Abstract
Carbonic anhydrase VI (CA VI), encoded by type A transcripts of the gene Car6, is a secretory product of salivary glands and is found in the enamel pellicle. Because higher caries prevalence is associated with lower salivary concentrations of CA VI in humans, we tested whether CA VI protects enamel surfaces from caries induced by Streptococcus mutans, using Car6−/− mice, in which salivary CA VI expression is absent. We detected aberrant Car6 type A transcripts in Car6−/− mice, likely targets for nonsense-mediated mRNA decay. Expression of the intracellular stress-induced isoform of CA VI encoded by type B transcripts was restricted to parotid and submandibular glands of wild type mice. The salivary function of Car6−/− mice was normal as assessed by the histology and protein/glycoprotein profiles of glands, salivary flow rates and protein/glycoprotein compositions of saliva. Surprisingly, total smooth surface caries and sulcal caries in Car6−/− mice were more than 6-fold and 2-fold lower than in wild type mice after infection with S. mutans strain UA159. Recoveries of S. mutans and total microbiota from molars were also lower in Car6−/− mice. To explore possible mechanisms for increased caries susceptibility, we found no differences in S. mutans adherence to salivary pellicles, in vitro. Interestingly, higher levels of Lactobacillus murinus and an unidentified Streptococcus species were cultivated from the oral microbiota of Car6−/− mice. Collective results suggest salivary CA VI may promote caries by modulating the oral microbiota to favor S. mutans colonization and/or by the enzymatic production of acid within plaque.
Keywords: carbonic anhydrase, saliva, Streptococcus mutans, oral colonization, nonsense-mediated decay, knockout mice
1. Introduction
As the portal for ingestion of water and food nutrients, the oral cavity is constantly under attack by microbiota from the external environment. The flow of saliva combined with salivary microbial-interacting constituents and with factors that can buffer and re-mineralize tooth surfaces function together to help protect the hard oral surfaces from mineral dissolution (i.e., caries) by acidogenic microorganisms such as Streptococcus mutans. S. mutans is the organism most consistently associated with dental caries worldwide, but is further linked to bacterial endocarditis [1] and to atheromatous plaques [2], suggesting an interconnection between oral infections and cardiovascular diseases. Hence, efforts to understand and treat oral diseases have potential ramifications to serious systemic disease processes.
First discovered in ovine saliva [3], carbonic anhydrase VI (CA VI) is the only secretory isozyme of the CA gene family. It is also found in other secretory systems such as lacrimal glands [4, 5], tracheobronchial glands [6] and nasal glands where it may function in olfaction [7]. It is also found in high concentrations in colostrum, suggesting a role in the development of the alimentary tract [8]. In the diverse system of salivary glands CA VI is produced in the parotid and submandibular glands [9] as well as minor salivary glands of the tongue, including von Ebner’s glands [10]. Although many carbonic anhydrase isoforms are key enzymes for pH regulation in tissues and biological fluids, CA VI does not appear to regulate the pH of whole saliva, but instead may function in oral microenvironments [11]. For example, CA VI within von Ebner’s gland secretions bathing taste receptors of the circumvallate and foliate papillae [10] may function in the growth and development of taste buds [12–14].
CA VI is also a component of the enamel pellicle, a thin layer of proteins between teeth enamel and overlying bacterial plaque [15]. A higher prevalence of caries is associated with lower concentrations of CA VI in the saliva of human subjects, thus raising the hypothesis that CA VI serves to protect enamel surfaces from caries, possibly through the removal of bacterial derived hydrogen ions within the microenvironment near the enamel surface by catalyzing the interaction of hydrogen ions with salivary bicarbonate ions to form CO2 and H2O [16]. An attractive model to test this hypothesis are mice in which targeted deletion of the gene encoding CA VI, Car6, display an absence of CA VI expression in salivary glands as confirmed in Western blots and by immunohistochemistry [17, 18]. The targeted deletion in Car6−/− mice removes Car6 exon 3 and part of exon 4, leaving the 3′-end of this latter exon. Both exons are normally incorporated into the two known isoforms of CA VI expressed by the Car6 gene, the secreted enzyme (type A) and an intracellular form (type B) [19]. Type B transcripts use a promoter within intron 1, are stress-induced in mouse NIH 3T3 fibroblasts and were initially detected in salivary tissue, although the type of salivary tissue was not specified [19]. Expression of the type B isoform by the three different major salivary glands in mice is therefore unclear, as is whether its deleted expression alters salivary function. Moreover, it is not known whether the transcriptional machinery in Car6−/− mice reads through the inserted neomycin cassette to reach the remaining exon 4 splice site and, if so, whether it is utilized during pre-mRNA splicing to create an aberrant translated message that may disrupt salivary function.
In the current study, we assessed whether the loss of Car6 gene expression has a significant impact on the cellular structure of the major salivary glands and on salivary constituents and flow. Furthermore, consequences from the absence of CA VI on the functions of saliva related to protection against caries development were tested, both in vitro and in vivo.
2. Materials and methods
2.1 Materials
Unless indicated, all materials were from Sigma Chemical Co., St. Louis, MO. All kits were used according to manufacturer’s instructions.
2.2 Animals and collection of glands
The University of Florida IACUC committee approved all animal procedures. del in the C57BL/6 genetic background has been described previously [17, 18]. Animals were anesthetized by CO2 inhalation and killed by sectioning the aorta. Mice were genotyped as described previously [18]. Unless indicated, tissues were excised, blotted on filter paper, flash frozen in liquid nitrogen and stored at −80 °C.
