Abstract
Neutral lipids are a diverse family of hydrophobic biomolecules that have important roles in cellular biochemistry of all living species but have in common the property of charge neutrality. A large component of neutral lipids are the glycerolipids composed of triacylglycerols, diacylglycerols, and monoacylglycerols that can serve as cellular energy stores as well as signaling molecules. Another abundant lipid class in many cells is the cholesterol esters that are on one hand sterols and the other fatty acyl lipids, but in either case are neutral lipids involved in cholesterol homeostasis and transport in the blood. The analysis of these molecules in the context of lipidomics remains challenging because of their charge neutrality and the complex mixtures of molecular species present in cells. Various techniques have been used to ionize these neutral lipids prior to mass spectrometric analysis including electron ionization, atmospheric chemical ionization, electrospray ionization and matrix assisted laser desorption/ionization. Various approaches to deal with the complex mixture of molecular species have been developed including shotgun lipidomics and chromatographic-based separations such as gas chromatography, reversed phase liquid chromatography, and normal phase liquid chromatography. Several applications of these approaches are discussed.
Keywords: glycerolipids, electrospray ionization, electron ionization, neutral lipid, atmospheric pressure ionization, mass spectrometry, matrix assisted laser desorption/ionization, normal phase, reversed phase, triacylglycerols, diacylglycerols, cholesterol esters
1. Introduction
Glycerolipids (GLs) constitute a class of abundant lipids present in all cells of animal and plant origin and play a variety of important biochemical functions from energy storage to being precursor pools for signaling molecules released as a result of occupation of membrane receptors. Major GL examples in mammalian cells are triacylglycerols (TAGs), diacylglycerols (DAGs), and monoacylglycerols (MAGs). Cholesterol esters (CEs) are also neutral lipids (NLs) present in virtually all animal cells as a part of the complex regulation of cholesterol homeostasis [1]. Both CE and TAGs are some of the most abundant lipids found circulating in the plasma of animals, typically present in amounts that would be equivalent to millimolar concentrations. Yet these molecules are not soluble in water and do not achieve these concentrations, but are carried by specialized lipoprotein particles [2]. In cells, these lipids are typically stored within lipid bodies where they serve as sources of fatty acids that are oxidized by β-oxidation for ATP generation [3] and cholesterol transport processes [4]. The GLs are abundant components in seeds and vegetable oils, such as linseed oil, castor oil, and olive oil. Analytical methods continue to be published concerning the analysis of these commercially important GLs. Discussion of the analysis of these oils will be limited, but rather the techniques that have been employed and developed for the analysis of NLs found at much lower levels in animal cells is the major focus of this review.
In spite of the relatively simple structure of these NLs, analysis has remained a major challenge, even analysis by mass spectrometry. In part this is due to the fact that these are chemically neutral molecules and that ionization is required for these lipids to be analyzed by mass spectrometry. The techniques of electron ionization (EI), matrix assisted laser desorption/ionization (MALDI), electrospray ionization (ESI), and atmospheric pressure chemical ionization (APCI) can generate ions from these molecules, but the ion yield for neutral lipids is lower for these latter three techniques when compared to preionized lipid molecules such as phospholipids or free carboxylic acids. The general mechanism for formation of ions from NLs is by attachment of a charged species (e.g. H+, NH4+, Na+, Li+, acetate−) or removal of a proton (e.g. loss of H+) in the mass spectrometer ion source. This formation of a charged ion is a gas-phase chemical event proportional to the affinity of the lipid molecule to the attachment ion or gas phase acidity/ basicity of the NL. This additional requirement for ionization has implications in the choice of mass spectrometry that is employed for analysis. APCI and ESI are perhaps more sensitive methods for NL analysis when compared to EI, however there have not been definitive studies comparing all ionization methods employed for NL analysis.
A second and equally difficult challenge is that NLs are present in cells as complex mixtures of closely related species that differ by fatty acyl chains esterified to either glycerol or cholesterol. Just from simple probability theory, if there are ten fatty acyl groups that could be esterified to glycerol, this would lead to 550 molecular species, not even taking into account enantiomers. In some systems the number of potential fatty acids can be as high as 20, which would result in 4,200 potential molecular species. Thus, the analytical challenge is to (1) identify each molecular species within the context of a large number of closely related compounds and (2) assess the amount of each of these unique molecular species in a sample. Assignment of the position of each fatty acyl group as to the carbon atom to which it is esterified on the backbone of glycerol (sn-1, sn-2, or sn-3 using the IUPAC systematic nomenclature) is an additional challenge, along with the stereochemical assignment at sn-2, since most naturally occurring glycerides are chiral. Furthermore, unsaturated or polyunsaturated NLs have the additional structural challenges of determination of double bond position and geometry. For some organisms, branched chain and otherwise derivatized fatty acyl groups exist that pose additional structural difficulties.
1.1 Electron Ionization
The first methods developed for TAG analysis using mass spectrometry involved application of electron ionization [5] as a means to generate ions. EI is highly energetic and a singular feature of the EI mass spectrum is the very low abundance, in some cases total absence, of a molecular ion, M+˙ (Figure 1). During EI, a M+˙ typically fragments to yield [M-RnCO2]+, [RnCO+128]+, [RnCO+74]+, and [RnCO]+ fragment products where n corresponds to the fatty acyl esterified at the 1,2, or 3 glycerol carbon atoms [6]. These fragment ions are used to generate quantitative data and to reconstruct the original molecular structure of the TAG. A major advantage of EI has been the ease of directly interfacing the gas chromatograph to the mass spectrometer (GC/MS) and the enormous resolving power of this ancillary technique. This chromatographic resolution can be used to separate a large number of molecular species and in this way retention time and EI mass spectral data can define the molecular species [7]. Unfortunately, GC/MS requires that the TAG molecules become volatile for this type of mass spectrometric analysis, and therefore, very high GC column temperatures, which favor TAG thermal decomposition, must be applied [8]. For many high molecular weight TAGs, thermal degradation proceeds at a faster rate than volatilization, thus limiting GC/MS application.
Figure 1.
