Abstract
The structural assignment of new natural product molecules supports research in a multitude of disciplines that may lead to new therapeutic agents and or new understanding of disease biology. However, reports of numerous structural revisions, even of recently elucidated natural products, inspired the present survey of techniques used in structural misassignments and subsequent revisions in the context of constitutional or configurational errors. Given the comparatively recent development of marine natural products chemistry, coincident with the modern spectroscopy, it is of interest to consider the relative roles of spectroscopy and chemical synthesis in the structure elucidation and revision of those marine natural products which were initially misassigned. Thus, a tabulated review of all marine natural product structural revisions from 2005 to 2010 is organized according to structural motif revised. Misassignments of constitution are more frequent than perhaps anticipated by reliance on HMBC and other advanced NMR experiments, especially considering the full complement of all natural products. However, these techniques also feature prominently in structural revisions, specifically of marine natural products. Nevertheless, as is the case for revision of relative and absolute configuration, total synthesis is a proven partner for marine, as well as terrestrial, natural products structure elucidation. It also becomes apparent that considerable ‘detective work’ remains in structure elucidation, in spite of the spectacular advances in spectroscopic techniques.
Keywords: Structure revision, marine natural products, total synthesis, structure elucidation, NMR spectroscopy
1. Introduction
Natural products research has evolved into a multifaceted discipline at the interface of organic and analytical chemistry, biochemistry, biology, ecology and pharmaceutical sciences. As small molecules, which facilitate dynamic biological processes, natural products are also pivotal to the more recently designated fields of chemical and systems biology.1 Therefore, the structural assignment of a new natural product molecule potentially provides the foundation for research in a multitude of disciplines, ultimately generating new therapeutic agents and/or new understanding of disease biology. The development of modern spectroscopic techniques has transformed the often painstaking structure assignment process, which previously relied on chemical degradation or derivatization followed by partial or total synthesis prior to the 1960’s.2 Subsequently, specialization in the spectroscopic structural assignment of natural products enabled the field of natural products chemistry to mature. Notably, it was only in this current spectroscopic era that the field of marine natural products chemistry took shape, with the advent of SCUBA allowing exploration of shallow underwater reef systems. One might then presume that the age-old partner of natural products chemistry, chemical synthesis, has had little application to the structural assignment of marine natural products. Indeed, today the processes of terrestrial and marine natural product isolation and structural determination are frequently streamlined and expeditious due to the evolving state-of-the-art in chromatographic3 and spectroscopic4, 5 technologies. Consequently, the focus of natural products chemists has shifted from merely describing the chemical properties of newly isolated metabolites to investigating their biological properties, and also harnessing the biosynthetic capacity of the producing organisms for combinatorial biosyntheses of new analogues. Further opportunities of where and how to find new natural products, or their cryptic biosynthesis genes, are being addressed most recently by mining of microbial genomic data.6 While the role of chemical synthesis in the pharmaceutical application and biosynthetic studies of marine natural products is easily recognized, the contribution of chemical synthesis to structure elucidation may be less apparent. Furthermore, the enduring fallibility of all structural assignment tools may be forgotten in view of the spectacular advances in analytical chromatographic and spectroscopic techniques, as well as chemical synthesis.
In 2005, Nicolaou and Snyder2 published an inspiring and fairly comprehensive review of natural product structural revisions that were made possible by total synthesis over the period of 1990 to 2004. As the authors follow the evolution of natural products chemistry intertwined with techniques in chemical degradation, derivatization and total synthesis, they articulate amazement at the unexpectedly large number (over 300) and profoundness of structural misassignments made in relatively recent years. Structural misassignments were noted in virtually every compound class and included both constitutional and configurational errors in molecules of all sizes and degrees of complexity. Nicolaou and Snyder also discuss the ramifications of structural misassignments using high profile historical examples to highlight the potential to propose erroneous biosynthetic pathways for entire classes of compounds, and the costs associated with extensive detective work to revise a misassigned structure. Finally, the syntheses and collaborative reassignments of the complex natural products azaspiracid-1 and diazonamide A are discussed. The latter example in particular illustrates the development of new synthetic methodologies in the pursuit of molecular structures. A detailed and comprehensive 2009 review by Maier7 extends this discussion of the role of total synthesis in the structural revision of natural products, highlighting a sustained rate of structural reassignments from 2005 to 2009. Maier also breaks down the types or sources of misassignments into subcategories of incorrect formula, constitution (planar connectivity), double bond configuration, absolute configuration, and one or several stereocenters assigned incorrectly, providing comprehensive treatment of numerous case studies.
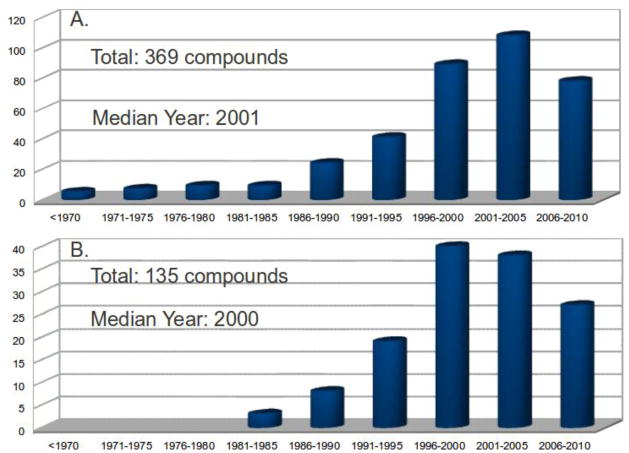
These two reviews on the application of chemical (particularly total) synthesis to natural product structure elucidation, which are inclusive of both terrestrial and marine-derived metabolites, prompted our broad analysis and comparison between terrestrial and marine products, of the spectroscopic or chemical methods used in initial structural misassignments, detection of those misassignments and structural revisions. As has been highlighted in previous reviews, detection and revision of misassignments often constitute distinct and challenging steps, although they may be part of an iterative process. Given the comparative “youth” of marine natural products chemistry, contemporary with the development of powerful spectroscopic methodologies, the role of total synthesis in the assignment of marine-derived structures was of specific interest. In addition, tabulation of revised marine natural product structures according to structural motif misassigned (within the two broad categories of constitution and configuration) draws attention to the limitations of particular techniques in certain structural contexts. The actual rate of structural misassignments remains elusive given the difficulty of estimating the total number of new natural products reported each year. However, Figure 1A shows that the number of all reported structural misassignments climaxed in the period 2001–2005, while the number of marine natural product misassignments was highest in the period 1996–2000 (Figure 1B).
Figure 1.
A. Numbers of all natural products misassigned per 5-year period; B. Numbers of marine natural products misassigned per 5-year period.
2. Sources of natural product structural misassignments
The considerable number of natural product structural misassignments reported each year elicits numerous questions within the two broad contexts of structure and corresponding assignment method. Consideration of structural elements most prone to misassignment and the techniques associated with these misassignments is undoubtedly an informative and educational pursuit. However, it may also provide deeper insight into the analytical process of structure elucidation to benefit the evolution of computer-assisted structure elucidation (CASE).8 CASE mimics the human expert in making a logical inference of the most probable structure based on well-established correlations between spectral and structural features.
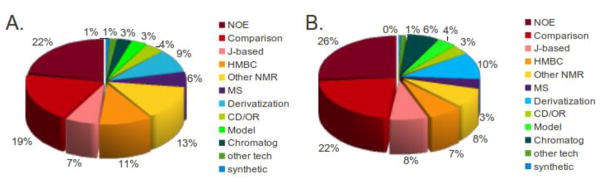
Given the relative ease of compiling data on the techniques used in structural assignments, Figure 2 presents the original techniques used for all misassigned natural products that were subsequently revised successfully (Figure 2A), as well as the subset that were of marine origin (Figure 2B).9 The proportion of errors attributable to different techniques seems likely to reflect the frequency of use of those techniques. Thus, NOE data is the basis for the majority of structural misassignments (22% of total, 26% of marine), followed closely by NMR comparisons, and HMBC and other NMR-derived data, for both marine and terrestrial natural products. The high frequency of NOE-associated errors implies that configurational misassignments predominate over constitutional errors. Similarly, NMR comparisons (of 1H and 13C data) are used to infer configuration, as well as constitution, and may also propagate errors disproportionately when one considers that a majority of natural products occur in structural series, often based on detailed spectral analysis and assignment of only the major congener. However, a significant portion of all structural misassignments (11%) stem specifically from interpretation of HMBC data. This implies errors in bond connectivity of the planar structure (constitutional errors). Indeed, considering all natural products, constitutional errors (45%) rival configurational errors (55%), although to a lesser extent in the subset of marine natural products (33% and 67%, respectively). The logical follow-up questions elicited by these over-view analyses concern the nature of the constitutional or configurational errors and the associated techniques that were used. For the following discussion we focus on structural revisions of marine natural products in the period from 2005 to 2010, according to type of structural misassignment, as summarized in Tables 1 to 9. Remarkably, a large number (78) of the revised structures presented in these tables were first elucidated (misassigned) within the last decade.
Figure 2.
A. Techniques used in the misassignment of all natural product structures revised during 2005 – 2010; B. Techniques used in the misassignment of marine natural product structures revised during 2005 – 2010. Piechart key: NOE – NOE, NOESY, ROESY; Comparison – NMR comparison; J-based – coupling constant analysis; Other NMR – mostly 1D NMR, but includes COSY, HSQC, etc; MS – any MS technique except LC-MS; Derivatization – Marfey’s, Mosher’s, peptide hydrolysis, chemical correlation; CD/OR – circular dichroism or optical rotation; Model – any computational modeling; Chromatog – any TLC identification to HPLC (often in combination with “Derivatization”); Other tech – IR, UV, biosynthetic considerations, etc; Synthesis – any of partial synthesis, model synthesis, semi-synthesis.
Table 1.
Marine natural product structural revisions of cyclization (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Chloroaurone10(2001) Brown alga |
1H &13C NMR HMQC HMBC |

|
Total Synthesis11 |
 Aspergione A12 (2003) Fungus |
HMBC ROESY J-based |
 Ustusorane C |
HMBC ROESY J-based14 |
 Aspergione B12 (2003) Fungus |
HMBC ROESY J-based |
 Pseudodeflectusin |
HMBC ROESY J-based13 |
 Pyrostatin A15(1995) Sponge |
HMBC |

|
Total Synthesis16 |
 Pyrostatin B (1995) Sponge |
HMBC |

|
Total Synthesis16 |
 Spongotine B17 (2005) Sponge |
COSY |

|
MS NMR18 |
 Laurendecumenyne B19 (2007) Red alga |
Comparison |
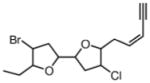
|
Comparison20 |
 Elatenyne21 (1986) Red alga |
NMR |

|
Total Synthesis22 |
 “Enyne from Lyngbya”23 (1993) Cyanobacteria |
Comparison |

|
Total Synthesis22 |
 Peribysin C162 (2004) Fungus |
HMBC NOE |

|
EI-MS 13C Prediction NMR comparison163 |
 Peribysin D162 (2004) Fungus |
HMBC NOE |

|
EI-MS 13C Prediction NMR comparison163 |
|
Irciformonin A164 (2006) Sponge |
Artifact |
|
Deriv.165 |
 Sipholenol I166 (2007) Sponge |
NMR |