2.3 Caries experimental protocol and scoring
The caries protocol was similar to that reported previously [20] with modifications. Briefly, timed pregnancies were established using twenty breeding pairs of heterozygous (+/−) Car6 mice. Females were negative for indigenous S. mutans as determined by streaking oral swabs on Mitis Salivarius agar (Becton, Dickinson and Co., Sparks, MD) with 1% Tellurite solution (Becton, Dickinson and Company), 20% sucrose, and 0.2 units/ml bacitracin (MSB) [21]. Pups were marked for identification with ear clips at 14 days of age and genotyped. At 16–17 days of age, each dam with pups were transferred to a BSL2 suite of the vivarium in microisolator cages containing a wire bottomed insert and a thin layer of corn-cob bedding underneath. S. mutans UA159 from a frozen low-passage aliquots were grown overnight in Brain Heart Infusion medium + 0.5% glucose (BHI; Becton, Dickinson and Co.) and concentrated to approximately 1010 CFU/ml by centrifugation. The dams and pups were then inoculated by oral swabbing, which delivers about 10 μl (108 CFU) of the concentrated solution. The diet was converted to powdered Diet 2000 (56% sucrose) with 5% sucrose water. Pups and dams were re-inoculated each of the next two days. At 21 days of age, pups were weaned and caged in pairs with non-littermates of the same sex. Pups were screened for S. mutans colonization 5 days after the initial infection by plating oral swabs on MSB. Non-infected mice were inoculated and subsequently tested as before. All were positive for S. mutans colonization. Mice were weighed weekly. Mice were sacrificed seven weeks after the initial inoculation. The left and right mandibles from twelve mice in each group were removed aseptically to assay for bacterial colonization of the molars (see below). The skulls and remaining mandibles were defleshed over an 18–24 h period by Dermestid beetles (Dermestes maculates) as described by Tanzer [22]. Buccal, lingual and proximal surfaces of all molars were scored for smooth surface caries with the aid of an Olympus dissecting microscope (Model SZX16). The teeth were then stained with murexide (0.024% w/v in 70% ethanol) and prepared for sectioning along the midline in order to score sulcal caries and to determine severity scores of proximal surfaces. Skulls and mandibles were prepared for sectioning by embedding in Embed-It Low Viscosity Epoxy (Polysciences, Warrington, PA) to a level about 1 mm below the enamel crown. Epoxy blocks were mounted in a custom made vice attached to a Narishige micromanipulator (Narishige International USA, Inc, East Meadow, NY) and the exposed teeth sectioned with a double-sided diamond-coated abrasive disk (0.15 mm thick, Abrasive Technology, Lewis Center, OH) attached to a hand piece (Foredom model 44T, Foredom Electric Co., Bethel, CT) and motor unit (Foredom Model SM; 18,000 RPM). The hand piece is mounted on a ball-bearing slider that restricts movement of the abrasive disk along the aligned midline of the three molars. The teeth were sectioned under a light flow of water.
Scoring was according to Larson’s modification of the Keyes’ scoring system and was conducted by a single calibrated and blinded examiner [23, 24]. The linear evaluations of enamel lesions were expressed as E, while severities were expressed as Ds, Dm and Dx (slight dentin involvement, moderate dentin involvement or penetrates the dentin, respectively). To stabilize variances, caries scores were expressed as proportions of their maximum possible values (124 for smooth surface caries and 56 for sulcal surface caries) and then the arcsine of the square root of the proportions calculated, as described previously [20].
2.4 Recoveries of S. mutans and total bacteria from molars
The mandibles from twelve mice in each caries group (wild type and Car6−/−) were aseptically extracted and underwent dissection to remove loose tissue on the bone near the molars. The bone was sectioned about 2 mm anterior to the first molar and 2 mm posterior to the third molar. Each pair of prepared mandible/molars from each mouse were sonicated on ice in 5 ml of ice-cold sterile 0.9% saline using three pulses of 10 s with 30 s intervals, at 50% power with a Branson Digital Sonifier 250. Sonicates from two non-cagemates were pooled. From each 10 ml we removed 200 μl to prepare 10-fold serial dilutions that were plated (50 μl per 60 mm plate) onto MSB agar and onto blood agar to estimate colonization of the molars by S. mutans (MSB agar) and by total bacteria (blood agar). Colony forming units (CFU) were then determined from 48 h cultures.
2.5 Preparation of cDNA
Preparation was as described previously [25]. Briefly, frozen tissues or growing cell cultures were homogenized directly in TRIzol® Reagent (Invitrogen, Carlsbad, CA), total RNA isolated using standard protocols and then treated with DNase I using Ambion’s DNA-free™ Reagent Kit (Applied Biosystems, Foster City, CA). Removal of genomic DNA was verified by the inability of the sample to amplify the mouse microsatellite marker D1Mit46 (UniSTS 116254). DNase I treated RNA was reverse transcribed with random primers (High Capacity cDNA Archive kit, Applied Biosystems), resultant cDNA purified (QIAquick® PCR purification system, Qiagen, Valencia, CA) and quantified (Quant-iT™ dsDNA HS assay kit, Invitrogen).
2.6 RT-PCR of Car6 transcripts
PCR reactions (10 ng cDNA) were: 3 min at 94 °C; 40 cycles of 30 s at 94 °C, 30 s at 52 °C and 90 s at 72 °C followed by 1 min at 72 °C. Products were resolved in 1.5 % agarose gels made in TAE buffer. Primers were from Invitrogen and included: Car6 type A, exon 1, 5′-UTR, forward (5′-TGCATTCAGGGCTACAGCATC-3′) and exon 8, 3′-UTR, reverse (5′-GGGTTAATCAGAGACTCGCTGAAG-3′); Car6 type B, exon 2, 5′-UTR, forward (5′-TGCTGGGCTTAGTTTAGAGCTTTC-3′) and the same reverse primer as for type A; and beta-actin forward (5′-CACCTTCCAGCAGATGTG-3′) and reverse (5′-AAATCCTGAGTCAAAAGCG-3′). Reactions were run in the FailSafe PCR System (Epicentre, Madison, WI), buffers G, B and C for type A, type B and beta-actin primers, respectively. PCR products were confirmed by direct sequencing of gel-purified bands (QIAquick Gel Extraction Kit; Qiagen). NIH 3T3 mouse fibroblasts were grown in Dulbecco modified Eagle medium in the presence of 10% fetal bovine serum to 80–90% confluency then treated with 2 μg/ml tunicamycin (Sigma) for 10 h before isolation of RNA. Sequencing was performed at the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) using Applied Biosystems 3100 Genetic Analyzer ABI and Big Dye version 3.1 chemistry.