Positive ion electron ionization (EI) mass spectrum of a triglyceride molecular species that contains palmitoyl, linoleoyl, and oleoyl chains in the molecule. The correct position of fatty acid chains is unknown, but groups are labeled as shown in the annotation of the ions. Adapted from [7] with permission from the American Chemical Society.
Another application of GC/MS has been with the analysis of fatty acids that are esterified to the glycerol backbone or cholesterol and liberated by hydrolysis to free fatty acids. The free fatty acids are then derivatized to improve thermal stability and volatility, followed by GC/MS analysis. This is a very straightforward approach for identification of fatty acids, including structural details such as the position of double bonds present in the acyl chain or methyl group branching, and to quantitate total TAGs. This technique suffers from the lack of information relevant to the exact molecular species structure from the standpoint of the position from which the fatty acyl groups were released from the glycerol backbone [6,9]. The GC/MS approach remains one of the most widely used techniques for the analysis of these NLs in food oils [10] where sensitivity of the ionization method is not a significant issue. Electron ionization and GC/MS techniques have been used to measure diacylglycerols [11] and monoacylglycerols [12].
1.2 Atmospheric pressure chemical ionization
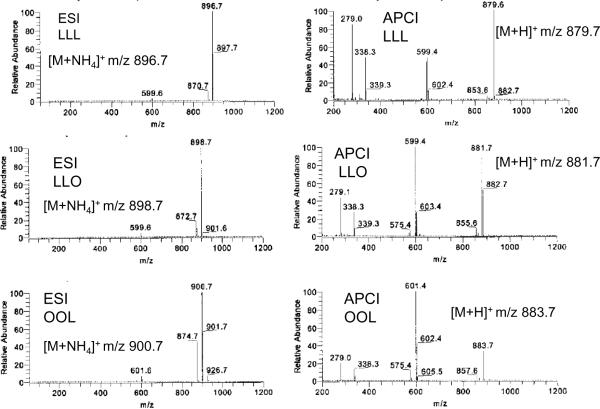
Ionization of GL by APCI results in abundant production of proton attachment ions [M+H]+ likely due to the cumulative proton affinities of the three carboxylate groups that make up the fatty acyl ester moieties. The APCI process is a spray ionization method similar in many respects to electrospray except that the production of abundant protons is in a high voltage or discharge region. These protons can attach to analyte molecules present in the spray and thereby ionize the analytes by proton attachment. APCI is typically more energetic than ESI so that decomposition of [M+H]+ ions often results during the ionization event with the resultant formation of product ions that typically correspond to diglycerol [DAG]+ ions [13]. These can be quite abundant (Figure 2). For those TAGs containing only a single fatty acyl group (R) only one [RR]+ product ion results in the API mass spectrum (Figure 2), but for a mixed fatty acyl TAG with two different fatty acyl groups (R1 and R2), [DAG]+ ions can be found that correspond to [R1R2]+, [R1R1]+, or [R2R2]+. For the TAG with three different fatty acyl groups (R1, R2, R3), the three [DAG]+ ions will correspond to [R1R2]+, [R2R3]+, and [R1R3]+. Studies of the formation of these [DAG]+ ions from TAGs of known structure revealed that the loss of the fatty acid from the sn-2 position is a somewhat less efficient event compared to the loss of the fatty acyl substituent found at the sn-1(3) position [14]. This observation has been the basis for using APCI to deduce the fatty acyl positions in a TAG species from the APCI mass spectrum. Mathematical calculations of the fatty acyl group position based on ion abundances of the [DAG]+ ions and [DAG]+ ion ratios have been described [15,16].
Figure 2.
Comparison of electrospray (ESI) and APCI ionization of triacylyglycerol molecular species. Adapted from [59]. (A) ESI-MS spectrum of ammoniated trilinolein (LLL); (B) ESI-MS spectrum of ammoniated 1,2-dilinoleoyl-3-oleoyl glycerol (LLO); (C) ESI-MS spectrum of ammoniated 1,2-dioleoyl-3-linoleoyl glycerol (OOL); (D) APCI-MS spectrum of LLL; (E) APCI-MS spectrum of LLO; (F) APCI-MS spectrum of OOL.
Another advantage of APCI is that this ionization technique is readily coupled to liquid chromatography so that separation of individual molecular species by reverse phase chromatographic conditions is readily implemented. While the interpretation of APCI spectra of a pure molecule appears straightforward, it is certainly not when mixtures of molecular species are ionized at the same time. The assignment of each [DAG]+ ion to a single molecular species becomes difficult if not impossible. To a large extent the power of chromatography overcomes this fundamental problem and furthermore, separation of species facilitates the use of ion abundances of the [M+H]+ and the [DAG]+ ions as a quantitative measure of the amount of TAG present in a biological system.
A closely related method is atmospheric pressure photo ionization (APPI) where a highly energetic photon is used to effect generation of charged ion species in the CI plasma [17] with generation of the [M+H]+ from TAG analytes. APPI has been reported to be more sensitive than APCI, but few applications have been reported to date.
Qualitative and quantitative analysis of TAGs in complex biological mixtures have also been reported using negative ion APCI. In these studies, ammonia gas was employed as the nebulizing gas and the ion chemistry yields an ammonia anion, [NH2]−, as a result of high voltage discharge. This anion is a sufficiently strong gas phase base to remove a proton from a TAG to form [M-H]− [18]. The collisional activation of these triglyceride anions results in abundant [M-H-R1,2,3COOH-100]− and [R1,2,3COO]− anions. Again, the formation of the DAG-like anion from the loss of the free acid from the sn-2 position [M-H-R2COOH-100]− was typically found to be of lower abundance compared to [M-H-R1,3COOH-100]−. The regioisomeric structure of TAGs can be deduced from the fatty acid carboxylate anions [R1,2,3COO]− generated after collision induced decomposition, along with the abundance of the DAG-like anions. Computer algorithms have been published to automatically process such data as well as obtain quantitative information about the abundance of each TAG molecular species [19]. The ability to interpret these results as to structure and quantity required adequate separation of molecular species, which is achieved by online reversed phase chromatography.