|
X-ray NMR167 |
 Eusynstyelamide145 (1994) Tunicate |
NMR |

|
NMR Comparison146 |
 Kasarin147(2000) Bacteria |
HMBC |

|
Synthetic Model NMR148 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Deriv. =derivatization.
Table 9.
Marine natural product structural revisions of double bond geometry (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Iejimalide A138(1988) Tunicate |
NOE |

|
NOE NMR139 |
 Iejimalide C140(1991) Tunicate |
Comparison |

|
Comparison139 |
 Iejimalide D140(1991) Tunicate |
Comparison |

|
Comparison139 |
 Scleritodermin A141 (2004) Sponge |
NOE Deriv. Chromatog. |

|
Total Synthesis142 |
|
Mycothiazole143(1988) Sponge |
J-based |

|
Comparison144 |
 Tridachiahydropyrone C223(2000) arotamer of B Mollusk |
Comparison |
 Oxytridachiahydropyrone |
Total Synthesis191 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
2.1. Misassignments of constitution of marine natural products
The constitutional misassignments of marine natural products may be arranged into categories following the recurring themes of errors in cyclization (Table 1), regioisomerism (Table 2), substituent identity (Table 3), hydrocarbon chains (Table 4), and symmetry of dimerization (based on incorrect formula, Table 5).
Table 2.
Marine natural product structural revisions of regioisomers (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Subereaphenol B24(2008) Sponge |
HSQC HMBC |

|
HSQC HMBC25 |
 Subereaphenol C24 (2008) Sponge |
HSQC HMBC |

|
HSQC HMBC25 |
 Aspernigrin A26 (2004) Fungus |
HMBC |

|
X-ray NOE27 |
 Pyranonigrin26 (2004) Fungus |
ROESY |

|
Chemical Reactivity CD predict.28 |
 Spiroleucettadine29 (2004) Sponge |
HMBC ROESY |

|
13C Predict. X-ray30 |
 Obtusallene V136 (2008) Red alga |
13C Predict. |

|
X-ray137 |
 Obtusallene VI136 (2008) Red alga |
13C Predict. |

|
Model Synthesis Comparison137 |
 Obtusallene VII136 (2008) Red alga |
13C Predict. |

|
Model Synthesis Comparison137 |
 Neopetrosiamide168 (2005) Sponge |
NOE |

|
Total Synthesis169 |
 Asbestinin 10170(1993) Coral |
Comparison |

|
Deriv. NMR171 |
 Asbestinin 20172(1994) Coral |
Deriv. NMR |

|
Deriv. NMR171 |
 Asbestinin 21172(1994) Coral |
Comparison |

|
Deriv. NMR171 |
 Largamide A173(2006) Cyanobacterium |
HMBC Marfey’s |

|
HMBC NOE Deriv. Chromatog.174 |
 Largamide B173 (2006) Cyanobacterium |
HMBC Marfey’s |

|
HMBC NOE Deriv. Chromatog.174 |
 Largamide C173(2006) Cyanobacterium |
HMBC Marfey’s |

|
HMBC NOE Deriv. Chromatog.174 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Predict. = predictions based on molecular modeling. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
Table 3.
Marine natural product structural revisions of substituents (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Almazole D31 (1996) Red alga |
Comparison |

|
Total Synthesis32 |
 Alcyonin33 (1988) |
MS NMR |

|
Total Synthesis34 |
 hydroxy-bromosphaerodiol B35(1987) Bacterium |
MS |

|
X-ray36 |
 Bromoditerpene35 (1987) Red alga |
HR-MS 1H NMR |
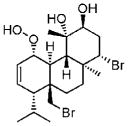
|
X-ray36 |
 Perthamide C37 (2009) Sponge |
J-based Deriv. |

|
Partial Synthesis Comparison38 |
 Eudistomidin B175 (1990) Tunicate |
NOE CD |

|
Total Synthesis176, 177 |
 Psammaplin I178(2003) Sponge |
IR 13C NMR |

|
MS Mol. Mod.179 |
 11-acetoxy-4-deacetoxyasbestinin F172(1994) Coral |
MS |

|
Deriv. NMR171 |
 Plakevulin A180(2003) Sponge |
Contamination |

|
Total Synthesis181 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization.
Table 4.
Marine natural product structural revisions of hydrocarbon chains (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
|
Pyrinodemin A39 (2001) Sponge |
EIMS |
|
Total Synthesis40 |
|
Batzelladine F41 (1997) Sponge |
Comparison NOE DCI-MS |
|
Total Synthesis42 |
|
Oceanapiside (1999)43 Sponge |
MS |
|
Deriv. MS44 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. DCI = Desorption Chemical Ionization.
Table 5.
Marine natural product structural revisions of dimerization (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Zamamistatin45 (2001) Sponge |
NOESY ESI HR-MS |
 See next entry |
Synthetic model compounds46 |
 Zamamistatin46 (2001) Sponge |
Synthetic model compounds |

|
IR NMR and OR47 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table.
Constitutions of cycles (Table 1)
The assignment of small cycles is challenging since direction of coherence around a ring is not necessarily discernible. A common constitutional error is differentiation of 5- versus 6-membered cycles by analysis of HMBC data. Several cases of an exocyclic olefin on a 5-membered ring as opposed to a 6-membered ring with endocyclic bond are evident in Table 1, for example, chloroaurone,10, 11 aspergiones A and B,12–14 pyrostatins A and B,15, 16 and spongotine B.17, 18 Another problematic theme is differentiation of fused 6-membered rings from two 5-membered rings linked by a single bond, as seen in the examples of the red algal metabolites laurendecumenyne B,19, 20 elatenyne21, 22 and Laurenciaenyne.22, 23 In these three compounds, the observed 13C NMR shifts of the oxygen-bearing carbons were more consistent with a 2,2′-bifuranyl moiety (> 76 ppm) than the proposed pyrano[3,2-b]pyran (< 76 ppm).7 Consideration of these structures elicits the question of whether these errors may be attributed to the assignment of observed HMBC correlations as 4-bond connections. An alternative scenario is that key 3-bond HMBC correlations were absent, presumably because they were outside the coupling constant range detectable in a standard HMBC experiment optimized for 3JCH = 8 Hz.
Regioisomers (Table 2)
As noted above, the assignment of small cycle connectivity can be ambiguous, and this issue extends to the placement of substituents on cycles. The difficulty of assigning substitution patterns on cycles is exacerbated especially by a paucity of 1H to provide HMBC correlations, as in highly substituted phenolic compounds (e.g. subereaphenols B and C24, 25), in tandem with possible signal overlap. In some instances, such as aspernigrin A,26, 27 a single HMBC correlation may theoretically differentiate two isomers. However, in cases such as pyranonigrin,26, 28 spiroleucettadine29, 30 and almazole D (Table 3),31, 32 careful consideration of the chemical shift values in the assigned structure may have provided the only clue to the structural misassignment.
Substituent identity (Table 3)
The identity of heteronuclear substituents may only be inferred indirectly from NMR data. Thus, the assignment of these substituents is often reliant on mass spectrometry (MS), and is subject to error depending on the stability of the substituent and the ionization technique used. For example, alcyonin,33, 34 12S-hydroxy-bromosphaerodiol B35, 36 and the red algal bromoditerpene35, 36 were misassigned with hydroxy rather than hydroperoxy subsituents, despite the acquisition of high resolution (HR) MS data. In the case of perthamide C,37, 38 sulfone and amide substituents went undetected until negative mode HR electrospray ionization (ESI) MS was employed.
Hydrocarbon chains (Table 4)
MS is again the traditional tool of choice in assigning saturated and unsaturated alkyl chains, based on the observed molecular ion and fragmentation patterns. However, these assignments remain a considerable challenge. Initial interpretation of the MS fragmentation data obtained for sponge metabolites pyrinodemin A39, 40 (electron ionization MS) and batzelladine F41, 42 (desorption chemical ionization MS) did not locate the olefin, or provide the correct chain lengths, respectively. Pyrinodemin A yielded an MS fragmentation pattern that was not exclusive to the proposed structure,40 while batzelladine F assignment was complicated by the presence of both an alkyl tether (between the two guanidine moieties) and a side chain.42 Misplacement of the keto group on C-11 in the hydrocarbon chain of oceanapiside resulted from charge-localized fragmentation of the Li+ adduct by MALDI (matrix-assisted laser desorption ionization) MS/MS.43 Finally, MS analysis of the Baeyer–Villiger oxidation products obtained from the oceanapiside ketone relocated the keto group to C-18, consistent with the related rhizocalins.44
Dimerization (Table 5)
The unusual structure of the sponge metabolite zamamistatin required repeated revision to arrive at the correct assignment.45–47 The more common scenario is that an NMR-derived monomeric structure is proven to exist as a dimer by MS. However, in this case, the tentative molecular ion peak at m/z 700.8 was attributable to the tendency for the isonitrile to dimerize under the conditions of MS analysis, and was finally shown to disappear upon dilution, leaving a peak at m/z 361.9.47 Thus, structural revision finally assigned zamamistatin as the known compound aeroplysinin-1, exemplifying the extensive effort that may be required to revise even misassigned known structures.
2.2. Misassignments of configuration of marine natural products
Configurational errors may be readily classified as misassignments of either absolute (Table 6) or relative configuration, the latter comprising the majority of misassigned marine natural products tabulated here (Tables 7–10). Misassignments of relative configuration have been sorted according to whether one or multiple stereocenters, or double bond geometries were misassigned. Structural themes emerging in the misassignment of multiple stereocenters then become evident.
Table 6.
Marine natural product structural revisions of absolute configuration (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Amphilactene derivative50(1999) Sponge |
Mosher’s |

|
Mosher’s51 |
 Amphilactene derivative50(1999) Sponge |
Comparison |

|
X-ray51 |
 Fusapyrone128 (1994) Fungus |
1D & 2D NMR |

|
HMBC NMR Comparison129 |
 Deoxyfusapyrone128 (1994) Fungus |
1D & 2D NMR |

|
HMBC NMR Comparison129 |
|
Plakolide A182(2004) Sponge |
CD |
|
Total Synthesis183 |
 4-keto-clonostachydiol184(2006) Fungus |
Deriv. OR |

|
Total Synthesis185 |
Theyear in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Predict. = predictions based on molecular modeling. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
Table 7.
Marine natural product structural revisions of single stereocenters (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Carriebowmide53 (2008) Cyanobacterium |
Marfey’s |

|
Marfey’s54 |
 Bisebromoamide55(2009) Cyanobacterium |
Marfey’s |

|
Total Synthesis56 |
 Callipeltin E57(2002) Sponge |
Marfey’s |

|
Total Synthesis58 |
 Brevenal61 (2005) Dinoflagellate |
NOE |

|
Total Synthesis62 |
 Solandelactone A63 (1996) Hydrozoan |
HMQC COSY Comparison NOE |

|
Total Synthesis64 |
 Solandelactone B63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone C63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone D63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone E63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone F63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone G63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Solandelactone H63 (1996) Hydrozoan |
Comparison NOE |