2.7 Comparison of Indigenous Oral Bacteria
The oral cavities of Car6 +/+ and Car6 −/− littermates were swabbed with sterile cotton applicators pre-wetted with sterile saline and applicator tips vortexed vigorously in 1mL of BHI medium. Each medium was diluted 3-, 10-, and 100-fold. Duplicate sheep blood agar plates (100 mm) were then streaked with 50 μl of each dilution. Plates were incubated 24 h at 37 °C under aerobic (air - 5% CO2) and anaerobic conditions. Total recovered CFUs for each of the six distinct colony morphotypes were calculated from the 24 h and 48 h plates. Isolate plates were then prepared from the aerobic plates from each of three mice of each genotype by streaking colonies representative of each colony morphotype on blood agar and culturing under aerobic conditions. A single colony from each isolate plate was subjected to colony PCR [26] using degenerate primers as reported by Paster et al. [27] that are complementary to the 5′- and 3′-ends of the 16S rRNA gene (D88, 5′-GAGAGTTTGATYMTGGCTCAG-3′; E94, 5′-GAAGGAGGTGWTCCARCCGCA-3′). PCR conditions were: 5 min at 95 °C; 5 cycles of 1 min at 95 °C, 1 min at 47 °C and 2 min at 68 °C; 25 cycles of 1 min at 95 °C, 1 min at 53 °C and 2 min at 68 °C; followed by 10 min at 68 °C. Reactions (25 μl) were run using the AccuPrime Taq DNA Polymerase High Fidelity kit (Invitrogen) with 100 μM primers. PCR products were purified from 1.2% agarose gels and sequenced directly, as described above for Car6 transcripts, using the same primers as for PCR as well as the internal primers E341F - 5′-CCTACGGGAGGCAGCAG-3′ and E907R - 5′-CCGTCAATTCMTTTRAGTTT-3′ [28]. High quality sequences from each colony were aligned using ClustalW alignment (MacVector, v12.0.3; MacVector, Inc, Cary, NC) to yield each full-length sequence. The three full-length sequences for each colony morphotype from each mouse genotype were compared and a consensus sequence derived by ClustalW. Consensus sequences were compared to two sets of 16S rRNA gene reference sequences, Human Oral Microbiome Database (HOMD) [29] 16S rRNA RefSeq and sequences of the Ribosome Database Project, release 10 [30]. Alignments were performed via the BLAST server at HOMD (www.homd.org/index.php).
2.8 SDS-PAGE
Frozen glands were immediately weighed (Mettler MT-5 microbalance; Mettler Toledo, Columbus, OH) and 10–20 mg wet weight of tissue placed in 300 to 500 μl of the appropriate sample buffer supplied for each gel type (Bis-Tris or Tricine, see below for gel types). Samples were then sonicated (4 × 10 s pulses at 30 s intervals) with a Branson Digital Sonifier 250 at 30% amplitude (Branson Ultrasonic Corp., Danbury, CT), boiled 10 min and centrifuged (10,000g × 10 min) at 4°C. Supernatant volumes equivalent to 150–200 μg wet weight of original tissue were applied directly to either 4%–12% NuPAGE Bis-Tris gradient gels (Invitrogen) or 10%–20% Novex Tricine Gels (Invitrogen). Gels were stained for proteins with Coomassie blue using standard protocols, or to assay highly glycosylated glycoproteins the gels were stained with Alcian blue as described previously [31]. Digital images of gels were obtained with a Scion Grayscale 1394 Digital Camera (Fotodyne, Hartland, WI).
2.9 Histochemistry
Animals were anesthetized by CO2 inhalation, the superior and inferior vena cava exposed and cut, then the vasculature perfused via the aorta for 3 min with ice-cold PBS containing 1 unit/ml heparin at a rate of 5 ml/min. The perfusate was then switched to 4% paraformaldehyde in PBS for 20 min. Tissues were excised and incubated in the same fixative for 5 h with rocking, washed three times in PBS for 10 min, then embedded in paraffin. Sections (5 μm) were de-paraffinized and stained with hematoxylin and eosin. Images were captured with a Micropublisher 5.0 RTV digital camera (Q Imaging, Pleasanton, CA) using QCapture Pro software (v5.01, Q Imaging) on a Leica DM LB2 microscope (Leica Microsystems, Bannockburn, IL) equipped with 100 W high-pressure mercury lamp and Chroma fluorescent filter cubes (Chroma Technology Corp., Rockingham, VT).
2.10 Timed collection of stimulated whole saliva
Mice (> 6 weeks of age) were weighed and then anesthetized (ketamine, 75 mg/kg, plus xylazine, 8 mg/kg, IP). Saliva secretion was stimulated by subQ injection of 5 mg/kg pilocarpine/0.5 mg/kg isoproterenol (resting saliva flow is too low to measure in mice). Each mouse was positioned on its side with its head slightly down and the pool of saliva collecting next to the cheek transferred to a 1.5 ml centrifuge tube on ice at regular intervals using a 200 μl pipette. A new collection tube was incorporated at 5 min intervals over a 20 min period. Mice were then euthanized by CO2 asphyxiation and exsanguinated. Tubes were centrifuged (2,000 × g × 4 min) to pellet debris and the volume of saliva in each tube was measured by drawing the supernatant into a tip attached to a calibrated adjustable pipette. Saliva secretion is expressed as μl (5 min) −1 (g body weight) −1.
2.11 Adherence of S. mutans to saliva-coated hydroxyapatite-discs
Adherence assays incorporated the cell permeable DNA-binding stain (SYTO-13, Invitrogen) to quantify bacteria bound to hydroxyapatite discs (0.25 in. diameter circular discs, Clarkson Chromatography Products Inc., South Williamsport, PA) positioned in the bottom of wells of 96-well plates (CulturPlate-96 Black, Perkin Elmer, Waltham, MA). To limit interference from indigenous bacteria within saliva samples, we used S. mutans UA159 containing the ΩKm element integrated into the gtfA gene, thus conferring kanamycin resistance (kindly provided by Dr. RA Burne). Disruption of gtfA gene expression does not alter carbohydrate metabolism except for melibiose and raffinose and was shown not to effect caries development in rats [32]. Saliva samples streaked onto blood agar containing 1 mg/ml kanamycin failed to produce colonies under aerobic or anaerobic conditions (not shown). All wells were washed three times with adherence buffer (AB) consisting of 50 mM KCl, 1.0 mM KPO4, 1.0 mM CaCl2 and 0.1 mM MgCl2, pH 6.5. Whole stimulated saliva samples were collected on ice as described above and stored at −80 °C. Thawed saliva from a single animal was used for each assay. Saliva was brought to an appropriate volume with AB containing 1mg/ml kanamycin (ABK) using 15 strokes of a siliconized ground-glass homogenizer and this suspension used to prepare additional saliva dilutions in ABK by conventional pipetting. Wells containing hydroxyapatite discs received either 100 μl diluted saliva or 100 μl ABK (no saliva controls). Plates were incubated 1 h at 37 °C on a rotary shaker (Titer Plate Shaker, Lab-Line Instruments Inc., Melrose, IL) at 300 rpm. Discs were washed twice with 200 μl ABK. All washing steps were with an 8-channel electronic pipette, setting 3 (BIOHIT, Helsinki, Finland).