1.3 Electrospray Ionization
Ionization by the electrospray process requires attachment of a charged ion to the neutral TAG, DAG, MAG, or CE. The most frequently used methods to form these attachment ions are to include alkali metal salts or ammonium salts in the spray solvent. The inclusion of such salts often satisfies the additional requirement of having the organic solvent conduct electricity to permit charging of a spray nozzle to complete the electrochemical cell [20]; and importantly, these cations adduct the NL to form a NL cation during electrospray. Abundant molecular species such as [M+NH4]+, [M+Na]+, or [M+Li]+ typically undergo little fragmentation during this ionization process, although if conditions are chosen inappropriately, [M+H]+ ions can be observed as ion source decomposition products of [M+NH4]+. The abundance of these molecular ion species is proportional to the quantity of neutral lipid present in the complex biological extract, but the direct relationship is confounded by subtle effects such as the exact number of carbon atoms in the fatty acyl chains and total number of double bonds, both of which affect ionization efficiency [21]. Structural information such as fatty acyl composition and regioisomeric identification can be obtained by subsequent tandem mass spectrometry of these molecular ion species. The ion chemistries of [M+NH4]+ [22, 23] and [M+Li]+ [24, 25] decomposition pathways have been well described. In general the loss of the sn-2 fatty acyl group following collisional activation of these molecular ion species is less favored than the loss of the sn-1(3) groups, which permits regioisomeric assignment [24]. Determination of the exact fatty acyl groups that constitute a TAG, requires MS3 while MS2 is sufficient to describe the fatty acyl components for a DAG species [22].
For complex mixtures of neutral lipids found in animal cells more sophisticated approaches have been developed to deal with the large number of components in each neutral lipid class. One of the most challenging problems has been the identification of the exact molecular species that are present at each single mass-to-charge ratio corresponding to the monoisotopic TAG. Under ESI conditions, the same monoisotopic mass will be generated for any molecular ion species having the same elemental composition, but still differ by the exact fatty acyl chains. Analysis of TAGs isolated from RAW cells revealed that in some cases the observed [M+NH4]+ ions were a mixture of 3–10 different species that could be differentiated by MS3 (Table 1)[22]. For this reason it has been useful to define isobaric TAG and DAG molecular ions as to the total number of acyl carbon atoms and total number of double bonds in the fatty acyl chains. For example, a 52:2 TAG molecular species could be composed of any combination of 52 total fatty acyl carbon atoms that have two total double bonds in their combined acyl groups. This would include a (16:0/18:1/18:1) TAG as well as a (14:0/18:2/20:0) TAG. Two different approaches were developed to deal with these complex issues. The first is shotgun lipidomics and the second a chromatography-based separation lipidomics. Both approaches continue to evolve and improve and both have strengths and weaknesses.
Table 1.
Mass spectrometric identification of triacylglycerols (TAG) molecular species from RAW 264.7 cells consistent with fatty acyl identification from MS3 data and molecular weight [22].
| TAG (48:0)1 | TAG (48:1) | TAG (48:2) | TAG (49:0) | TAG (49:1) | TAG (49:2) |
|---|---|---|---|---|---|
| 14:0/16:0/18:0 | 14:0/16:0/18:1 | 14:0/16:0/18:2 | 14:0/15:0/20:0 | 13:0/16:0/20:1 | 13:0/16:0/20:2 |
| 14:0/17:0/17:0 | 14:0/16:1/18:0 | 14:0/16:1/18:1 | 14:0/16:0/19:0 | 13:0/17:0/19:1 | 13:0/17:1/19:1 |
| 15:0/15:0/18:0 | 14:0/17:0/17:1 | 14:0/17:1/17:1 | 14:0/17:0/18:0 | 13:0/18:0/18:1 | 13:0/18:1/18:1 |
| 15:0/16:0/17:0 | 14:1/16:0/18:0 | 14:1/16:0/18:1 | 15:0/15:0/19:0 | 14:0/15:0/20:1 | 14:0/15:0/20:2 |
| 16:0/16:0/16:0 | 14:1/17:0/17:0 | 14:1/16:1/18:0 | 15:0/16:0/18:0 | 14:0/16:0/19:1 | 14:0/16:1/19:1 |
| 15:0/15:0/18:1 | 14:1/17:1/17:0 | 15:0/17:0/17:0 | 14:0/17:0/18:1 | 14:0/17:1/18:1 | |
| 15:0/15:1/18:0 | 15:0/15:0/18:2 | 16:0/16:0/17:0 | 14:0/17:1/18:0 | 14:0/17:0/18:2 | |
| 15:0/16:0/17:1 | 15:0/15:1/18:1 | 14:1/17:0/18:0 | 14:1/17:1/18:0 | ||
| 15:0/16:1/17:0 | 15:0/16:1/17:1 | 15:0/15:0/19:1 | 14:1/17:0/18:1 | ||
| 15:1/16:0/17:0 | 15:1/15:1/18:0 | 15:0/16:0/18:1 | 15:0/15:1/19:1 | ||
| 16:0/16:0/16:1 | 15:1/16:0/17:1 | 15:0/16:1/18:0 | 15:0/16:0/18:2 | ||
| 15:1/16:1/17:1 | 15:0/17:0/17:1 | 15:0/16:1/18:1 | |||
| 16:0/16:1/16:1 | 15:1/16:0/18:0 | 15:0/17:1/17:1 | |||
| 15:1/17:0/17:0 | 15:1/16:0/18:1 | ||||
| 16:0/16:0/17:1 | 15:1/16:1/18:0 | ||||
| 16:0/16:1/17:0 | 15:1/17:0/17:1 | ||||
| 16:0/16:1/17:1 | |||||
| 16:1/16:1/17:0 |
Numbers in parentheses correspond to total fatty acyl carbon atoms/total double bonds.