|
Total Synthesis64 |
 Ritterazine B65(1995) Tunicate |
NOE Comparison |

|
Partial Synthesis66 |
 Ritterazine F67(1995) Tunicate |
NOE Comparison |

|
Partial Synthesis66 |
 Netamine E68 (2006) Sponge |
NOE |

|
Total Synthesis69 |
 Netamine G68 (2006) Sponge |
NOE |

|
Total Synthesis69 |
 Briarellin J70 (2003) Coral |
NOESY |

|
Total Synthesis71 |
 Dictyosphaeric acid A72(2004) Fungus |
NOE |

|
Total Synthesis73 |
 Manzacidin B74(1991) Sponge |
J-based Comparison |

|
Total Synthesis75 |
 Agelasine C76 (1984) Sponge |
CD Comparison NMR |

|
Total Synthesis77 |
 Epi-agelasine C78(1984) Sponge |
CD Comparison NMR |

|
Total Synthesis77 |
 Peyssonol A79 (1994) Red alga |
13C NMR |

|
Total Synthesis80 |
 Isoepitaondiol81 (1992) Brown alga |
Comparison |

|
X-ray82 |
 Suberoretisteroid B83 (2000) Gorgonian |
1H NMR |

|
NMR Comparison84 |
 Lanostane-type triterpenoid85(1994) Green alga |
Comparison |

|
X-ray NMR86 |
 Amphidinolide W87 (2002) Dinoflagellate |
Derivatization J-based Mosher |

|
Total Synthesis88 |
 Aspergillide A89(2008) Fungus |
Mosher’s NOE |

|
X-ray90 |
 Aspergillide B89 (2008) Fungus |
Mosher’s NOE |

|
Total Synthesis91, 92 X-ray90 |
|
Schulzeine A93 (2004) Sponge |
Deriv. Comparison |
|
Total Synthesis94 |
|
Amphidinol 3186 (1999) Dinoflagellate |
Ozonolysis Derivatization Chiral HPLC |
|
Combinatorial synthesis of fragments187 |
 Gymnangiamide188 (2004) Hydrozoan |
Degradation Deriv. Marfey’s |

|
Total Synthesis189 |
 Tridachiahydropyrone190 (1996) Mollusk |
NOE |

|
Total Synthesis191 |
 dihydrodiscorhabdin A192 (2009) Sponge |
J-based Mol. Mod. |

|
CD Semi-synthesis193 |
 Obtusallene V135 (2000) Red alga |
Mol. Mod. NMR Comparison |

|
13C Predict.136 |
 Obtusallene VI135(2000) Red alga |
Mol. Mod. NMR Comparison |

|
13C Predict.136 |
 Obtusallene VII135 (2000) Red alga |
Mol. Mod. NMR Comparison |

|
13C Predict.136 |
 2-formamido-6-axene194 (1989) Sponge |
NOE |

|
NOE Comparison195 |
 Isothiocyanato-6-axene194(1989) Sponge |
NOE |

|
NOE Deriv.195 |
 Obyanamide196 (2002) Cyanobacterium |
Degradation Chromatog. |

|
Total Synthesis197 |
|
Dragonamide198(2001) Cyanobacterium |
Degradation OR |
|
Total Synthesis199 |
 Oxazinin-2200(2001) Mussel |
ROESY |

|
Total Synthesis201 |
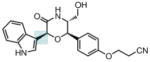 Oxazinin-1200(2001) Mussel |
ROESY |

|
Comparison201 |
 Papuamide B202(1999) Sponge |
Degradation Chromatog. |

|
Total Synthesis203 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Predict. = predictions based on molecular modeling. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
Absolute configuration (Table 6)
In the majority of cases, the assignment of absolute configuration necessitates chemical degradation and/or derivatization, which may be followed by comparison of chromatographic retention and/or spectroscopic properties with standards or model compounds. Various Mosher’s ester and Marfey’s chiral derivatization methods are widely used examples of this general empirical approach. The destructive and/or laborious procedures necessary for most assignments of absolute configuration mean that spectroscopic comparisons of new metabolites with known assigned compounds to extrapolate absolute configuration is common and may lead to extensive propagation of errors. Given the frequently limited quantities of marine natural products isolated, errors resulting from these approaches almost exclusively await detection and revision by total synthesis. Thus, they may be more common than is evident from the structural revisions reported to date. Chiroptical spectroscopy in the form of circular dichroism (CD) or optical rotatory dispersion (ORD) is an attractive alternative for the analysis of configuration that may not require modification of the isolated natural product. However, similar to Mosher and Marfey’s analyses, interpretation of these data relies largely on application of empirically derived principles, such as the octant rules, or comparison with data for analogous compounds of known conformation and configuration.48 In either of these indirect approaches, errors can be sustained and propagated. The use of CD has traditionally been confined to consideration of the sign (but not magnitude) of the Cotton effects, which provides information only about the sign of a torsion angle around a bond, in other words the absolute conformation of a molecule. The determined absolute conformation needs to be correlated to the predicted absolute configuration by a second CD argument (e.g. magnitude and position of bands) or an alternative analytical method, such as conformational analysis, NMR, or X-ray, to avoid erroneous assumptions of quasi–axial or quasi–equatorial conformations of ring substituents, for example.48 In other words, one must be certain of the molecular conformation in order to derive the absolute configuration by chiroptical spectroscopy. Notably, an additional achiral substituent on an aromatic moiety may even lead to inversion of the sign of a Cotton effect.48 Mosher’s method49 is an empirically derived correlation of configuration and NMR chemical shifts for diastereomericphenylacetate ester derivatives (commonly α-methoxy-α-trifluoromethylphenylacetate, MTPA) that is widely used for small-scale analyses of secondary alcohols, amines and chiral α-substituted carboxylic acids. In this method, it is important to carefully examine models of the expected diastereomeric esters to ensure that the experimentally observed chemical shift changes are consistent with the predicted orientation and thus shielding effect of the phenyl ring. However, the sponge-derived amphilectenes50, 51 were misassigned simply due to overlapping 1H NMR chemical shifts in the CDCl3 spectrum of the Mosher ester of an alcohol-containing congener. Subsequent analysis of the more dispersed spectrum obtained in pyridine-d5 for this compound, and also crystallization of the isobutene-containing amphilectene, permitted structural revision of this series.51
Marfey’s method, using the chiral derivatizing agent 1-fluoro-2,4-dinitrophenyl-5-L-alaninamide (FDAA, Marfey’s reagent) or 1-fluoro-2,4-dinitrophenyl-5-L-leucinamide (modified Marfey’s reagent), also requires little material and is particularly effective for the configurational assignment of peptidic natural products. Regular C18 HPLC is used to compare retention times of the diastereomeric Marfey’s derivatized standards and the unknown. Importantly, neutral Marfey’s reagent reacts rapidly (within 1 h), stoichiometrically, and without significant racemization, with the α-amino group of amino acids.52 The predominant source of errors arising from Marfey’s analyses appears to be misassignment or close overlap of HPLC peaks (representing different amino acids) for the natural product hydrolysates and/or the Marfey’s derivatized standards (e.g. carriebowmide,53, 54 bisebromide55, 56 and callipeltin E,57, 58 Table 7). It is often impossible to gain resolution of all residues in the hydrolysate, as well as the stereochemical standards, under one set of HPLC conditions. Changes in retention time for all residues must then be tracked under several different solvent conditions. These challenges warrant the consistent use of LC-MS for Marfey’s analyses of peptide hydrolysates. Coinjection of standards with the natural product hydrolysate could also be performed more routinely, as well as standardization of the concentrations of injected derivatives. In addition, although the derivatization reaction with Marfey’s reagent is mild, the potential for racemization under the harsh conditions of acid hydrolysis (reflux in 6N HCl for 12–24 h) of the natural product should be recognized. For example, methionine, proline, arginine and lysine are more prone to racemization under these conditions than are threonine, serine, leucine, isoleucine and valine.59 Thus, any configurational analysis that relies on initial hydrolysis of the natural product before derivatization is subject to potential errors.
With sufficient sample, X-ray crystallography is considered a gold standard for assignment of absolute configuration, when the sample is amenable to crystallization and contains either a sufficiently heavy atom for unequivocal determination or a known chiral center. However, an unlikely scenario is worth noting here. In the assignment,60 total synthesis lead to a diastereomeric product that was derivatized for cystallization. Crystals of the minor (“unwanted”) diastereomer were unwittingly selected for X-ray crystallography, and resulted in an incorrect revision of the absolute configuration for the natural product.
Relative configuration (Tables 7–9)
Not unexpectedly, interpretation of NOE data and spectroscopic comparison contribute the majority of relative configurational misassignments of marine natural products. Misassignments of single stereocenters (Table 7) near to correctly assigned centers are associated with interpretation of NOE data for structurally diverse compounds. For example, this is the case for brevenal,61, 62 solandelactones A-H,63, 64 ritterazines B65, 66 and F,67 netamines E and G,68, 69 briarellin J,70, 71 and dictyosphaeric acid A.72, 73 Consideration of the source of these errors highlights the need for careful evaluation of all possible alternative molecular models that adhere to NOE constraints. The starting conformation of the subject molecule is critical, as is a consideration of the predicted distances between atoms (NOE’s may be observed between protons within 5Å). These factors support the use of computational modeling for all applications of NOE data to the assignment of configuration. Even use of physical models may readily result in overlooking possible free rotation about an atom in a linear chain or alternate conformations of a ring system, as well as over- or underestimated interatomic distances. Once a putative configuration is assigned, it is clearly essential to examine molecular models closely for all potentially observable NOEs based on interatomic distances, and to match these to the NMR data. In this way, errors arising from assignments of configuration based on single NOE correlations may be avoided. Other common single stereocenter misassignments occur at ring junctions, as in the cases of manzacidin B,74, 75 agelasine C76, 77 and its epimer,78 peyssonol A,79, 80 isoepitaondiol,81, 82 suberoretisteroid B83, 84 and the green algal lanostane-type triterpenoid85, 86 (Table 7), and are caused primarily by NMR spectral overlap. Assignment is also difficult for an isolated substituent on a macrocycle or extended chain, perhaps too far from the point of derivatization in a flexible system. Examples of this circumstance include amphidinolide W,87, 88 aspergillides A and B89–92 and schulzeine A93, 94 (Table 7).
The presence of COSY-like peaks in ROESY and NOESY spectra necessitates careful attention to the phase of the cross peaks in these spectra. The long ROESY spin-lock pulse has the same effect as the last 90° pulse in a COSY experiment, causing coherence transfer between J-coupled spins to appear as weak antiphase cross peaks. Direct TOCSY effects are caused in a similar manner to COSY-like peaks in ROESY experiments since the TOCSY experiment relies on a similar spin-lock. While these TOCSY peaks are also antiphase to direct ROE cross peaks, multistep coherence transfers involving both ROESY and TOCSY (ROE-TOCSY or TOCSY-ROE) can lead to false ROEs that are in phase with direct (true) ROEs.95
Misassignments of multiple stereocenters in a broad range of marine natural products (Table 8) are also most frequently associated with NOE data. Structural motifs misassigned on the basis of NOEs include epoxides (calafianin,96–98 symbiodinolide99, 100), cyclopropyl rings (clavosolide A,101, 102 laurentristich-4-ol103, 104), bridges (vannusals A and B105–107), fused ring junctions (itomanallene A,108, 109 asperdimin,110, 111 aplysiallene112, 113), polyethers (aplysiol B,114, 115 azaspiracids116–119), vicinal polyols (amphidinolide H2,120, 121 hyrtiosterol,122, 123 pericosine A124, 125), sugars (callipeltoside C;126, 127 fusapyrone and deoxyfusapyrone,128, 129 Table 6) and macrolides (palmerolide A,130–132 neopeltolide133, 134).
Table 8.
Marine natural product structural revisions of multiple stereocenters (2005–2010).a
| Proposed Structure | Initial Structure Determination Method | Revised Structure | Structure Revision Method |
|---|---|---|---|
 Calafianin96(2000) Sponge |
NOE |

|
Total Synthesis97, 98 |
 Symbiodinolide99 (2007) Dinoflagellate |
Degradation NOE J-based |

