarraf-Yazdi S, Sullenger BA, Clary BM. Targeted inhibition of αvβ3 integrin with an RNA aptamer impairs endothelial cell growth and survival. Biochem. Biophys. Res. Commun. 2005;338:956–963. doi: 10.1016/j.bbrc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 74.Higashimoto Y, Yamagishi S, Nakamura K, Matsui T, Takeuchi M, Noguchi M, Inoue H. In vitro selection of DNA aptamers that block toxic effects of AGE on cultured retinal pericytes. Microvasc. Res. 2007;74:65–69. doi: 10.1016/j.mvr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 75.Marro ML, Daniels DA, McNamee A, Andrew DP, Chapman TD, Jiang MS, Wu Z, Smith JL, Patel KK, Gearing KL. Identification of potent and selective RNA antagonists of the IFN-gamma-inducible CXCL10 chemokine. Biochemistry. 2005;44:8449–8460. doi: 10.1021/bi048145w. [DOI] [PubMed] [Google Scholar]
- 76.Williams KP, Liu XH, Schumacher TN, Lin HY, Ausiello DA, Kim PS, Bartel DP. Bioactive and nuclease-resistant L-DNA ligand of vasopressin. Proc. Natl. Acad. Sci. USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Missailidis S, Hardy A. Aptamers as inhibitors of target proteins. Expert Opin. Ther. Pat. 2009;19:1073–1082. doi: 10.1517/13543770903042337. [DOI] [PubMed] [Google Scholar]