1.3.1 Shotgun lipidomics
The shotgun approach that has been largely developed by Han and Gross [26] employs formation of [M+Li]+ of NLs and takes advantage of the unique behavior of these alkali metal adducts after collisional activation in the mass spectrometer to generate an ion corresponding to the neutral loss of each of the fatty acyl groups as a free carboxylic acid. The central workflow of this approach has been to carry out an organic solvent extraction of the lipids, such as a Bligh/Dyer method [27], and then add LiOH to the extract. This crude lipid extract is then directly electrosprayed into the mass spectrometer for a period of time during which a number of neutral loss scans after collisional activation are performed over the mass range of interest, e.g. m/z 700–1000 (Figure 3). During this time the spray must be very constant since variation in the spray properties could affect ion yield during a particular neutral loss scan experiment. Each exact neutral loss mass experiment is constructed from the known mass-loss for a specific fatty acyl group and thus, from each precursor ion [M+Li]+, it is possible to deduce those fatty acyl substituents that are present in the molecular ion species [M+Li]+. The observed ion currents are used to generate quantitative data after C-13 isotope corrections and potential corrections from a species two daltons lower in mass, which would contribute two C-13 isotopes at this precursor mass. The corrected abundance for each fatty acyl group is then summed [28] and a standard curve is used to calculate the quantity of TAG present. This method does not correct for the abundance of the [M+Li-R2COOH]+ ion, which has been reported in many cases to be lower in absolute abundance than the corresponding neutral loss signals for fatty acyl losses [M+Li-R1,3COOH]+ [24]. Identification of multiple species at each nominal mass is determined mathematically from the neutral loss spectra that yield this precursor ion. Unfortunately, an unexpected fatty acyl group, for example, an odd chain or oxidized fatty acyl group, would not be detected by this method unless one preselects these type fatty acyl groups to examine in the neutral loss scan list. In addition, the larger number of fatty acyl neutral loss scans included in the neutral loss scan list, the longer the electrospray of the crude extract must be carried out while scanning over the mass range of interest. Nonetheless, this approach has been widely used by others [29] to reveal significant biochemistry related to TAG and DAG biochemistry and by Han and Gross for both in vitro and in vivo studies [30,31]. Variations of the shotgun approach have used ammonium ion adducts of NL [32] and sodiated ions [33] for discrimination of isomeric TAGs.
Figure 3.
Shotgun lipidomic approach used for the analysis of triacylglycerol molecular species isolated from mouse myocardium [26]. Analysis after addition of LiOH and neutral loss scanning in positive-ion mode of specific fatty acyl groups indicated. All neutral loss (NL) scans displayed are normalized to the base peak in the individual scan. With permission from Wiley.
The analysis of cholesterol and CEs by shotgun lipidomics has been described using several approaches including derivatization. In a straightforward approach using the exact mass of cholesterol with an orbitrap mass spectrometer, the abundance of the ammonium ion adduct of cholesterol at m/z 404.3892 was measured and used to calculate the quantity of cholesterol after infusion [34]. The cholesteryl esters were quantitated from the MS/MS product ion abundance of m/z 369.3521 after ratioing to the internal standard. A similar approach using d7-cholesterol as internal standard was used in a lipidomics study [35]. Prior derivatization of cholesterol to cholesteryl acetate was employed as a strategy to make use of precursor ion scanning to generate the same ion for both cholesterol and cholesteryl esters [36,37]. Free cholesterol was determined using the d7-cholesterol internal standard and CE(17:0) and CE(22:0) as internal standards for the esters using precursors of m/z 369.3 to detect these esters [36]. Another derivatization strategy involved reacting the lipid extract with methoxyacetic acid after treatment with a carbodiimide to effect esterification of the free alcohol moiety in cholesterol [31,38]. The cholesterol methoxyacetate Li+ adduct was detected by precursor ion scanning for m/z 97, an abundant ion observed after collisional activation of this cholesterol derivative. The resulting signal was converted into abundance information using a standard curve to determine the exact quantity of cholesterol. This approach was also used to quantitate CEs after saponification of a second aliquot of the lipid extract to yield free cholesterol. The difference between the saponified value for cholesterol and the free cholesterol quantity was used to calculate the esterified cholesterol [38]. However analysis of cholesterol ester molecular species was not possible since both free cholesterol and CEs were converted to methoxyacetate derivatives.
1.3.2 Chromatographic separation-based lipidomics
The second major approach to analyze neutral lipids such as CE, TAG, and MAG makes use of the facile interface of HPLC with ESI as a method to pre-separate neutral lipids from an extract as well as to ionize the NL prior to mass spectrometric analysis. The chromatographic separation can be either a normal phase (NP) or reversed phase (RP) process, and both approaches have distinctive advantages and disadvantages. Normal phase chromatography is used to separate neutral lipids based on polarity so that relatively crude lipid extracts can be separated into CEs, TAGs, DAGs, and MAGs with very different retention times. However, very little separation of molecular species within each of these classes, if any at all, is realized [39]. Reversed phase chromatography is well suited to separate molecular species of lipids due to the lipophilicity of the fatty acyl components, but not lipids according to their polar classes. Thus, it is easy to separate neutral lipids roughly as to the fatty acyl chain lipophilicity. However complete separation of all molecular species has not been reported for NLs isolated from a cell extract. Both isocratic and gradient elution have been used to profile neutral lipids from biological extracts using RP chromatography separation, for example, the analysis of neutral lipids from yeast [40] and from animal tissues [41]. Separation of crude extracts containing lipids ranging from phospholipids to TAGs by RP chromatography is illustrated in Figure 4. Since RP separates by lipophilicity, the molecular species of all phospholipids and DAGs elute over the same retention time range [41]. The TAGs elute after the mobile phase was changed to that with a higher concentration of organic solvent.
Figure 4.

Separation of a complex lipid mixture of lipids from mouse liver by on line reverse-phase chromatography, high resolution LC/ESI-QTOF MS analysis. The lipid mixtures of mouse liver were eluted in the following order: PLs =DAGs > SMs= Cers > TAGs. The 2D map was constructed with X (retention time) and Y (m/z value) axes, and abundance of individual TAGs was indicated by single color density (originally adjusted by color gradient). With permission from Elsevier [41].