|
Degradation Partial Synthesis Comparison100 |
 Clavosolide A101 (2002) Sponge |
J-based ROESY |

|
Total Synthesis102 |
 Laurentristich-4-ol103 (2005) Red alga |
NOE |

|
Total Synthesis104 |
 Vannusal A105 (1999) Ciliate |
NOE J-based |

|
Total Synthesis106, 107 |
 Vannusal B105 (1999) Ciliate |
NOE J-based Comparison |

|
Total Synthesis106, 107 |
 Itomanallene A108(2002) Red alga |
NOE |
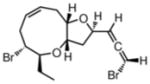
|
Total Synthesis109 |
 Asperdimin110 (2004) Fungus |
Comparison |

|
Total Synthesis111 |
 Aplysiallene112 (1985) Sea Hare |
OR NOE Comparison |

|
Total Synthesis113 |
 Aplysiol B114 (2010) Red alga |
NOE J-based Mol. Mod. |

|
Biogenesis NOE115 |
 Azaspiracid-1116(1998) Mussel |
NOE J-based |

|
Total Synthesis117 |
 Azaspiracid-2118 (1999) Mussel |
NOE Comparison |

|
Total Synthesis119 |
 Azaspiracid-3118(1999) Mussel |
NOE Comparison |

|
Total Synthesis119 |
 Amphidinolide H2120 (2002) Dinoflagellate |
NOE |

|
Total Synthesis121 |
 Hyrtiosterol122(2004) Coral |
NOE |

|
NOE123 |
 Pericosine A124 (1997) Fungus |
NOE J-based |

|
Total Synthesis125 |
 Callipeltoside C126 (1996) Sponge |
ROESY NOE |

|
Total Synthesis127 |
 Palmerolide A130 (2006) Tunicate |
Mosher J-based NOE |

|
Total Synthesis131, 132 |
 Neopeltolide133(2007) Sponge |
NOE |

|
Total Synthesis134 |
 Agardhilactone204(1996) Red alga |
Derivatization NMR |

|
Total Synthesis205 |
 Palau’amine206 (1993) Sponge |
ROESY J-based |

|
ROESY Mol. Mod. J-based207 |
 Bromopalau’amine208(1998) Sponge |
Comparison |

|
Comparison209 |
 Dibromopalau’amine208(1998) Sponge |
Comparison |

|
ROESY209 |
 Bromostyloguanidine208(1998) Sponge |
Comparison |
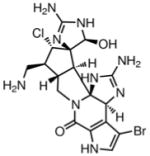
|
Comparison209 |
 Dibromostyloguanidine208(1998) Sponge |
Comparison |

|
Comparison209 |
|
Malyngamide U210 (2003) Cyanobacteria |
J-based NOESY |
|
Total Synthesis211 |
 Lucentamycin A212 (2007) Bacterium |
Marfey’s |

|
Total Synthesis213 |
 Nakiterpiosin214 (2004) Sponge |
NOE |

|
Total Synthesis215 |
 Kulokekahilide-2216 (2004) Mollusk |
Marfey’s |

|
Total Synthesis217 |
 Muurolane-derivative218(1997) Bacterium |
NOE J-based |

|
X-ray219 |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE219 |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE |
 Muurolane-derivative220(2007) Bacterium |
Comparison |