Low-passage frozen cell aliquots were cultured overnight in BHI containing 1 mg/ml kanamycin. Cells in suspension were centrifuged (5,000g × 15 min), resuspended in pre-warmed semi-defined biofilm medium (BM) containing 1% glucose and 1 mg/ml kanamycin, and then incubated 4 h at 37 °C in air-5% CO2 [33]. Pelleted cells were resuspended in 10 ml ABK to a concentration of 3 × 109 cells/ml and sonicated to de-chain the cells using three pulses of 15 s with 60 s intervals, at 60% power with a Branson Digital Sonifier 250. Cells were pelleted and resuspended in ABK to an OD600 of 0.75 (109 cells/ml). Aliquots (100 μl) of this cell suspension were added to half of the freshly washed discs. The other half received 100 μl ABK and served as “saliva-only” background controls. Plates were incubated 1 h at 37 °C with rotary shaking as described above. Hydroxyapatite discs were then washed twice at 37 °C with 200 μl of AB for 1 min with rotary shaking at 300 rpm and 100 μl AB added to each well. To develop a standard curve of cell numbers versus fluorescence, 100 μl of the concentrated cell suspension and appropriate 10-fold dilutions were added to wells without discs. To quantify bacteria, 100 μl of 10 μM SYTO-13 was added to each well, plates incubated in the dark for 25 min at room temperature and fluorescence read (488nm ex/509nm em) using a Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT) at sensitivity 70. All experimental conditions were performed in triplicate. The mean fluorescent reading for each experimental condition (saliva + cells), after subtracting the mean for the appropriate saliva-only controls, were converted to cell numbers based on the standard curve. Values for each experimental condition were then normalized to the mean for cells bound to discs without saliva (100%). The correlation coefficients of linear regressions of standard curves were >0.998. Mean fluorescence of the saliva-only background controls ranged from 22–35 percent of the mean for each associated experimental condition (saliva + cells).
2.12 Statistical Comparisons
Statistical comparisons between wild type and Car6−/− mice were conducted using the unpaired, two tailed t-test with Prism 5.0 software (GraphPad Software, Inc., San Diego, CA).
3. Results
3.1 Expression of type A and B Car6 transcripts in major salivary glands of wild type and Car6−/− mice
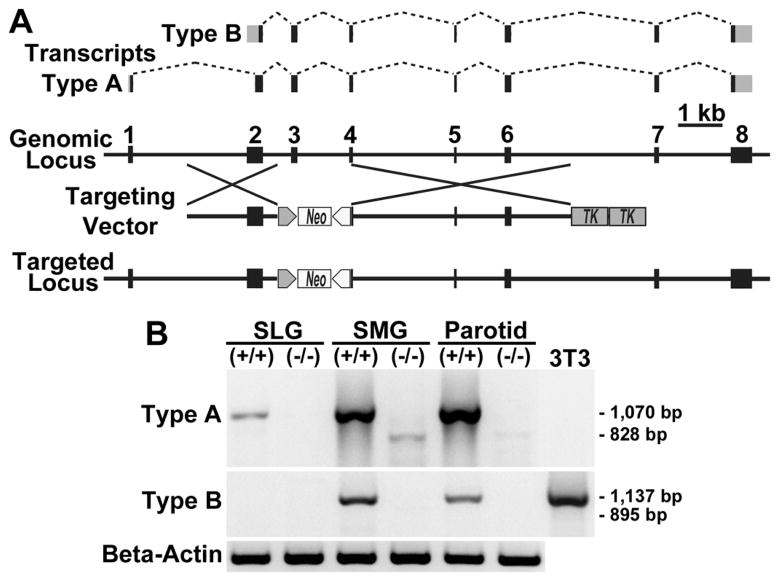
Shown in Fig. 1A is an exon map of transcripts type A and B compared to the genomic Car6 locus. Also shown is a map of the targeting vector and resultant targeted locus used in creation of Car6−/− mice. Transcription of the targeted locus is expected to produce transcripts without exons 3 and 4, due to deletion of exon 3 and loss of the 3′-splice site of exon 4. As shown in Fig. 1B, PCR products for type A transcripts (1,070 bp) are expressed in all three major salivary glands of wild type mice. Interestingly, products for aberrant type A transcripts (those missing sequences from exons 3 and 4; 828 bp) are detectable at much lower levels in parotid and submandibular glands of Car6−/− mice, thus verifying that the sequence remaining from exon 4 after targeted deletion is not spliced. Products for type B transcripts (1137 bp) are detected only in parotid and submandibular glands of wild type mice, whereas aberrant type B transcripts (895 bp predicted size) are not detected in the glands of Car6−/− mice.
Fig. 1.
Car6 transcripts in the major salivary glands of wild type and Car6−/− mice. A: Exon usage for transcripts of type A and type B are shown. Gray areas represent untranslated regions. Below is a diagram of the genomic locus and its modification by the targing vector to create the targeted locus in which exon 3 and part of exon 4 are deleted. The neo and thymidine kinase (TK) cassettes are under control of the phosphoglycerate kinase promoter. See Pan et al. [17] for targeting vector details. B: RT-PCR of random primed cDNA from the three major salivary glands from wild type (+/+) and Car6−/− mice to probe for Car6 transcripts using primer pairs targeting the distinct 5′-UTRs of type A and B transcripts, with the reverse primer in exon 8. Primers to type A transcripts produce the expected 1,070 bp products in all three glands from wild type mice. Products of 828 bp are detected in parotid and submandibular glands of Car6−/− mice and from subsequent sequencing correspond to aberrant type A transcripts with exons 3 and 4 omitted. When probed for type B transcripts, products are detected in only submandibular and parotid glands of wild type mice. Tunicamycin-treated NIH 3T3 cells serve as a positive control for type B transcripts [19]. Beta-actin controls are shown below. SLG, sublingual gland; SMG, submandibular gland.