Reversed phase HPLC separation of molecular species of TAGs isolated from olive oil was used for the determination of regioisomers of individual molecular species. The chromatographic system was optimized for TAG separation since there were few interfering polar lipids in these commercial oils. It was found that the Na+ adducts gave the most reliable quantitative analysis of specific molecular species because the loss of the sn-2 fatty acyl group [M+Na-R2COOH]+ fragment ion was consistently in lower abundance than the signal for the neutral loss of R1,3COOH [41]. A similar approach using only ammonium ion adducts has been reported for quantitation of 10 molecular species of TAGs from cod liver oil [42].
Several groups have employed NP separation as the basic approach to elute relatively pure neutral lipid classes sequentially to the electrospray ion source. For these studies, an electrolyte is mixed into the HPLC effluent stream after the NP separation column in order to change the mobile phase into one that can conduct electricity. These electrolytes drive the adduct ion formation process, but do not interfere with the NP separation. Kallo [43] reported over 450 TAGs species present in butter fat using a NP-ESI/MS/MS approach. In this case, some separation of molecular species was realized in the NP column, which facilitated analysis.
NP separation was chosen as the platform for the analysis of CEs, TAGs, MAGs, and DAGs in the same HPLC run (Figure 5) from our laboratory [44, 45]. The least polar of these lipids were the naturally occurring CEs, which eluted within the first few minutes of the HPLC run. Since these were the least efficiently ionized by ESI, a precursor ion scanning method (MS/MS) was employed to increase sensitivity during the first 5 minutes of this HPLC run. All CEs readily undergo collisional activation as a [M+NH4]+ ions to yield m/z 369.3, the cholesteryl cation [46]. Therefore, precursor ion scanning for m/z 369.3 will reveal each of the CE molecular species present in the biological extract. A stable isotope labeled CE was used as internal standard to facilitate quantitation of each of these CE species in a straightforward isotope dilution approach [39].
Figure 5.
Normal phase LC/MS analysis (base peak chromatogram) of the neutral lipids extracted from bone marrow cells differentiated in culture to macrophages, then derivatized with difluorophenyl isocyanate to the urethane derivative of the free hydroxyl moiety. Regions of the elution of neutral lipids are indicated. Imp1 is a mixture of polysiloxanes and Imp2 is the oxidized form of a plastic antioxidant (tris(2,4-ditert-butylphenyl)phosphate) and Imp3 is the elution of phthalate esters. With permission of Elsevier [44].
Analysis of TAGs and related triradylglycerides [47] was accomplished by forming the ammonium ion adduct to generate [M+NH4]+ ions, which were detected with the instrument automatically reset during the analytical run to carry out normal MS operation and scanning from m/z 500–1000 [39]. The quantity of isobaric molecular species was calculated using standard curves for the abundance of [M+NH4]+ from reference standards relative to that of an added internal standard. No attempt was made to further define constituents that made up each observed mass-to-charge ratio as isobaric molecular species. Thus isobaric molecular species were defined as those that contain the same number of acyl carbon atoms and the same number of total double bonds. However, this NP chromatography does provide excellent separation of alkylether triradylglycerides [39] from the normal triester TAGs and, in a rather straightforward fashion, these triradyl species could be identified. Shotgun lipidomics cannot detect such species from direct mass spectral data and therefore are typically not reported as components in an extract.
The analysis of DAGs has been complicated even if LC/MS separations are employed because the behavior of these lipid species during collision activation of the corresponding adduct ion is less than ideal. Product ion spectra of the ammonium adduct ion are complicated by the facile loss of H2O [22, 39], an effect that can be minimized by studying the sodiated molecular ion species of the diglyceride [48]. An alternative strategy has been to derivatize the free hydroxyl group in a manner that facilitates molecular species identification. DAGs had been derivatized to a quaternary ammonium structure using betaene to substantially enhance formation of [M+H]+ ion of the DAG derivative [49]. We have shown that efficient formation of the 2,4-difluorophenylurethane derivative of DAGs (and MAGs) while these are still in the crude lipid extract [44], yields a product that is not only readily ionized as the ammonium ion adduct, but also separated into 1,2-DAG derivatives and 1,3-DAG derivatives for regioisomer identification when normal phase chromatography is employed (Figure 5). The same approach simultaneously forms a dual urethane adduct of MAGs and the 1(3)-MAGs and 2-MAGs are readily separated by the NP chromatography. Interestingly, this single chromatographic separation of the neutral lipids can be readily adapted to these urethane derivatives by switching analysis to a neutral loss scan during the elution of the urethane derivatives [44]. Under these conditions, a neutral loss of 190 u identifies the elution of the urethane derivatives, since this neutral loss mass is a common feature of the collisional activation of these urethane derivatives. This neutral loss signal can then be used as a quantitative measure of both DAGs and MAGs. The NP LC/MS and NP LC/MS/MS approach with urethane derivatization has been used to measure TAGs and DAGs in the murine macrophage lipidome [50].
An additional feature of this NP chromatographic approach is that rather unique species, which might be unexpected or difficult to predict if one is using a neutral loss approach, can be identified. This is particularly true for the oxidation of CEs where collisional activation of oxidized CEs still yields the abundant ion at m/z 369.3, if the oxidation of the CEs occurs on the fatty acyl portion. Additionally, precursor ion scanning for m/z 369 + 16 or m/z 369 + 32 will reveal those oxidized CE species where oxidation of the cholesterol nucleus occurred. Using this technique numerous oxidized forms of CEs have been identified in human atherosclerotic plaques where oxidation occurred not only on the fatty acyl groups, corresponding to linoleate, arachidonate, and docosahexanoate oxidation, but also oxidation of the cholesterol nucleus itself [51].
1.4 Matrix assisted laser desorption/ionization (MALDI)
MALDI has been widely used to analyze neutral lipids but most examples have focused on profiling the TAGs present in various biological systems, including NL components in serum [52–54]. The sodium attachment ion is the typically observed ion species if the sample is extracted form a biological tissue and the [M+Na]+ molecular species undergoes a facile loss of each fatty acid (as a sodium salt) forming a diglyceride-like positive ion [55, 56]. A major advantage of MALDI has been the speed at which data can be collected from many samples when compared to techniques to define molecular species using chromatographic separations. In a direct comparison of MALDI/MS analysis to LC-APCI/MS of fat bodies from insects, the data set generated by both techniques was found to be quite comparable and each method was able to define numerous species [57]. Even though only molecular weight information can be gleaned from MALDI/MS data, this technique can be quite useful in detecting oxidation and dimerization of TAGs [58]. The use of MALDI/MS and MALDI/MS/MS for the analysis of NL will likely increase, because of the ease and rapidity in obtaining useful information from the former technique and the structural details relevant to NL molecular species obtained with tandem mass spectrometry approach.