|
Comparison NOE219 |
 Dolastatin 19221(2004) Mollusk |
NOE |

|
Total Synthesis222 |
The year in which the initial structure was published is in parentheses. Only the structure determination methods used for the portion of the structure that is erroneous are mentioned in this table. Predict. = predictions based on molecular modeling. Mol. Mod. = use of geometry optimized structure. Deriv. = derivatization. Chromatog. = chiral HPLC or GC with or without derivatization.
Double bond geometries are accessible through NOEs, and NMR comparisons of coupling constants and chemical shifts. The latter may be altered significantly by different heteroatom substituents, as for the bromo-substituted obtusallenes V–VII135–137 (Tables 2 and 7). Similarly, NOE data analyses (iejimalides,138–140 scleritodermin A,141, 142 Table 9) may be ambiguous for endocyclic double bonds if the structural constraints of the ring are not well defined. In the case of mycothiazole,143, 144 the side-chain double bond was misassigned based on an estimated 3JH,H coupling constant from the only resolved multiplet.141
3. Detection and revision of marine natural product misassignments
Consideration of the sources of structural misassignments leads to the question of how these errors are detected and what techniques are used in revisions of particular structural features. It is reasonable to assume that most errors are detected and revised by orthogonal, complementary approaches to those used in the original assignments. Initial detection of structural misassignments may be due to re-isolation of the same metabolite, or its close analogue, and identification of the error in the process of compound dereplication. Indeed, rigorous dereplication of assumed known compounds is an important exercise for its potential to either validate or question assigned structures. Moreover, a careful re-examination of structure in the compound dereplication process should also consider whether there are inconsistencies from a biosynthetic perspective, which is an important consideration in the de novo structure elucidation process. However, partial or total synthesis of complex and/or biologically active structures is most often the first indication of a structural misassignment. Subsequent revision of a configurationally complex structure may require the synthesis of numerous diastereomers.2
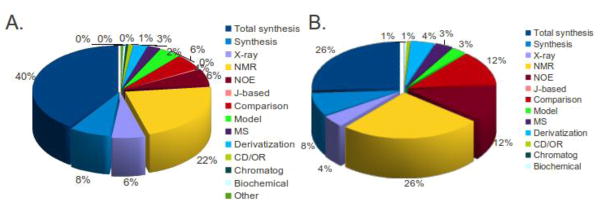
Total synthesis accounts for the overwhelming majority of natural product structural revisions (40%, Figure 3A), but this is not mirrored for marine natural products (26%, Figure 3B). Nondestructive and relatively sensitive NMR methods, although the greatest source of incorrect assignments, still provide a major contribution to marine structural revisions (38% for NOE and other NMR combined), exceeding that of total and partial syntheses (34% combined), which may require degradation of limited or unavailable natural products for comparisons.
Figure 3.
A. Techniques used in the detection and revision of all natural product structures (2005 – 2010); B. Techniques used in the detection and revision of marine natural product structures (2005 – 2010).
As alluded to in section 2.1, the significant number of misassignments associated with interpretation of HMBC data (Figure 2) highlight a subtle problem. Unless all theoretically possible HMBC correlations are observed, some alternative structures are indistinguishable on the basis of HMBC experiments alone. In these circumstances, one needs to “recognize” the alternative structure(s) and actively discredit these using another technique. It also becomes important to examine a designated structure for all possible two- and three-bond HMBC correlations. Those correlations expected but not observed experimentally indicate that further investigation may be necessary. Finally, the existence of 4-bond HMBC correlations needs to be taken into consideration in certain molecular classes (e.g. heteroaromatics).
Several marine natural product structure elucidations elicited our interest because of the use of NMR spectroscopy in the original misassignment as well as in the detection of the misassignment and subsequent structural revision. For example, the structure proposed in 1994 for the ascidian metabolite eusynstyelamide (Table 1) was based on extensive 1D and 2D NMR analyses.145 While, it was noted that dehydration of the proposed α-keto amide hydrate did not occur in aprotic solvents, this was presumed to occur during FABMS since the highest molecular weight ion observed was m/z 787, assigned as [M+H-H2O]+. In 2009, three metabolites named eusynstyelamides A–C were isolated from a different Eusynstyelaspecies.146 Comparison of the previously reported data sets for eusynstyelamide with those of the newly assigned compounds, revealed almost identical data for eusynstyelamide and eusynstyelamide A ([M+H]+, m/z 787). A detailed analysis of the re-assessment of the NMR data for eusynstyelamide, and its reassignment as eusynstyelamide A, is presented in the report of eusynstyelamides A–C.146 In particular, a small 3-bond HMBC correlation observed in the spectrum for eusynstyelamide A, one that was not likely observed in the 1994 spectrum for eusynstyelamide, would have represented a 6-bond connection for the original structure.
Kasarin (Table 1) is a fungal metabolite from the zoanthid coral symbiont Hyphomycetessp. The β-lactam skeleton was originally assigned from 13C- and 15N-HMBC data, although it was noted that the connectivity between the lactam α-carbon and the methoxy-bearing nitrogen could not be established.147 Re-isolation of kasarin by the same research group permitted the application of INADEQUATE, as well as 15N- and 13C-HMBC experiments. In this second analysis,148 the methoxy substituent could be re-assigned to the original β-lactam nitrogen (N-1) on the basis of a 15N-HMBC correlation, and the isopropyl 1H showed a 3-bond 13C-HMBC correlation to the lactam carbon, as well as a 2-bond correlation to the imino carbon on which it is located. To differentiate between the 6-membered pyrazinone heterocycle and an alternative 5-membered oxazolone, a model pyrazinone was synthesized. The spectral data (including IR and UV) for kasarin were much more consistent with the data for this pyrazinone derivative than with those of commercially available oxazolones which contain a significantly deshielded lactone carbon. Finally, ammonolysis of kasarin yielded a degradation product in which the malonyl substituent was eliminated. The total synthesis of kasarin is also in progress by the same research group.148
Interestingly, in the original assignment of pyranonigrin A, both regioisomers shown in Table 2 were considered, and their differentiation recognized as a challenge given the amino and oxymethine HMBC correlations permeating throughout the dihydropyrrole ring.26 Finally, the incorrect 7-keto dihydropyrrole structure (numbering according to the revised structure) was assigned on the basis of a strong ROESY correlation between the amino and oxymethine protons. In the reassignment of pyranonigrin A,28 initiated due to its re-isolation from the fermentation broth of the marine fungus Aspergillusniger, an 15N-HMBC permitted assessment of the pyrrolenitrogen chemical shift as more consistent with an amide N (δ 126.6 ppm) than the hemiaminal N (~ δ 80 ppm) of the original proposed structure. In addition, it was noted that the carbonyl 13C NMR shift (δ 174 ppm) is more consistent with a lactam carbon than the isolated ketone of the original structure (predicted ~ 190 ppm). Nevertheless, given the potential for keto-enol tautomerism in the molecule, three other possible regioisomers (around positions 5–7) had to be considered. Unequivocal assignment of the structure shown was enabled finally by acetylation of the two hydroxyl substituents, and subsequent methanolysis of the diacetate. Direct substitution of an acetate by a methoxy group can only have occurred in the regioisomer assigned (Table 2).28
4. Misassigned marine natural product structures not yet revised
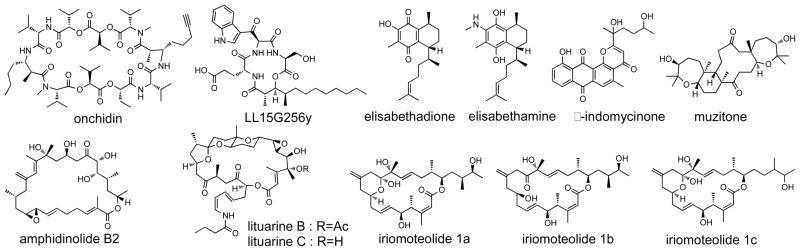
Tables 1–9 present only those structures that have been revised successfully. In some cases, this multi-step process required repeated revisions (e.g. zamamistatin), or was substantially extended beyond the first report of structural misassignment. Misassigned natural products (Figure 4) for which revised constitutions have yet to be presented include sponge-derived muzitone,149 the anti-inflammatory gorgonian metabolites elisabethadione and elisabethamine,150 iriomoteolides 1a–1c,151–155 and likely marine actinomyceteanthraquinone δ-indomycinone.156 In all of these cases, total synthesis of the proposed structures revealed discrepancies between the spectroscopic data for the synthetic and natural products. Attempted total synthesis of elisabethamine revealed that this aminohydroquinone structure is unstable in air and is readily oxidized to the quinone.150 In several of these cases, the lack of readily available authentic natural product sample precludes definitive structural revision, and therefore awaits re-isolation of the natural product or a close analogue. Total synthesis also revealed misassignment of the relative configurations for amphidinolide B2,157 lituarines B and C from a sea pen,158 marine fungal metabolite LL15G256γ,159 and the molluscandepsipeptideonchidin160 (Figure 4).Thus, total synthesis has been instrumental in the detection of marine natural product structural misassignments.
Figure 4.
Misassigned marine natural product structures not yet revised (detected 2005 – 2010).
5. Concluding Remarks