3.2 Phenotypic characterization of the salivary function of Car6−/− mice
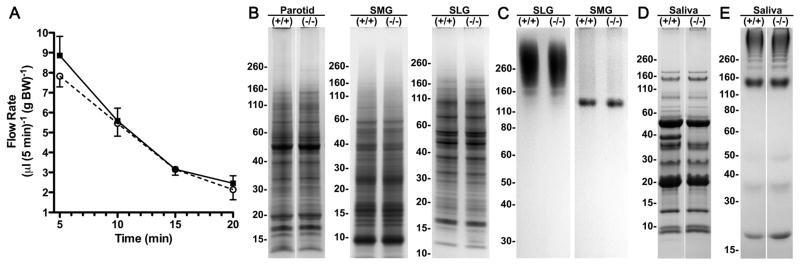
As shown in Fig. 2, deletion of CA VI expression has no apparent effect on the fine morphology of acinar and ductal cell types in the three major salivary glands. In addition, stimulated secretion of whole saliva by wild type and Car6−/− mice are equivalent, indicating salivary function with respect to fluid flow is not affected (Fig. 3A). We further find no apparent differences in protein profiles in the three major glands by SDS-PAGE (Fig. 3B). Expression of the high molecular weight mucin, Muc19, by sublingual glands and the low molecular weight mucin, Muc10, by submandibular glands are unaffected in Car6−/− mice (Fig. 3C). Apparent in Fig. 3D is the markedly similar expression profiles of proteins in whole stimulated saliva from wild type and Car6−/− mice, albeit for the absence of a major band at 38 kDa, consistent with previous Western blots demonstrating no detectable expression CA VI of similar size in parotid and submandibular glands [18]. We also found the consistent lower expression in saliva samples from Car6−/− mice of a light band of unknown identity at about 63 kDa. Fig. 3E provides confirmation that the composition of highly glycosylated glycoproteins in whole stimulated saliva is not altered in Car6−/− mice. The absence of CA VI in Car6−/− mice combined with no significant disruptions in salivary functions (i.e., glandular protein/glycoprotein profiles, glandular cell morphologies, salivary flow and saliva protein/glycoprotein composition) allows for the assessment of the contribution of salivary CA VI to caries development in mice challenged with S. mutans.
Fig. 2.
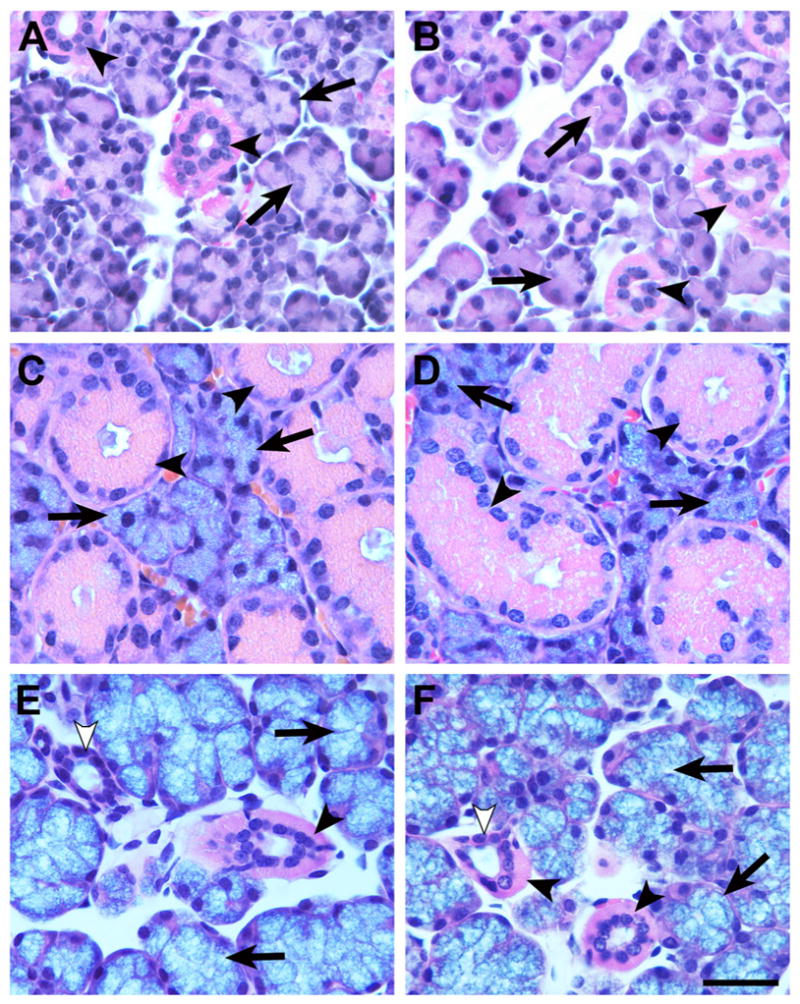
Hematoxylin and eosin stained paraffin sections (5 μm) of the three major salivary glands, parotid (A and B), submandibular (C and D) and sublingual (E and F), from wild type (A, C, E) and Car6−/− (B, D, F) mice. Acinar cells and ductal cells in each of the three gland types display similar morphologies in wild type and Car6−/− mice. A and B: Parotid serous acinar cells (arrows) and striated ducts (arrowheads). C and D: Seromucous acinar cells (arrows) and granular ducts (arrowheads) of submandibular glands. E and F: Sublingual mucous acinar cells (arrows), striated ducts (black arrowheads) and intercalated ducts (white arrowheads). Scale bar in F = 30 μm.
Fig. 3.
Comparisons between wild type and Car6−/− mice in salivary flow and by protein and glycoprotein expression in saliva and the three major salivary glands. (A) Flow of stimulated whole saliva by wild type (closed squares) and Car6−/− (open circles) mice. See Materials and Methods for details. Values are means ± SD from five animals of each genotype. Comparisons of the mean values between the two groups at each time point indicated no statistical difference (P > 0.05). (B–E) SDS-PAGE of homogenates from the major glands and from saliva samples of wild type (+/+) and Car6−/− mice. Mobilites of molecular size markers are indicated to the left of each gel panel. (B) Homogenates from parotid (150 μg wet wt.), submandibular (SMG; 200 μg wet wt.) and sublingual (SLG; 150 μg wet wt.) glands run on 10–20% gradient gels and stained with Coomassie blue. (C) Homogenates from SMGs (150 μg wet wt.) and SLGs (150 μg wet wt.) glands run on 4–12% gradient gels and stained with Alcian blue to detect high molecular weight Muc19 mucin glycoproteins in SLGs and the smaller Muc10 in SMGs. Parotid gland homogenates do not stain with Alcian blue (not shown). (D) Stimulated whole saliva samples (6 μl/lane) from female mice run on a 10–20% gradient gel and stained with Coomassie blue. Absence in the Car6−/− sample is a prominent band at 38 kDa, consistent with the absence of CA VI. A faint band at approximately 63 kDa appears down-regulated in Car6−/− mice saliva. (E) Stimulated whole saliva samples (9 μl) were run on a 4–12% gradient gel and stained with Alcian blue.