2. Conclusions
Both shotgun lipidomics and chromatographic-based separation lipidomics have emerged as powerful techniques to profile neutral lipids, including CEs, MAGs, DAGs, and triradylglycerides present in biological extracts. Advantages and disadvantages of each approach make it difficult to unambiguously define the best technique. However, from both techniques, important information can be gleaned relative to the biochemistry of these neutral lipid species. A remaining challenge for both techniques is improved quantitative analysis in terms of accuracy of the data, which is now problematic due to many reasons, including the multiple isobaric species present at a mass-to-charge ratio for each molecular species and the importance of subtle structural changes that alter ionization efficiency in each NL species. Equally important is the lack of many reference standards from which one can generate standard curves for a particular molecular species. Nevertheless, analysis of these neutral lipids is emerging as an important component of lipidomic studies to provide insight into the exact biochemical events and complex biochemical interactions taking place at the cellular level.
Research Highlights
-
-
Electron ionization and GC/MS of TAGs
-
-
APCI and APCI/LC/MS analysis of neutral lipids
-
-
Electrospray ionization of cholesterol esters and glycerolipids
-
-
Shotgun lipidomics
-
-
Chromatography-based lipidomics with examples of reversed phase and normal phase LC/MS/MS
Acknowledgements
This work was supported, in part, by the Lipid Maps large scale collaborative grant (GM069338) from the Institute of General Medical Sciences of the National Institutes of Health.
Abbreviations
- GL
glycerolipids
- NLs
neutral lipids
- DAG
diacylglycerol
- TAG
triacylglycerol
- MAG
monoacylglycerol
- CE
cholesterol esters
- MALDI
matrix-assisted laser desorption/ionization
- ESI
electrospray ionization
- EI
electron ionization
- APCI
atmospheric pressure chemical ionization
- APPI
atmospheric pressure photo ionization
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Goldstein JL, Brown MS. The LDL receptor. Arterioscler Thromb Vasc Biol. 2009;29:431–438. doi: 10.1161/ATVBAHA.108.179564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wanders RJ, van Roermund CW, Visser WF, Ferdinandusse S, Jansen GA, van den Brink DM, Gloerich J, Waterham HR. Peroxisomal fatty acid α- and β-oxidation in health and disease: New insights. Adv. Exp. Med. Biol. 2003;544:293–302. doi: 10.1007/978-1-4419-9072-3_37. [DOI] [PubMed] [Google Scholar]
- [4].Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- [5].Hites RA. Mass spectrometry of triglycerides. Methods Enzymol. 1975;35:348–359. doi: 10.1016/0076-6879(75)35175-6. [DOI] [PubMed] [Google Scholar]
- [6].Murphy RC. Mass Spectrometry of Lipids. In: Snyder F, editor. The Handbook of Lipid Research. Vol.7. Plenum Press; New York: 1993. [Google Scholar]
- [7].Moldoveanu SC, Chang Y. Dual Analysis of Triglycerides from Certain Common Lipids and Seed Extracts. J Agric Food Chem. 2011 Feb 23; doi: 10.1021/jf104114p. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [8].Ruiz-Samblás C, González-Casado A, Cuadros-Rodríguez L, García FP. Application of selected ion monitoring to the analysis of triacylglycerols in olive oil by high temperature-gas chromatography/mass spectrometry. Talanta. 2010;82:255–260. doi: 10.1016/j.talanta.2010.04.030. [DOI] [PubMed] [Google Scholar]
- [9].Griffiths WJ. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom Rev. 2003;22:81–152. doi: 10.1002/mas.10046. [DOI] [PubMed] [Google Scholar]
- [10].Myher JJ, Kuksis A, Marai L, Sandra P. Identification of the more complex triacylglycerols in bovine milk fat by gas chromatography-mass spectrometry using polar capillary columns. J Chromatogr. 1988;452:93–118. doi: 10.1016/s0021-9673(01)81440-0. [DOI] [PubMed] [Google Scholar]
- [11].Hubbard WC, Hundley TR, Oriente A, MacGlashan DW., Jr. Quantitation of 1-stearoyl-2-arachidonoyl-sn-3-glycerol in human basophils via gas chromatography-negative ion chemical ionization mass spectrometry. Anal Biochem. 1996;236:309–321. doi: 10.1006/abio.1996.0172. [DOI] [PubMed] [Google Scholar]
- [12].Balogh G, Péter M, Liebisch G, Horváth I, Török Z, Nagy E, Maslyanko A, Benko S, Schmitz G, Harwood JL, Vígh L. Lipidomics reveals membrane lipid remodelling and release of potential lipid mediators during early stress responses in a murine melanoma cell line. Biochim Biophys Acta. 2010;1801:1036–1047. doi: 10.1016/j.bbalip.2010.04.011. [DOI] [PubMed] [Google Scholar]
- [13].Byrdwell WC. Atmospheric pressure chemical ionization mass spectrometry for analysis of lipids. Lipids. 2001;36:327–346. doi: 10.1007/s11745-001-0725-5. [DOI] [PubMed] [Google Scholar]
- [14].Laakso P, Voutilainen P. Analysis of triacylglycerols by silver-ion high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. Lipids. 1996;31:1311–1322. doi: 10.1007/BF02587918. [DOI] [PubMed] [Google Scholar]
- [15].Byrdwell WC. The bottom-up solution to the triacylglycerol lipidome using atmospheric pressure chemical ionization mass spectrometry. Lipids. 2005;40:383–417. doi: 10.1007/s11745-006-1398-9. [DOI] [PubMed] [Google Scholar]
- [16].Cvacka J, Krafková E, Jiros P, Valterová I. Computer-assisted interpretation of atmospheric pressure chemical ionization mass spectra of triacylglycerols. Rapid Commun Mass Spectrom. 2006;20:3586–3594. doi: 10.1002/rcm.2770. [DOI] [PubMed] [Google Scholar]