Natural product structure elucidation has been described as routine and no longer the art that it once was.161 However, while it may have evolved considerably since its origins, it is safe to say that the art of structure elucidation has not yet achieved perfection, and that the partnership between natural products chemistry and synthetic organic chemistry is still very much in order. Although chemical degradation and derivatization may have been somewhat less involved in the constitutional assignment of marine-derived versus terrestrial natural products, total synthesis plays a critical role in marine natural product structure elucidation. Furthermore, it is clear that significant detective work using a variety of orthogonal techniques, while keeping in mind biosynthetic considerations, is still necessary for unequivocal assignment, not only of configuration, but also of constitution. The ramifications of structural misassignments in reallocation of resources become evident when almost simultaneous reports of a structural revision are published by different research groups, when sequential revisions of structure are necessary before the correct structure is achieved, or when total syntheses of multiple diastereomers for comparison with the natural product must be accomplished. It is also evident that all methodologies used for structure elucidation can generate errors, and thus structure elucidation of a new natural product should always be accomplished by the most rigorous methods available to the natural products chemist. Similarly, presumed known compounds re-isolated from new collections or cultures should be assigned unequivocally as such by orthogonal techniques and in light of current biosynthetic knowledge and understanding. Structural misassignments continue to be reported for even recently reported marine natural products, and thus, it seems that the increasingly high-field magnets and sensitive probes do not necessarily attenuate the rate of structural misassignments. Rather, they permit the attempted structure elucidation of increasingly limited quantities of minor components from natural products extracts, as well as larger molecules of greater structural complexity. Therefore, total synthesis of natural products will surely continue to be central to confirmation of natural product structure assignment, as well as providing material for biological testing towards pharmaceutical development, and investigations of biosynthetic pathways.
Structural revisions of even recently elucidated natural products continue to be reported.
A tabulated review of marine natural product structural revisions from 2005 to 2010 is presented.
Techniques used in constitutional/configurational misassignments and revisions are surveyed.
Chemical synthesis remains critical to the modern art of natural product structure elucidation.
Acknowledgments
This work was supported by the OSU College of Pharmacy (McPhail) and NIH grant NS 053398 (W.H.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- 1.Carlson EE. ACS Chem Biol. 2010;5:639. doi: 10.1021/cb100105c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolaou KC, Snyder SA. Angew Chem Int Ed. 2005;44:1012. doi: 10.1002/anie.200460864. [DOI] [PubMed] [Google Scholar]
- 3.Zhao H-Y, Jiang J-G. Mini-Rev Med Chem. 2010;10:1223. doi: 10.2174/13895575110091223. [DOI] [PubMed] [Google Scholar]
- 4.Molinski TF. Curr Opin Biotech. 2010;21:819. doi: 10.1016/j.copbio.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Svatos A. Trends Biotech. 2010;28:425. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Challis GL. Microbiol. 2008;154:1555. doi: 10.1099/mic.0.2008/018523-0. [DOI] [PubMed] [Google Scholar]
- 7.Maier ME. Nat Prod Rep. 2009;26:1105. doi: 10.1039/b809658a. [DOI] [PubMed] [Google Scholar]
- 8.Elyashberg M, Williams AJ, Blinov K. Nat Prod Rep. 2010;27:1296. doi: 10.1039/c002332a. [DOI] [PubMed] [Google Scholar]
- 9.Searches of Scifinder Scholar database used the following strings: structure revision, s. r., structure correction, structure misassignment, stereochemical revision, stereochemical reassignment, stereochemical correction, stereochemical misassignment, configurational revision, configurational reassignment, configurational correction, configurational misassignment.
- 10.Atta-Ur-Rahman, Choudhary MI, Hayat S, Khan AM, Ahmed A. Chem Pharm Bull. 2001;49:105. doi: 10.1248/cpb.49.105. [DOI] [PubMed] [Google Scholar]
- 11.Venkateswarlu S, Panchagnula GK, Gottumukkala AL, Subbaraju GV. Tetrahedron. 2007;63:6909. [Google Scholar]
- 12.Lin W, Brauers G, Ebel R, Wray V, Berg A, Sudarsono, Proksch P. J Nat Prod. 2003;66:57. doi: 10.1021/np020196b. [DOI] [PubMed] [Google Scholar]
- 13.Saito F, Kuramochi K, Nakazaki A, Mizushina Y, Sugawara F, Kobayashi S. Eur J Org Chem. 2006;21:4796. [Google Scholar]
- 14.Kuramochi K, Saito F, Nakazaki A, Takeuchi T, Tsubaki K, Sugawara F, Kobayashi S. Biosci Biotechnol Biochem. 2010;74:1635. doi: 10.1271/bbb.100241. [DOI] [PubMed] [Google Scholar]
- 15.Aoyama T, Kojima F, Imada C, Muraoka Y, Naganawa H, Okami Y, Takeuchi T, Aoyagi TJ. Enzyme Inhib. 1995;8:223. doi: 10.3109/14756369509020129. [DOI] [PubMed] [Google Scholar]
- 16.Castellanos L, Duque C, Zea S, Espada A, Rodríguez J, Jiménez C. Org Lett. 2006;8:4967. doi: 10.1021/ol062087k. [DOI] [PubMed] [Google Scholar]
- 17.Bao B, Sun Q, Yao X, Hong J, Lee CO, Sim CJ, Im KS, Jung JH. J Nat Prod. 2005;68:711. doi: 10.1021/np049577a. [DOI] [PubMed] [Google Scholar]
- 18.Bao B, Sun Q, Yao X, Hong J, Lee CO, YCH, Jung JH. J Nat Prod. 2007;70:2. doi: 10.1021/np060206z. [DOI] [PubMed] [Google Scholar]
- 19.Ji NY, LX M, Li K, Wang BG. J Nat Prod. 2007;70:1499. doi: 10.1021/np0701172. [DOI] [PubMed] [Google Scholar]
- 20.Ji NY, Li XM, Li K, Wang BG. J Nat Prod. 2010;73:1192. [Google Scholar]
- 21.Hall JG, Reiss JA. Aust J Chem. 1986;39:1401. [Google Scholar]
- 22.Sheldrake HM, Jamieson C, Burton JW. Angew Chem Int Ed. 2006;45:7199. doi: 10.1002/anie.200602211. [DOI] [PubMed] [Google Scholar]
- 23.Wright AD, König GM, De Nys R, Sticher O. J Nat Prod. 1993;56:394. [Google Scholar]
- 24.Abou-Shoer MI, Shaala LA, Youssef DTA, Badr JM, Habib AM. J Nat Prod. 2008;71:1464. doi: 10.1021/np800142n. [DOI] [PubMed] [Google Scholar]
- 25.Shaker KH, Zinecker H, Ghani MA, Imhoff JF, Schneider B. Chem Biodivers. 2010;7:2880. doi: 10.1002/cbdv.200900277. [DOI] [PubMed] [Google Scholar]
- 26.Hiort J, Maksimenka K, Reichert M, Perovi-Ottstadt S, Lin WH, Wray V, Steube K, Schaumann K, Weber H, Proksch P, Ebel R, Mller WEG, Bringmann G. J Nat Prod. 2004;67:1532. doi: 10.1021/np030551d. [DOI] [PubMed] [Google Scholar]
- 27.Ye YH, Zhu HL, Song YC, Liu JY, Tan RX. J Nat Prod. 2005;68:1106. doi: 10.1021/np050059p. [DOI] [PubMed] [Google Scholar]
- 28.Schlingmann G, Taniguchi T, He H, Bigelis R, Yang HY, Koehn FE, Carter GT, Berova N. J Nat Prod. 2007;70:1180. doi: 10.1021/np070175n. [DOI] [PubMed] [Google Scholar]
- 29.Ralifo P, Crews P. J Org Chem. 2004;69:9025. doi: 10.1021/jo048789+. [DOI] [PubMed] [Google Scholar]
- 30.White KN, Amagata T, Oliver AG, Tenney K, Wenzel PJ, Crews P. J Org Chem. 2008;73:8719. doi: 10.1021/jo800960w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.N’Diaye I, Guella G, Mancini I, Pietra F. Tetrahedron Lett. 1996;37:3049. [Google Scholar]
- 32.Miyake F, Hashimoto M, Tonsiengsom S, Yakushijin K, Horne DA. Tetrahedron. 2010;66:4888. [Google Scholar]
- 33.Kusumi T, Uchida H, Ishitsuka MO, Yamamoto H, Kakisawa H. Chem Lett. 1988:1077. [Google Scholar]
- 34.Corminboeuf O, Overman LE, Pennington LD. J Org Chem. 2009;74:5458. doi: 10.1021/jo9010156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cafieri F, De Napoli L, Fattorusso E, Santacroce C. Phytochem. 1987;26:471. [Google Scholar]
- 36.Smyrniotopoulos V, Quesada A, Vagias C, Moreau D, Roussakis C, Roussis V. Tetrahedron. 2008;64:5184. [Google Scholar]
- 37.Festa C, De Marino S, Sepe V, Monti MC, Luciano P, D’Auria MV, Debitus C, Bucci M, Vellecco V, Zampella A. Tetrahedron. 2009;65:10424. [Google Scholar]
- 38.Sepe V, D’Auria MV, Bifulco G, Ummarino R, Zampella A. Tetrahedron. 2010;66:7520. [Google Scholar]
- 39.Snider BB, Shi B. Tetrahedron Lett. 2001;42:1639. [Google Scholar]
- 40.Ishiyama H, Tsuda M, Endo T, Kobayashi J. Molecules. 2005;10:312. doi: 10.3390/10010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patil AD, Freyer AJ, Taylor PB, Carté B, Zuber G, Johnson RK, Faulkner DJJ. J Org Chem. 1997;62:1814. [Google Scholar]
- 42.Cohen F, Overman LE. J Am Chem Soc. 2006;128:2594. doi: 10.1021/ja0574320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nicholas GM, Hong TW, Molinski TF, Lerch ML, Cancilla MT, Lebrilla CB. J Nat Prod. 1999;62:1678. doi: 10.1021/np990190v. [DOI] [PubMed] [Google Scholar]
- 44.Mkarieva TN, Dmitrenok PS, Zakharenko AM, Denisenko VA, Guzii AG, Li R, Skepper CK, Molinski TF, Stonik VA. J Nat Prod. 2007;70:1991. doi: 10.1021/np0704811. [DOI] [PubMed] [Google Scholar]
- 45.Takada N, Watanabe R, Suenaga K, Yamada K, Ueda K, Kita M, Uemura D. Tetrahedron Lett. 2001;42:5265. [Google Scholar]
- 46.Hayakawa I, Teruya T, Kigoshi H. Tetrahedron Lett. 2006;47:155. [Google Scholar]
- 47.Kita M, Tsunematsu Y, Hayakawa I, Kigoshi H. Tetrahedron Lett. 2008;49:5383. [Google Scholar]
- 48.Snatzke G. Pure Appl Chem. 1979;51:769. [Google Scholar]
- 49.Dale JA, Mosher HS. J Am Chem Soc. 1973;95:512. [Google Scholar]
- 50.Ciavatta ML, Fontana A, Puliti R, Scognamiglio G, Cimino G. Tetrahedron. 1999;55:12629. [Google Scholar]
- 51.Ciavatta M, Gavagnin M, Manzo E, Puliti R, Mattia CA, Mazzarella L, Cimino G, Simpson JS, Garson MJ. Tetrahedron. 2005;61:8049. [Google Scholar]
- 52.Bhushan R, Brückner H. Amino Acids. 2004;27:231. doi: 10.1007/s00726-004-0118-0. [DOI] [PubMed] [Google Scholar]
- 53.Gunasekera SP, Ritson-Williams R, Paul VJ. J Nat Prod. 2008;71:2060. doi: 10.1021/np800453t. [DOI] [PubMed] [Google Scholar]
- 54.Jiménez JI, Vanasch T, Yoshada WY, Sakamoto B, Pörzgen P, Horgen FD. J Nat Prod. 2009;72:1573. doi: 10.1021/np900173d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Teruya T, Sasaki H, Fukazawa H, Suenaga K. Org Lett. 2009;11:5062. doi: 10.1021/ol9020546. [DOI] [PubMed] [Google Scholar]
- 56.Gao X, Liu Y, Kwong S, Xu Z, Ye T. Org Lett. 2010;12:3018. doi: 10.1021/ol101021v. [DOI] [PubMed] [Google Scholar]
- 57.Zampella A, Randazzo A, Borbone N, Luciani S, Trevisi L, Debitus C, D’Auria MV. Tetrahedron Lett. 2002;43:6163. [Google Scholar]
- 58.Çalimsiz S, Morales ÁI, Lipton MA. J Org Chem. 2006;71:6351. doi: 10.1021/jo060351h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manning JM. 1970;92:7449. doi: 10.1021/ja00728a033. [DOI] [PubMed] [Google Scholar]
- 60.Hiraoka S, Harada S, Nishida A. J Org Chem. 2010;75:3871. doi: 10.1021/jo1003746. [DOI] [PubMed] [Google Scholar]
- 61.Bourdelais AJ, Jacocks HM, Wright JLC, Bigwarfe PM, Jr, Baden DG. J Nat Prod. 2005;68:2. doi: 10.1021/np049797o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fuwa H, Ebine M, Bourdelais AJ, Baden DG, Sasaki M. J Am Chem Soc. 2006;128:16989. doi: 10.1021/ja066772y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seo Y, Cho KW, Rho J-R, Shin J, Kwon B-M, Bok S-H, Song J-I. Tetrahedron. 1996;52:10583. [Google Scholar]
- 64.Pietruszka J, Rieche ACM. Adv Synth Catal. 2008;350:1407. [Google Scholar]
- 65.Fukuzawa S, Matsunaga S, Fusetani N. J Org Chem. 1995;60:608. doi: 10.1021/jo970091r. [DOI] [PubMed] [Google Scholar]