3.3 Caries development and recovery of bacteria
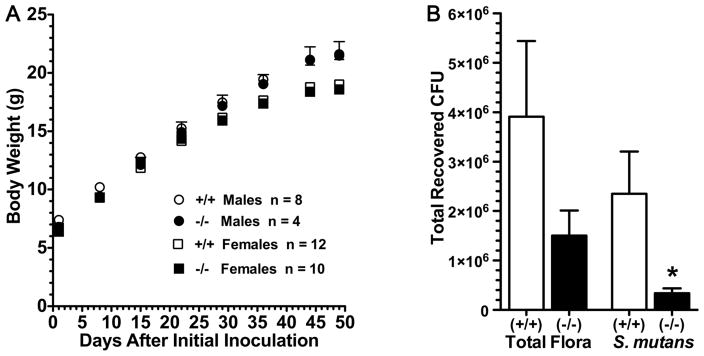
Shown in Table 1 are results of caries development in Car6−/− and wild type mice infected with S. mutans UA159. Of the 20 breeding pairs of heterozygous mice that were mated, 11 females became pregnant and delivered a total of 69 pups. Of these, 21 were of the wild type phenotype but only 15 were homozygous mutants. One pup died prior to initiating the caries protocol, thus the Car6−/− group contained 14 mice. Nevertheless, we find significant and surprising differences between the two strains. Total smooth surface caries is more than 6-fold lower in Car6−/−mice and the proportional differences between the two genotypes increases further with increasing levels of caries severities (i.e., Ds, Dm and Dx). In wild type mice the localization of smooth surface caries was mostly confined to buccal and lingual surfaces with a trend towards higher levels on lingual surfaces. This same trend continued in Car6−/− mice, but lingual and buccal caries scores were more than 5-fold and 7-fold lower, respectively, compared to wild type mice. Proximal smooth caries was low in wild type mice and nonexistent in Car6−/− mice. Total sulcal caries was more than 2-fold less in Car6−/− mice and as with smooth surface caries, the differences between the two strains of mice became more pronounced with increasing severity scores. Weight gains by males and females during the caries experiment were markedly similar between the two genotypes and all mice consistently appeared healthy and were active (Fig. 4A). The lower caries levels in Car6−/− mice are consistent with the recoveries of S. mutans and total microbiota after sonication of mandibular molars (Fig. 4B). Recovered S. mutans levels were 7-fold lower in Car6−/− mice compared to the wild type. The mean total cultivable microbiota was less in Car6−/− mice, but given the large variations in CFU values obtained, this difference is not statistically significant.
Table 1.
Comparisons of the development caries and their severities on molar smooth and sulcal surfaces between wild type and Car6−/− mice.
| Wild Type (+/+) N = 20 |
Car6 (−/−) N = 14 |
|
|---|---|---|
| SMOOTH SURFACES | ||
| Total - E | 6.60 (2.04) | 1.00 (0.49) † |
| Total - Ds | 3.75 (1.36) | 0.43 (0.25) † |
| Total - Dm | 1.95 (0.87) | 0.14 (0.14) |
| Total - Dx | 1.00 (0.56) | 0 (0) |
| Buccal E | 2.65 (1.11) | 0.36 (0.23) † |
| Buccal Ds | 1.45 (0.63) | 0.21 (0.21) † |
| Buccal Dm | 0.75 (0.35) | 0.14 (0.14) |
| Buccal Dx | 0.40 (0.23) | 0 (0) |
| Lingual E | 3.35 (0.77) | 0.64 (0.34) † |
| Lingual Ds | 1.80 (0.56) | 0.21 (0.15) † |
| Lingual Dm | 0.90 (0.40) | 0 (0) |
| Lingual Dx | 0.40 (0.24) | 0 (0) |
| Proximal E | 0.60 (0.36) | 0 (0) |
| Proximal Ds | 0.50 (0.35) | 0 (0) |
| Proximal Dm | 0.30 (0.21) | 0 (0) |
| Proximal Dx | 0.20 (0.14) | 0 (0) |
| SULCAL SURFACES | ||
| E | 10.65 (2.29) | 4.71 (0.98) † |
| Ds | 8.90 (2.34) | 2.21 (0.53) † |
| Dm | 4.30 (1.39) | 0.50 (0.27) † |
| Dx | 2.65 (1.05) | 0.07 (0.07) † |
Values are means (SE) of Larson’s modified Keyes’ scores. N = number of mice in each group. Total smooth surface caries is the sum of buccal, lingual and proximal caries. E, lesion at least in white opaque enamel; Ds, lesion extends at least into dentine; Dm, dentine is exposed or missing; Dx, dentine is missing.
P < 0.05 compared to wild type.
Fig. 4.
Body weight and microbiota recoveries from mice in the caries experiment. (A) Body weights (means ± SEM) of male (circles) and female (squares) mice at weekly intervals during the experiment. (B) Recoveries of total microbiota and S. mutans from sonication of mandibular molars. Recoveries by CFU (colony forming units) of S. mutans on MSB agar and total bacteria on blood agar. Values are means (SE) from 6 pairs of mandibles prepared from each group. *; P < 0.05 for Car6−/− mice compared to the wild type (+/+).