- [17].Cai SS, Syage JA. Atmospheric pressure photoionization mass spectrometry for analysis of fatty acid and acylglycerol lipids. J Chromatogr A. 2006;1110:15–26. doi: 10.1016/j.chroma.2006.01.050. [DOI] [PubMed] [Google Scholar]
- [18].Leskinen HM, Suomela JP, Kallio HP. Quantification of triacylglycerol regioisomers by ultra-high-performance liquid chromatography and ammonia negative ion atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:1–5. doi: 10.1002/rcm.4346. [DOI] [PubMed] [Google Scholar]
- [19].Kurvinen JP, Rua P, Sjövall O, Kallio H. Software (MSPECTRA) for automatic interpretation of triacylglycerol molecular mass distribution spectra and collision induced dissociation product ion spectra obtained by ammonia negative ion chemical ionization mass spectrometry. Rapid Commun Mass Spectrom. 2001;15:1084–1091. doi: 10.1002/rcm.340. [DOI] [PubMed] [Google Scholar]
- [20].Van Berkel GJ. Electrolytic deposition of metals on to the high-voltage contact in an electrospray emitter: implications for gas-phase ion formation. J Mass Spectrom. 2000;35:773–783. doi: 10.1002/1096-9888(200007)35:7<773::AID-JMS4>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- [21].Li X, Evans JJ. Examining the collision-induced decomposition spectra of ammoniated triglycerides as a function of fatty acid chain length and degree of unsaturation. I. The OXO/YOY series. Rapid Commun Mass Spectrom. 2005;19:2528–2538. doi: 10.1002/rcm.2087. [DOI] [PubMed] [Google Scholar]
- [22].McAnoy AM, Wu CC, Murphy RC. Direct qualitative analysis of triacylglycerols by electrospray mass spectrometry using a linear ion trap. J Am Soc Mass Spectrom. 2005;16:1498–1509. doi: 10.1016/j.jasms.2005.04.017. [DOI] [PubMed] [Google Scholar]
- [23].Gakwaya R, Li X, Wong YL, Chivukula S, Collins EJ, Evans JJ. Examining the collision-induced decomposition spectra of ammoniated triglycerides. III. The linoleate and arachidonate series. Rapid Commun Mass Spectrom. 2007;21:3262–3268. doi: 10.1002/rcm.3208. [DOI] [PubMed] [Google Scholar]
- [24].Hsu FF, Turk J. Electrospray ionization multiple-stage linear ion-trap mass spectrometry for structural elucidation of triacylglycerols: assignment of fatty acyl groups on the glycerol backbone and location of double bonds. J Am Soc Mass Spectrom. 2010;21:657–669. doi: 10.1016/j.jasms.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hsu FF, Turk J. Structural characterization of triacylglycerols as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisionally activated dissociation on a triple stage quadrupole instrument. J Am Soc Mass Spectrom. 1999;10:587–599. doi: 10.1016/S1044-0305(99)00035-5. [DOI] [PubMed] [Google Scholar]
- [26].Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- [27].Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- [28].Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- [29].Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mancuso DJ, Sims HF, Yang K, Kiebish MA, Su X, Jenkins CM, Guan S, Moon SH, Pietka T, Nassir F, Schappe T, Moore K, Han X, Abumrad NA, Gross RW. Genetic ablation of calcium-independent phospholipase A2gamma prevents obesity and insulin resistance during high fat feeding by mitochondrial uncoupling and increased adipocyte fatty acid oxidation. J Biol Chem. 2010;285:36495–36510. doi: 10.1074/jbc.M110.115766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gross RW, Han X. Shotgun lipidomics of neutral lipids as an enabling technology for elucidation of lipid-related diseases. Am J Physiol Endocrinol Metab. 2009;297:E297–303. doi: 10.1152/ajpendo.90970.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Murphy RC, James PF, McAnoy AM, Krank J, Duchoslav E, Barkley RM. Detection of the abundance of diacylglycerol and triacylglycerol molecular species in cells using neutral loss mass spectrometry. Anal Biochem. 2007;366:59–70. doi: 10.1016/j.ab.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Herrera LC, Potvin MA, Melanson JE. Quantitative analysis of positional isomers of triacylglycerols via electrospray ionization tandem mass spectrometry of sodiated adducts. Rapid Commun Mass Spectrom. 2010;24:2745–2752. doi: 10.1002/rcm.4700. [DOI] [PubMed] [Google Scholar]
- [34].Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS One. 2009;4:e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sampaio JL, Gerl MJ, Klose C, Ejsing CS, Beug H, Simons K, Shevchenko A. Membrane lipidome of an epithelial cell line. Proc Natl Acad Sci U S A. 2011;108:1903–1907. doi: 10.1073/pnas.1019267108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liebisch G, Binder M, Schifferer R, Langmann T, Schulz B, Schmitz G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS) Biochim Biophys Acta. 2006;1761:121–128. doi: 10.1016/j.bbalip.2005.12.007. [DOI] [PubMed] [Google Scholar]
- [37].Schwudke D, Liebisch G, Herzog R, Schmitz G, Shevchenko A. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Methods Enzymol. 2007;433:175–191. doi: 10.1016/S0076-6879(07)33010-3. [DOI] [PubMed] [Google Scholar]
- [38].Cheng H, Jiang X, Han X. Alterations in lipid homeostasis of mouse dorsal root ganglia induced by apolipoprotein E deficiency: a shotgun lipidomics study. J Neurochem. 2007;101:57–76. doi: 10.1111/j.1471-4159.2006.04342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hutchins PM, Barkley RM, Murphy RC. Separation of cellular non-polar neutral lipids by normal phase chromatography and analysis by electrospray ionization mass spectrometry. J Lipid Res. 2008;49:804–813. doi: 10.1194/jlr.M700521-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shui G, Guan XL, Low CP, Chua GH, Goh JS, Yang H, Wenk MR. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol Biosyst. 2010;6:1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