- 66.Phillips ST, Shair MD. J Am Chem Soc. 2007;129:6589. doi: 10.1021/ja0705487. [DOI] [PubMed] [Google Scholar]
- 67.Fukuzawa S, Matsunaga S, Fusetani N. Tetrahedron. 1995;51:6707. [Google Scholar]
- 68.Sorek H, Rudi A, Gueta S, Reyes F, Martin MJ, Aknin M, Gaydou E, Vacelet J, Kashman Y. Tetrahedron. 2006;62:8838. [Google Scholar]
- 69.Yu M, Pochapsky SS, Snider BB. J Org Chem. 2008;73:9065. doi: 10.1021/jo801956w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ospina CA, Rodriguez AD, Ortega-Barria E, Capson TL. J Nat Prod. 2003;66:357. doi: 10.1021/np0204500. [DOI] [PubMed] [Google Scholar]
- 71.Crimmins MT, Mans MC, Rodríguez AD. Org Lett. 2010;12:5028. doi: 10.1021/ol102169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bugni TS, Janso JE, Williamson RT, Feng X, Bernan VS, Greenstein M, Carter VS, Maiese WM, Ireland CM. J Nat Prod. 2004;67:1396. doi: 10.1021/np049973t. [DOI] [PubMed] [Google Scholar]
- 73.Burns AR, McAllister GD, Shanahan SE, Taylor RJK. Angew Chem Int Ed. 2010;49:5574. doi: 10.1002/anie.201002416. [DOI] [PubMed] [Google Scholar]
- 74.Kobayashi J, Kanda F, Ishibashi M, Shigemori H. J Org Chem. 1991;56:4574. [Google Scholar]
- 75.Shinada T, Ikebe E, Oe K, Namba K, Kawasaki M, Ohfune Y. Org Lett. 2007;9:1765. doi: 10.1021/ol0704789. [DOI] [PubMed] [Google Scholar]
- 76.Nakamura H, Wu H, Ohizumi Y, Hirata Y. Tetrahedron Lett. 1984;25:2989. [Google Scholar]
- 77.Marcos IS, Garcia N, Sexmero MJ, Basabe P, Diez D, Urones JG. Tetrahedron. 2005;61:11672. [Google Scholar]
- 78.Hattori T, Adachi K, Shizuri Y. J Nat Prod. 1997;60:411. doi: 10.1021/np970527y. [DOI] [PubMed] [Google Scholar]
- 79.Talpir R, Rudi A, Kashman Y. Tetrahedron. 1994;50:4179. [Google Scholar]
- 80.Snyder SA, Treitler DS, Brucks AP. J Am Chem Soc. 2010;132:14303. doi: 10.1021/ja106813s. [DOI] [PubMed] [Google Scholar]
- 81.Rovirosa J, Sepulveda M, Quezada E, San-Martin A. Phytochem. 1992;31:2679. [Google Scholar]
- 82.Areche C, San-Martín A, Rovirosa J, Muñoz MA, Hernández-Barragán A, Bucio MA, Joseph-Nathan P. J Nat Prod. 2010;73:79. doi: 10.1021/np900553p. [DOI] [PubMed] [Google Scholar]
- 83.Subrahmanyam C, Kumar SR. J Chem Res (S) 2000;4:182. [Google Scholar]
- 84.Zhang W, Guo YW, Gavagnin M, Mollo E, Cimino G. Helv Chim Acta. 2005;88:87. [Google Scholar]
- 85.Govindan M, Abbas SA, Schmitz FJ, Lee RH, Papkoff JS, Slate D. J Nat Prod. 1994;57:74. doi: 10.1021/np50103a010. [DOI] [PubMed] [Google Scholar]
- 86.Jiang RW, Lane AL, Mylacraine L, Hardcastle KI, Fairchild CR, Aalbersberg W, Hay ME, Kubanek J. J Nat Prod. 2008;71:1616. doi: 10.1021/np800307h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimbo K, Tsuda M, Izui N, Kobayashi J. J Org Chem. 2002;67:1020. doi: 10.1021/jo016089o. [DOI] [PubMed] [Google Scholar]
- 88.Ghosh AK, Gong G. J Org Chem. 2006;71:1085. doi: 10.1021/jo052181z. [DOI] [PubMed] [Google Scholar]
- 89.Kito K, Ookura R, Yoshida S, Namikoshi M, Ooi T, Kusumi T. Org Lett. 2008;10:225. doi: 10.1021/ol702598q. [DOI] [PubMed] [Google Scholar]
- 90.Ookura R, Kito K, Saito Y, Kusumi T, Ooi T. Chem Lett. 2009;38:384. [Google Scholar]
- 91.Liu J, Xu K, He J, Zhang L, Pan X, She X. J Org Chem. 2009;74:5063. doi: 10.1021/jo900820f. [DOI] [PubMed] [Google Scholar]
- 92.Díaz-Oltra S, Angulo-Pachón CA, Kneeteman MN, Murga J, Carda M, Marco JA. Tetrahedron Lett. 2009;50:3783. [Google Scholar]
- 93.Takada K, Uehara T, Nakao Y, Matsunaga S, van Soest RWM, Fusetani N. J Am Chem Soc. 2004;126:187. doi: 10.1021/ja037368r. [DOI] [PubMed] [Google Scholar]
- 94.Bowen EG, Wardrop DJ. J Am Chem Soc. 2009;131:6062. doi: 10.1021/ja9005755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Claridge TDW. High-Resolution NMR Techniques in Organic Chemistry. Elsevier; New York: 2008. [Google Scholar]
- 96.Encarnacion RD, Sandoval E, Malmstrom J, Christophersen C. J Nat Prod. 2000;63:874. doi: 10.1021/np990489d. [DOI] [PubMed] [Google Scholar]
- 97.Ogamino T, Nishiyama S. Tetrahedron Lett. 2005;46:1083. [Google Scholar]
- 98.Ogamino T, Obata R, Tomoda H, Nishiyama S. Bull Chem Soc Jpn. 2006;79:134. [Google Scholar]
- 99.Kita M, Ohishi N, Konishi K, Kondo M, Koyama T, Kitamura M, Yamada K, Uemura D. Tetrahedron. 2007;63:6241. [Google Scholar]
- 100.Murata T, Sano M, Takamura H, Kadota I, Uemura D. J Org Chem. 2009;74:4797. doi: 10.1021/jo900546k. [DOI] [PubMed] [Google Scholar]
- 101.Rao MR, Faulkner DJ. J Nat Prod. 2002;65:386. doi: 10.1021/np010495l. [DOI] [PubMed] [Google Scholar]
- 102.Son JB, Kim SN, Kim NY, Lee DH. Org Lett. 2006;8:661. doi: 10.1021/ol052851n. [DOI] [PubMed] [Google Scholar]
- 103.Sun J, Shi D, Ma M, Li S, Wang S, Han L, Yang Y, Fan X, Shi J, He L. J Nat Prod. 2005;68:915. doi: 10.1021/np050096g. [DOI] [PubMed] [Google Scholar]
- 104.Chen P, Wang J, Liu K, Li C. J Org Chem. 2008;73:339. doi: 10.1021/jo7021247. [DOI] [PubMed] [Google Scholar]
- 105.Guella G, Dini F, Pietra F. Angew Chem Int Ed. 1999;38:1134. doi: 10.1002/(SICI)1521-3773(19990419)38:8<1134::AID-ANIE1134>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 106.Nicolaou KC, Ortiz A, Zhang H, Dagneau P, Lanver A, Jennings MP, Arseniyadis S, Faraoni R, Lizos DE. J Am Chem Soc. 2010;132:7138. doi: 10.1021/ja100740t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nicolaou KC, Ortiz A, Zhang H, Guella G. J Am Chem Soc. 2010;132:7153. doi: 10.1021/ja100742b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Suzuki M, Takahashi Y, Mitome Y, Itoh T, Abe T, Masuda M. Phytochem. 2002;60:861. doi: 10.1016/s0031-9422(02)00151-6. [DOI] [PubMed] [Google Scholar]
- 109.Jeong W, Kim MJ, Kim H, Kim S, Kim D, Shin KJ. Angew Chem Int Ed. 2010;49:752. doi: 10.1002/anie.200905826. [DOI] [PubMed] [Google Scholar]
- 110.Ovenden SPB, Sberna G, Tait RM, Wildman HG, Patel R, Li B, Steffy K, Nguyen N, Meuer-Grimes BM. J Nat Prod. 2004;67:2093. doi: 10.1021/np0497494. [DOI] [PubMed] [Google Scholar]
- 111.Pérez-Balado C, Rodríguez-Graña P, de Lera ÁR. Chem Eur J. 2009;15:9928. doi: 10.1002/chem.200901056. [DOI] [PubMed] [Google Scholar]
- 112.Suzuki M, Kurosawa E. Phytochem. 1985;24:1999. [Google Scholar]
- 113.Wang J, Pagenkopf BL. Org Lett. 2007;9:3703. doi: 10.1021/ol701797e. [DOI] [PubMed] [Google Scholar]
- 114.Manzo E, Gavagnin M, Bifulco G, Cimino P, Micco SD, Ciavatta ML, Guo YW, Cimino G. Tetrahedron. 2007;63:9970. [Google Scholar]
- 115.Ola ARB, Babey AM, Motti C, Bowden BF. Aust J Chem. 2010;63:907. [Google Scholar]
- 116.Satake M, Ofuji K, Naoki H, James KJ, Furey A, McMahon T, Silke J, Yasumoto T. J Am Chem Soc. 1998;120:9967. [Google Scholar]
- 117.Nicolaou KC, Koftis TV, Vyskocil S, Petrovic G, Tang W, Frederick MO, Chen DYK, Li Y, Ling T, Yamada YMA. J Am Chem Soc. 2006;128:2859. doi: 10.1021/ja054750q. [DOI] [PubMed] [Google Scholar]
- 118.Ofuji K, Satake M, McMahon T, Silke J, James KJ, Naoki H, Oshima Y, Yasumoto T. Nat Toxins. 1999;7:99. doi: 10.1002/(sici)1522-7189(199905/06)7:3<99::aid-nt46>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 119.Nicolaou KC, Frederick MO, Petrovic G, Cole KP, Loizidou EZ. Angew Chem Int Ed. 2006;45:2609. doi: 10.1002/anie.200600295. [DOI] [PubMed] [Google Scholar]
- 120.Kobayashi J, Shimbo K, Sato M, Tsuda M. J Org Chem. 2002;67:6585. doi: 10.1021/jo016222c. [DOI] [PubMed] [Google Scholar]
- 121.Fürstner A, Bouchez LC, Morency L, Funel JA, Liepins V, Porée FH, Gilmour R, Laurich D, Beaufils F, Tamiya M. Chem Eur J. 2009;15:3983. doi: 10.1002/chem.200802067. [DOI] [PubMed] [Google Scholar]
- 122.Youssef DTA, Singab ANB, van Soest RWM, Fusetani N. J Nat Prod. 2004;67:1736. doi: 10.1021/np049853l. [DOI] [PubMed] [Google Scholar]
- 123.Jia R, Guo YW, Mollo E, Gavagnin M, Cimino G. Helv Chim Acta. 2006;89:1330. [Google Scholar]
- 124.Numata A, Iritani M, Yamada T, Minoura K, Matsumura E, Yamori T, Tsuruo T. Tetrahedron Lett. 1997;38:8215. [Google Scholar]
- 125.Usami Y, Horibe Y, Takaoka I, Ichikawa H, Arimoto M. Synlett. 2006:1598. [Google Scholar]
- 126.Zampella A, D’Auria MV, Minale L, Debitus C, Roussakis C. J Am Chem Soc. 1996;118:11085. [Google Scholar]
- 127.Carpenter J, Northrup AB, Chung D, Wiener JJM, Kim S-G, MacMillan DWC. Angew Chem Int Ed. 2008;47:3568. doi: 10.1002/anie.200800086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Evidente A, Conti L, Altomare C, Bottalico A, Sindona G, Segre AL, Logrieco A. Nat Toxins. 1994;2:4. doi: 10.1002/nt.2620020103. [DOI] [PubMed] [Google Scholar]
- 129.Hiramatsu F, Miyajima T, Murayama T, Takahashi K, Koseki T, Shiono Y. J Antibiot. 2006;59:704. doi: 10.1038/ja.2006.94. [DOI] [PubMed] [Google Scholar]
- 130.Diyabalanage T, Amsler CD, McClintock JB, Baker BJ. J Am Chem Soc. 2006;128:5630. doi: 10.1021/ja0588508. [DOI] [PubMed] [Google Scholar]
- 131.Nicolaou KC, Guduru R, Sun Y-P, Banerji B, Chen DY-K. Angew Chem Int Ed. 2007;46:5896. doi: 10.1002/anie.200702243. [DOI] [PubMed] [Google Scholar]
- 132.Nicolaou KC, Sun Y-P, Guduru R, Banerji B, Chen DY-K. J Am Chem Soc. 2008;130:3633. doi: 10.1021/ja710485n. [DOI] [PubMed] [Google Scholar]
- 133.Wright AE, Botelho JC, Guzmán E, Harmody D, Linley P, McCarthy PJ, Pitts TP, Pomponi SA, Reed JK. J Nat Prod. 2007;70:412. doi: 10.1021/np060597h. [DOI] [PubMed] [Google Scholar]
- 134.Custar DW, Zabawa TP, Scheidt KA. J Am Chem Soc. 2008;130:804. doi: 10.1021/ja710080q. [DOI] [PubMed] [Google Scholar]
- 135.Guella G, Mancini I, Öztunç A, Pietra F. Helv Chim Acta. 2000;83:336. [Google Scholar]
- 136.Braddock DC, Rzepa HS. J Nat Prod. 2008;71:728. doi: 10.1021/np0705918. [DOI] [PubMed] [Google Scholar]
- 137.Braddock DC, Millan DS, Pérez-Fuertes Y, Pouwer RH, Sheppard RN, Solanki S, White AJP. J Org Chem. 2009;74:1835. doi: 10.1021/jo8026577. [DOI] [PubMed] [Google Scholar]
- 138.Kobayashi J, Cheng JF, Ohta T, Nakamura H, Nozoe S, Hirata Y, Ohizumi Y, Sasaki T. J Org Chem. 1988;53:6147. [Google Scholar]
- 139.Nozawa N, Tsuda M, Ishiyama H, Sasaki T, Tsuruo T, Kobayashi J. Bioorg Med Chem. 2006;14:1063. doi: 10.1016/j.bmc.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 140.Kikuchi Y, Ishibashi M, Sasaki T, Kobayashi J. Tetrahedron Lett. 1991;32:797. [Google Scholar]
- 141.Schmidt EW, Raventos-Suarez C, Bifano M, Menendez AT, Fairchild CR, Faulkner DJ. J Nat Prod. 2004;67:475. doi: 10.1021/np034035z. [DOI] [PubMed] [Google Scholar]
- 142.Liu S, Cu YM, Nan FJ. Org Lett. 2008;10:3765. doi: 10.1021/ol801419m. [DOI] [PubMed] [Google Scholar]
- 143.Crews P, Kakou Y, Quiñoà E. J Am Chem Soc. 1988;110:4365. [Google Scholar]