3.4 Adherence of S. mutans to saliva-coated hydroxyapatite discs
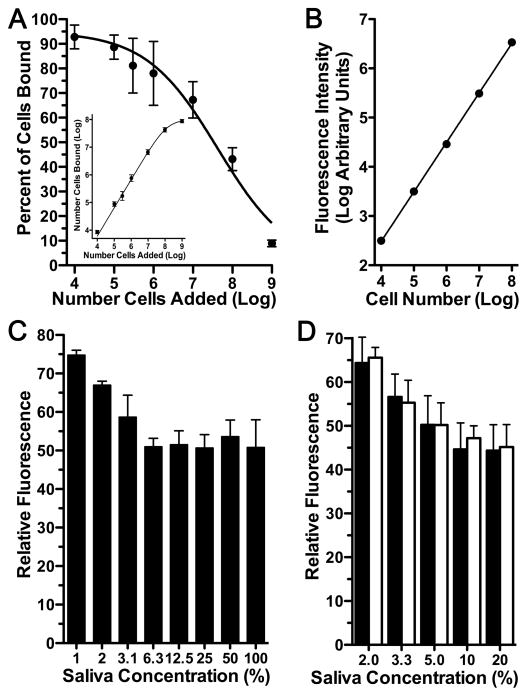
CA VI expression is associated with higher recoveries of S. mutans from molars as well as increased caries suggesting CA VI may promote the adherence of S. mutans to hard surfaces, possibly by direct interaction with S. mutans or indirectly through complexation of CA VI with other salivary constituents. To explore this hypothesis we developed a fluorescent microtiter plate assay to compare adherence of S. mutans to hydroxyapatite discs coated with whole stimulated saliva from wild type and Car6 −/− mice. It has long been established that S. mutans binds avidly to untreated hydroxyapatite, whereas formation of a salivary pellicle reduces total S. mutans adsorption sites by more than 50% [34]. In assay development, we therefore first determined cell loading per microtiter well to obtain near saturation binding to untreated hydroxyapatite while providing a wide dynamic range in quantifying a reduction in adherent cells. As shown in Fig. 5A, 108 cells per well resulted in more than a 2-fold excess of cells, a near saturating amount (Fig. 5A insert). In standard curves to convert fluorescence to cell numbers, 108 cells represent the upper end of a linear range that spans 4 log units (Fig. 5B). The adherence of S. mutans to hydroxyapatite discs was increasingly reduced by treatment with increasing concentrations of saliva from wild type mice (Fig. 5C). Interestingly, maximal inhibition of adherence was about 50%, consistent with whole human saliva [34], and was saturable at saliva concentrations of 6.3% and higher. As shown in Fig. 5D, there were no differences in S. mutans adherence between saliva samples from wild type and Car6 −/− mice at all saliva concentrations tested.
Fig. 5.
Microtiter plate and fluorescent-based assay of S. mutans adherence to hydroxyapatite discs treated with and without whole stimulated saliva from wild type and Car6−/− mice. (A) Comparisons of cell adherence to untreated hydroxyapatite with increasing cell loading (number of cells added) expressed as either the percent of cells added to wells or as the total number of cells bound (insert). Results are means (SEM) from 3 separate experiments. (B) A representative standard curve of fluorescence intensity due to increasing numbers of cells stained with SYTO-13 in wells without hydroxyapatite discs. (C) Fluorescence due to cells bound to hydroxyapatite discs treated with increasing concentrations of saliva from wild type mice. Fluorescence was normalized to that for cells bound to untreated hydroxyapatite (100%). Results are means (SEM) from 4 separate experiments. (D) Comparison of S. mutans adherence to hydroxyapatite discs treated with increasing concentrations of saliva from wild type (black bars) and Car6 −/− mice (open bars). Fluorescence was normalized to that for cells bound to untreated hydroxyapatite (100%). Results are means (SEM) from 6 separate experiments. P > 0.05 for wild type compared to Car6 −/− at each saliva concentration (unpaired, two tailed t-test).
3.5 Comparison of Indigenous Oral Bacteria
We further hypothesized that loss of CA VI in saliva may alter the indigenous oral microbiota, which may impact subsequent colonization by S. mutans. We thus carried out an initial assessment of the indigenous oral microbiota of each mouse genotype. Colonies of six distinct morphologies were observed consistently on blood agar plates under aerobic and anaerobic culture conditions after streaking oral swabs from mice of both genotypes. No additional colony morphologies were apparent with extended culture. Characteristics of each colony morphotype are given in Table 2. For taxonomic assignment of each colony morphotype, we conducted colony PCR of 16S rRNA gene sequences from single isolates cultured under aerobic conditions from three mice of each genotype. High quality sequences for each colony morphotype were at least 99.8% identical, regardless of host genotype. We therefore derived a consensus sequence from the three reads of each colony morphotype for each mouse genotype. Consensus sequences ranged from 1420 to 1472 base pairs and were aligned to sequences of the Ribosome Database Project (RDP), release 10, using the BLAST server at Human Oral Microbiome Database (HOMD). 16S rRNA gene sequences from both the hemolytic-grey colonies and the non-hemolytic white colonies of cocci most closely align to sequences from uncultured bacteria and hence do not have species designations, but are assigned to the genus Streptococcus. Sequences from the four other colony morphotypes each align with identities of 97.5% to 100% to cultured bacteria of known species. For comparisons, each consensus sequence was also aligned to the annotated HOMD 16S rRNA reference sequences from the human oral microbiome. Results are shown in Table 3. Because the highest scoring alignments were identical when comparing consensus sequences between wild type and Car6 −/− mice, only the lowest alignment score and percent identity for each pair of consensus sequences are given.
Table 2.
Characteristics of distinct colonies isolated on blood agar from oral swabs of wild type and Car6−/− mice.
| Colony Appearance | Cell Appearance | Gram Stain | Catalase Activity |
|---|---|---|---|
| Large Grey | Short thin rods | − | + |
| Large Yellow | Large cocci | + | + |
| Grey (α-Hemolytic) | Cocci - chains | + | − |
| Medium White | Cocci - chains | + | − |
| Medium White | Medium to long rods | + | − |
| Small Grey | Short rods | + | + |
Colony appearance on blood agar was identical under aerobic and anaerobic conditions. Cell appearance was determined under light microscopy (100x).
Table 3.