- [41].Ikeda K, Oike Y, Shimizu T, Taguchi R. Global analysis of triacylglycerols including oxidized molecular species by reverse-phase high resolution LC/ESI-QTOF MS/MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2639–2647. doi: 10.1016/j.jchromb.2009.03.047. [DOI] [PubMed] [Google Scholar]
- [42].Zeng YX, Araujo P, Du ZY, Nguyen TT, Frøyland L, Grung B. Elucidation of triacylglycerols in cod liver oil by liquid chromatography electrospray tandem ion-trap mass spectrometry. Talanta. 2010;82:1261–1270. doi: 10.1016/j.talanta.2010.06.055. [DOI] [PubMed] [Google Scholar]
- [43].Kalo P, Kemppinen A, Ollilainen V. Determination of triacylglycerols in butterfat by normal-phase HPLC and electrospray-tandem mass spectrometry. Lipids. 2009;44:169–195. doi: 10.1007/s11745-008-3247-5. [DOI] [PubMed] [Google Scholar]
- [44].Leiker TJ, Barkley RM, Murphy RC. Analysis of diacylglycerol molecular species in cellular lipid extracts by normal-phase LC-electrospray mass spectrometry. Intl J Mass Spectrom. doi: 10.1016/j.ijms.2010.09.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, Bandyopadhyay S, Jones KN, Kelly S, Shaner RL, Sullards CM, Wang E, Murphy RC, Barkley RM, Leiker TJ, Raetz CR, Guan Z, Laird GM, Six DA, Russell DW, McDonald JG, Subramaniam S, Fahy E, Dennis EA. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51:3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Duffin K, Obukowicz M, Raz A, Shieh JJ. Electrospray/tandem mass spectrometry for quantitative analysis of lipid remodeling in essential fatty acid deficient mice. Anal Biochem. 2000;279:179–188. doi: 10.1006/abio.1999.4452. [DOI] [PubMed] [Google Scholar]
- [47].Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- [48].Callender HL, Forrester JS, Ivanova P, Preininger A, Milne S, Brown HA. Quantification of diacylglycerol species from cellular extracts by electrospray ionization mass spectrometry using a linear regression algorithm. Anal Chem. 2007;79:263–272. doi: 10.1021/ac061083q. [DOI] [PubMed] [Google Scholar]
- [49].Li YL, Su X, Stahl PD, Gross ML. Quantification of diacylglycerol molecular species in biological samples by electrospray ionization mass spectrometry after one-step derivatization. Anal Chem. 2007;79:1569–1574. doi: 10.1021/ac0615910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown AH, Milne SB, Myers DS, Glass CK, Hardiman GT, Reichart D, Merrill AH, Sullards CM, Wang E, Murphy RC, Raetz CR, Garrett T, Guan Z, Ryan AC, Russell DW, McDonald JG, Thompson BM, Shaw WA, Sud M, Zhao Y, Gupta S, Maurya MR, Fahy E, Subramaniam S. A mouse macrophage lipidome. J Biol Chem. 2010;285:39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hutchins PM, Murphy RC. Peroxide bond driven dissociation of hydroperoxy-cholesterol esters following collision induced dissociation. J Am Soc Mass Spectrom. 2011 Feb; doi: 10.1007/s13361-011-0109-9. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wiesman Z, Chapagain BP. Determination of fatty acid profiles and TAGs in vegetable oils by MALDI-TOF/MS fingerprinting. Methods Mol Biol. 2009;579:315–336. doi: 10.1007/978-1-60761-322-0_16. [DOI] [PubMed] [Google Scholar]
- [53].Hidaka H, Hanyu N, Sugano M, Kawasaki K, Yamauchi K, Katsuyama T. Analysis of human serum lipoprotein lipid composition using MALDI-TOF mass spectrometry. Ann Clin Lab Sci. 2007;37:213–221. [PubMed] [Google Scholar]
- [54].Dannenberger D, Süss R, Teuber K, Fuchs B, Nuernberg K, Schiller J. The intact muscle lipid composition of bulls: an investigation by MALDI-TOF MS and 31P NMR. Chem Phys Lipids. 2010;163:157–164. doi: 10.1016/j.chemphyslip.2009.10.011. [DOI] [PubMed] [Google Scholar]
- [55].Benard S, Arnhold J, Lehnert M, Schiller J, Arnold K. Experiments towards quantification of saturated and polyunsaturated diacylglycerols by matrix-assisted laser desorption and ionization time-of-flight mass spectrometry. Chem. Phys. Lipids. 1999;100:115–125. [Google Scholar]
- [56].Asbury G. Reid, Al-Saad Khalid, Siems William F., Hannan Richard M., Hill Herbert H., Jr. Analysis of Triacylglycerols and Whole Oils by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry. J. Am. Soc. Mass Spectrom. 1999;10:983–991. [Google Scholar]
- [57].Kofronová E, Cvacka J, Vrkoslav V, Hanus R, Jiros P, Kindl J, Hovorka O, Valterová I. A comparison of HPLC/APCI-MS and MALDI-MS for characterising triacylglycerols in insects: species-specific composition of lipids in the fat bodies of bumblebee males. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3878–3884. doi: 10.1016/j.jchromb.2009.09.040. [DOI] [PubMed] [Google Scholar]
- [58].Picariello G, Paduano A, Sacchi R, Addeo F. Maldi-tof mass spectrometry profiling of polar and nonpolar fractions in heated vegetable oils. J Agric Food Chem. 2009;57:5391–5400. doi: 10.1021/jf9008795. [DOI] [PubMed] [Google Scholar]
- [59].Byrdwell WC, Neff WE. Dual parallel electrospray ionization and atmospheric pressure chemical ionization mass spectrometry (MS), MS/MS and MS/MS/MS for the analysis of triacylglycerols and triacylglycerol oxidation products. Rapid Commun Mass Spectrom. 2002;16:300–319. doi: 10.1002/rcm.581. [DOI] [PubMed] [Google Scholar]