- 144.Sonnenschein RN, Johnson TA, Tenney K, Valeriote FA, Crews P. J Nat Prod. 2006;69:145. doi: 10.1021/np0503597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Swersey JC, Ireland CM, Cornell LM, Peterson RW. J Nat Prod. 1994;57:842. doi: 10.1021/np50108a027. [DOI] [PubMed] [Google Scholar]
- 146.Tapiolas DM, Bowden BF, Abou-Mansour E, Willis RH, Doyle JR, Muirhead AN, Liptrot C, Llewellyn LE, Wolff CWW, Wright AD, Motti CA. J Nat Prod. 2009;72:1115. doi: 10.1021/np900099j. [DOI] [PubMed] [Google Scholar]
- 147.Suenaga K, Aoyama S, Xi W, Arimoto H, Yamaguchi K, Yamada K, Tsuji T, Yamada A, Uemura D. Heterocycles. 2000;52:1033. [Google Scholar]
- 148.Kita M, Miwa R, Widianti T, Ozaki Y, Aoyama S, Yamada K, Uemura D. Tetrahedron Lett. 2007;48:8628. [Google Scholar]
- 149.Boone MA, Tong R, McDonald FE, Lense S, Cao R, Hardcastle KI. J Am Chem Soc. 2010;132:5300. doi: 10.1021/ja1006806. [DOI] [PubMed] [Google Scholar]
- 150.Dai X, Wan Z, Kerr RG, Davies HML. J Org Chem. 2007;72:1895. doi: 10.1021/jo0618167. [DOI] [PubMed] [Google Scholar]
- 151.Tsuda M, Oguchi K, Iwamoto R, Okamoto Y, Kobayashi Ji, Fukushi E, Kawabata J, Ozawa T, Masuda A, Kitaya Y, Omasa K. J Org Chem. 2007;72:4469. doi: 10.1021/jo070414b. [DOI] [PubMed] [Google Scholar]
- 152.Tsuda M, Oguchi K, Iwamoto R, Okamoto Y, Fukushi E, Kawabata J, Ozawa T, Masuda A. J Nat Prod. 2007;70:1661. doi: 10.1021/np0702537. [DOI] [PubMed] [Google Scholar]
- 153.Xie J, Ma Y, Horne DA. Chem Commun. 2010;46:4770. doi: 10.1039/c0cc00628a. [DOI] [PubMed] [Google Scholar]
- 154.Fang L, Yang J, Yang F. Org Lett. 2010;12:3124. doi: 10.1021/ol1011423. [DOI] [PubMed] [Google Scholar]
- 155.Ghosh AK, Yuan H. Org Lett. 2010;12:3120. doi: 10.1021/ol101105v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tietze LF, Singidi RR, Gericke KM. Chem Eur J. 2007;13:9939. doi: 10.1002/chem.200700823. [DOI] [PubMed] [Google Scholar]
- 157.Lu L, Zhang W, Carter RG. J Am Chem Soc. 2008;130:7253. doi: 10.1021/ja803012n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smith ABI, Duffey MO, Basu K, Walsh SP, Suennemann HW, Fromont J. J Am Chem Soc. 2008;130:422. doi: 10.1021/ja078293k. [DOI] [PubMed] [Google Scholar]
- 159.Li S, Liang S, Xu Z, Ye T. Synlett. 2008:569. [Google Scholar]
- 160.Kobayashi S, Kobayashi J, Yazaki R, Ueno M. Chem Asian J. 2007;2:135. doi: 10.1002/asia.200600232. [DOI] [PubMed] [Google Scholar]
- 161.For references to quotes by several renowned chemists on this topic see Snyder and Nicolaou, 2005 (reference 2).
- 162.Yamada T, Iritani M, Minomura K, Kawai K, Numata A. Org Biomol Chem. 2004;2:2131. doi: 10.1039/b404459b. [DOI] [PubMed] [Google Scholar]
- 163.Koshino H, Satoh H, Yamada T, Esumi Y. Tetrahedron Lett. 2006;47:4623. [Google Scholar]
- 164.Shen YC, Lo KL, Lin YC, Khalil AT, Kuo YH, Shih PS. Tetrahedron Lett. 2006;47:4007. [Google Scholar]
- 165.Shen YC, Shih PS, Lin YS, Lin YC, Kuo YH, Kuo YC, Khalil AT. Helv Chim Acta. 2009;92:2101. [Google Scholar]
- 166.Jain S, Laphookhieo S, Shi Z, Fu LW, Akiyama S, Chen ZS, Youssef DT, van Soest RW, El Sayed KA. J Nat Prod. 2007;70:928. doi: 10.1021/np0605889. [DOI] [PubMed] [Google Scholar]
- 167.Jain S, Abraham I, Carvalho P, Kuang YH, Shaala LA, Youssef DTA, Avery MA, Chen ZS, El Sayed KA. J Nat Prod. 2009;72:1291. doi: 10.1021/np900091y. [DOI] [PubMed] [Google Scholar]
- 168.Williams DE, Austin P, Diaz-Marrero AR, Soest RV, Matainaho T, Roskelley CD, Roberge M, Andersen RJ. Org Lett. 2005;7:4173. doi: 10.1021/ol051524c. [DOI] [PubMed] [Google Scholar]
- 169.Liu H, Boudreau MA, Zheng J, Whittal RM, Austin P, Roskelley CD, Roberge M, Andersen RJ, Vederas JC. J Am Chem Soc. 2010;132:1486. doi: 10.1021/ja9102925. [DOI] [PubMed] [Google Scholar]
- 170.Rodríguez AD, Cóbar OM. Tetrahedron. 1993;49:319. [Google Scholar]
- 171.Ospina CA, Rodríguez AD. J Nat Prod. 2006;69:1721. doi: 10.1021/np060317y. [DOI] [PubMed] [Google Scholar]
- 172.Rodríguez AD, Cóbar OM, Martínez N. J Nat Prod. 1994;57:1638. doi: 10.1021/np50114a005. [DOI] [PubMed] [Google Scholar]
- 173.Plaza A, Bewley CA. J Org Chem. 2006;71:6898. doi: 10.1021/jo061044e. [DOI] [PubMed] [Google Scholar]
- 174.Matthew S, Paul VJ, Luesch H. Planta Med. 2009;75:528. doi: 10.1055/s-0029-1185332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kobayashi J, Cheng J-F, Ohta T, Nozoe S, Ohizumi Y, Sasaki T. J Org Chem. 1990;55:3666. [Google Scholar]
- 176.Ito T, Kitajima M, Takayama H. Tetrahedron Lett. 2009;50:4506. [Google Scholar]
- 177.Takahashi Y, Ishiyama H, Kubota T, Kobayashi J. Bioorg Med Chem Lett. 2010;20:4100. doi: 10.1016/j.bmcl.2010.05.071. [DOI] [PubMed] [Google Scholar]
- 178.Pina IC, Gautschi JT, Wang G-Y-S, Sanders ML, Schmitz FJ, France D, Cornell-Kennon S, Sambucetti LC, Remiszewski SW, Perez LB, Bair KW, Crews P. J Org Chem. 2003;68:3866. doi: 10.1021/jo034248t. [DOI] [PubMed] [Google Scholar]
- 179.McCulloch MWB, Coombs GS, Banerjee N, Bugni TS, Cannon KM, Harper MK, Veltri CA, Virshup DM, Ireland CM. Bioorg Med Chem. 2009;17:2189. doi: 10.1016/j.bmc.2008.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tsuda M, Endo T, Perpelescu M, Yoshida S, Watanabe K, Fromont J, Mikami Y, Kobayashi J. Tetrahedron. 2003;59:1137. [Google Scholar]
- 181.Kuramochi K, Saito F, Takeuchi R, Era T, Takemura M, Kobayashi J, Sakaguchi K, Kobayashi S, Sugawara F. Tetrahedron. 2006;62:8006. [Google Scholar]
- 182.Gunasekera SP, Isbrucker RA, Longley RE, Wright AE, Pomponi SA, Reed JK. J Nat Prod. 2004;67:110. doi: 10.1021/np030294c. [DOI] [PubMed] [Google Scholar]
- 183.Kanayama M, Nishiwaki K, Matsuo K. Heterocycles. 2006;68:233. [Google Scholar]
- 184.Lang G, Mitova MI, Ellis G, van der Sar S, Phipps RK, Blunt JW, Cummings NJ, Cole ALJ, Munro MHG. J Nat Prod. 2006;69:621. doi: 10.1021/np0504917. [DOI] [PubMed] [Google Scholar]
- 185.Han J, Su Y, Jiang T, Xu Y, Huo X, She X, Pan X. J Org Chem. 2009;74:3930. doi: 10.1021/jo900370a. [DOI] [PubMed] [Google Scholar]
- 186.Murata M, Matsuoka S, Matsumori N, Paul GK, Tachibana K. J Am Chem Soc. 1999;121:870. [Google Scholar]
- 187.Oishi T, Kanemoto M, Swasono R, Matsumori N, Murata M. Org Lett. 2008;10:5203. doi: 10.1021/ol802168r. [DOI] [PubMed] [Google Scholar]
- 188.Milanowski DJ, Gustafson KR, Rashid MA, Pannell LK, McMahon JB, Boyd MR. J Org Chem. 2004;69:3036. doi: 10.1021/jo0303113. [DOI] [PubMed] [Google Scholar]
- 189.Tone H, Buchotte M, Mordant C, Guittet E, Ayad T, Ratovelomanana-Vidal V. Org Lett. 2009;11:1995. doi: 10.1021/ol900184d. [DOI] [PubMed] [Google Scholar]
- 190.Gavagnin M, Mollo E, Cimino G, Ortea J. Tetrahedron Lett. 1996;37:4259. [Google Scholar]
- 191.Sharma P, Lygo B, Lewis W, Moses JE. J Am Chem Soc. 2009;131:5966. doi: 10.1021/ja900369z. [DOI] [PubMed] [Google Scholar]
- 192.El-Naggar M, Capon RJ. J Nat Prod. 2009;72:460. doi: 10.1021/np8007667. [DOI] [PubMed] [Google Scholar]
- 193.Grkovic T, Copp BR. Tetrahedron. 2009;65:6335. [Google Scholar]
- 194.He H, Faulkner DJ, Shumsky JS, Clardy J. J Org Chem. 1989;54:2511. [Google Scholar]
- 195.Wegerski CJ, Sonnenschein RN, Cabriales F, Valeriote FA, Matainaho T, Crews P. Tetrahedron. 2006;62:10393. [Google Scholar]
- 196.Williams PG, Yoshida WY, Moore RE, Paul VJ. J Nat Prod. 2002;65:29. doi: 10.1021/np0102253. [DOI] [PubMed] [Google Scholar]
- 197.Zhang W, Ma ZH, Mei D, Li CX, Zhang XL, Li YX. Tetrahedron. 2006;62:9966. [Google Scholar]
- 198.Jimenez JI, Scheuer PJ. J Nat Prod. 2001;64:200. doi: 10.1021/np000462q. [DOI] [PubMed] [Google Scholar]
- 199.Chen H, Feng Y, Xu Z, Ye T. Tetrahedron. 2005;61:11132. [Google Scholar]
- 200.Ciminiello P, Dell’Aversano C, Fattorusso E, Forino M, Magno S, Di Rosa M, Ianaro A. Eur J Org Chem. 2001;1:49. doi: 10.1021/jo001437s. [DOI] [PubMed] [Google Scholar]
- 201.Ciminiello P, Dell’Aversano C, Fattorusso E, Forino M, Magno S, Santelia FU, Moutsos VI, Pitsinos EN, Couladouros EA. Tetrahedron. 2006;62:7738. [Google Scholar]
- 202.Ford PW, Gustafson KR, McKee TC, Shigematsu N, Maurizi LK, Pannell LK, Williams DE, de Silva ED, Lassota P, Allen TM, Soest RV, Andersen RJ, Boyd MR. J Am Chem Soc. 1999;121:5899. [Google Scholar]
- 203.Xie W, Ding D, Zi W, Li G, Ma D. Angew Chem Int Ed. 2008;47:2844. doi: 10.1002/anie.200705557. [DOI] [PubMed] [Google Scholar]
- 204.Graber MA, Gerwick WH. Tetrahedron Lett. 1996;37:4635. [Google Scholar]
- 205.Miyaoka H, Hara Y, Shinohara I, Kurokawa T, Yamada Y. Tetrahedron Lett. 2005;46:7945. [Google Scholar]
- 206.Kinnel RB, Gehrken H-P, Scheuer PJ. J Am Chem Soc. 1993;115:3376. [Google Scholar]
- 207.Koeck M, Grube A, Seiple IB, Baran PS. Chem Int Ed. 2007;46:2320. doi: 10.1002/anie.200701798. [DOI] [PubMed] [Google Scholar]
- 208.Kinnel RB, Gehrken HP, Swali R, Skoropowski G, Scheuer PJ. J Org Chem. 1998;63:3281. [Google Scholar]
- 209.Buchanan MS, Carroll AR, Addepalli R, Avery VM, Hooper JNA, Quinn RJ. J Org Chem. 2007;72:2309. doi: 10.1021/jo062007q. [DOI] [PubMed] [Google Scholar]
- 210.McPhail KL, Gerwick WH. J Nat Prod. 2003;66:132. doi: 10.1021/np0204186. [DOI] [PubMed] [Google Scholar]
- 211.Li Y, Feng J-P, Wang W-H, Chen J, Cao X-P. J Org Chem. 2007;72:2344. doi: 10.1021/jo061456n. [DOI] [PubMed] [Google Scholar]
- 212.Cho JY, Williams PG, Kwon HC, Jensen PR, Fenical W. J Nat Prod. 2007;70:1321. doi: 10.1021/np070101b. [DOI] [PubMed] [Google Scholar]
- 213.Pal U, Ranatunga S, Ariyarathna Y, Valle JRD. Org Lett. 2009;11:5298. doi: 10.1021/ol902251c. [DOI] [PubMed] [Google Scholar]
- 214.Teruya T, Nakagawa S, Koyama T, Arimoto H, Kita M, Uemura D. Tetrahedron. 2004;60:6989. [Google Scholar]
- 215.Gao S, Wang Q, Chen C. J Am Chem Soc. 2009;131:1410. doi: 10.1021/ja808110d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nakao Y, Yoshida WY, Takada Y, Kimura J, Yang L, Mooberry SL, Scheuer PJ. J Nat Prod. 2004;67:1332. doi: 10.1021/np049949f. [DOI] [PubMed] [Google Scholar]
- 217.Takada Y, Umehara M, Nakao Y, Kimura J. Tetrahedron Lett. 2008;49:1163. [Google Scholar]
- 218.He K, Zeng L, Shi G, Zhao GX, Kozlowski JF, McLaughlin JL. J Nat Prod. 1997;60:38. doi: 10.1021/np960513c. [DOI] [PubMed] [Google Scholar]
- 219.Ding L, Pfoh R, Rühl S, Qin S, Laatsch H. J Nat Prod. 2009;72:99. doi: 10.1021/np8006843. [DOI] [PubMed] [Google Scholar]
- 220.Wu SJ, Fotso S, Li F, Qin S, Laatsch H. J Nat Prod. 2007;70:304. doi: 10.1021/np050358e. [DOI] [PubMed] [Google Scholar]
- 221.Pettit GR, Xu JP, Doubek DL, Chapuis JC, Schmidt JM. J Nat Prod. 2004;67:1252. doi: 10.1021/np030198b. [DOI] [PubMed] [Google Scholar]
- 222.Paterson I, Findlay AD, Florence GJ. Org Lett. 2006;8:2131. doi: 10.1021/ol060609q. [DOI] [PubMed] [Google Scholar]
- 223.Fu X, Hong EP, Schmitz FJ. Tetrahedron. 2000;56:8989. [Google Scholar]