Alignments of 16S rRNA gene sequences of colonies isolated on blood agar from oral swabs of wild type and Car6−/− mice.
| Colony | Reference Sequences | Genus | Species | GenBank Accession | Score (bits) | % Identities |
|---|---|---|---|---|---|---|
| Large Grey | RDP | Actinobacillus | muris | AF024526 | 2154 | 98.5 |
| HOMD | Haemophilus | aegyptius | M75044 | 1902 | 93.9 | |
| Large Yellow | RDP | Staphylococcus | saprophyticus | EU855227 | 2257 | 100 |
| HOMD | Staphylococcus | warneri | L37603 | 2169 | 98.3 | |
| Grey (α-Hemolytic) | RDP | Streptococcus | Unassigned | EU453973 | 2126 | 99.7 |
| HOMD | Streptococcus | sanguinis | AF003928 | 2076 | 96.7 | |
| White (Cocci) | RDP | Streptococcus | Unassigned | FJ893080 | 2090 | 97.7 |
| HOMD | Streptococcus | parasanguinis II | AY278635 | 1986 | 95.2 | |
| White (Rods) | RDP | Lactobacillus | murinus | AF157049.1 | 2261 | 100 |
| HOMD | Lactobacillus | salivarious | AF089108.2 | 1959 | 94.9 | |
| Small Grey | RDP | Corynebacterium | mastitidis | AY834747.1 | 2081 | 97.5 |
| HOMD | Corynebacterium | diphtheriae | X82059 | 1882 | 94.5 |
High quality consensus 16S rRNA sequences from colony PCR of single isolates under aerobic conditions from three mice of each genotype were aligned by the BLAST server at Human Oral Microbiome Database (HOMD) [29] to two sets of 16S rRNA reference sequences, HOMD 16S rRNA RefSeq and sequences of the Ribosome Database Project, release 10 [30]. Highest scoring alignments were identical between consensus sequences from wild type and Car6 −/− mice. Thus, only the lowest alignment score and percent identities for each pair of consensus sequences are given. Percent identities are the total nucleotide mismatches per total matching nucleotides, including gaps.
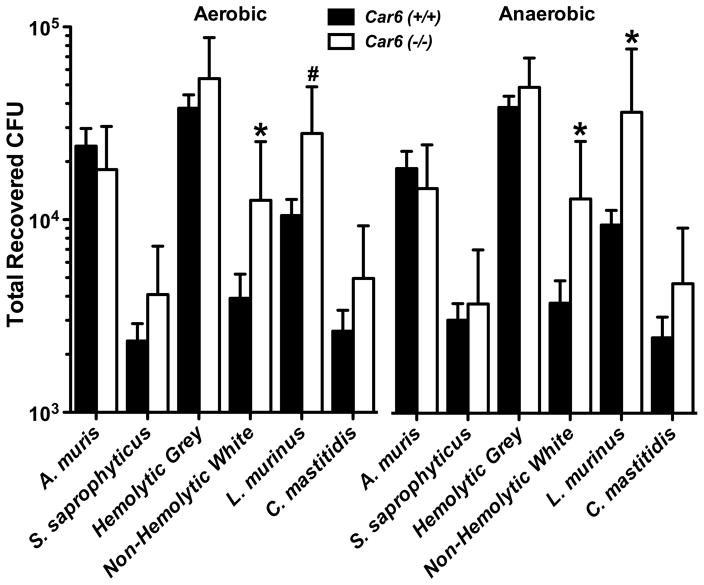
We further determined total recoveries of each colony morphotype from aerobic and anaerobic cultures from Car6 −/− and wild type mice. Results are shown in Fig. 6. There were no significant differences in the total recovered CFUs for each colony morphotype between the two culture conditions. Interestingly, the non-hemolytic white colonies of cocci and the colonies aligned to L. murinus were recovered at significantly higher levels from Car6 −/− mice versus wild type mice under each culture condition, although the significance was slightly less than 95% (P = 0.066) for L. murinus in aerobic cultures.
Fig. 6.
Total recoveries in colony forming units (CFUs) for each of six distinct colony morphotypes from the oral cavities of Car6 +/+ and Car6 −/− mice. Oral swabs were vortexed in BHI medium and 50 μl of three different dilutions streaked onto duplicate blood agar plates cultured under aerobic or anaerobic conditions. Each colony morphotype is identified by its taxonomic species assignment from the RDP database as given in Table 3, except for the hemolytic grey colonies and non-hemolytic white cocci colonies, both of which are assigned as members of the genus Streptococcus. Results are means (SE) from cultures of oral swabs from 11 Car6 +/+ and 9 Car6 −/− mice. *; P < 0.05 for Car6−/− compared to Car6 +/+. #; P = 0.066 for Car6−/− mice compared to Car6 +/+.
4. Discussion
In evaluation of Car6 gene expression in wild type mice we found predominant expression of type A transcripts in parotid and submandibular glands, consistent with the previous immunohistochemical localization of CA VI enzyme in serous acini of parotid glands and in seromucous acini of submandibular gland [18]. In contrast, expression in sublingual glands was sparse, suggesting it is not a product of the mucous acinar cell type that is predominant in this gland. Instead, it is likely a product of ductal cells as reported for sublingual glands of rats [35]. In Car6−/− mice we find evidence in parotid and submandibular glands for spliced type A transcripts resulting from transcriptional read-through of the inserted neo cassette, but without inclusion of the remaining exon 4 sequence. Splicing occurs from exon 2 directly to exon 5, and thus produces aberrant type A transcripts predicted to contain four premature termination codons within the sequences encoded by exons 5 and 6 (not shown). Such mRNA is a candidate for nonsense-mediated mRNA decay, in which aberrant transcripts are detected during the pioneer round of translation, are segregated to an intracellular pool associated with RNA decay machinery and do not produce protein [36]. Detection by RT-PCR of these aberrant transcripts is likely due to the high rate of Car6 transcription in acinar cells of submandibular and parotid glands and result in overloading of the RNA decay pathway. These aberrant transcripts do not result in protein production as evidenced by the absence in Car6−/− mice of proteins immunoreactive to a CA VI polyclonal antibody [18], nor in the current study do we detect an aberrant protein in saliva or the major salivary glands by SDS-PAGE.
Type B transcripts are detected in wild type mice at apparently low levels in parotid and submandibular glands, but not sublingual glands. The absence of aberrant type B transcripts in all three glands of Car6−/− mice suggests either the transcriptional machinery does not read through the neomycin cassette or that RNA decay processes in affected cells are able to efficiently degrade the lower levels of nonsense mRNA. In light of the regulated expression of type B transcripts by the ER stress-related transcription factor C/EBP homologous protein (CHOP) [19, 37], our detection of type B CA VI transcripts in glands of wild type mice may represent low levels of ER stress intrinsic to serous and seromucous salivary acinar cells during heightened and/or prolonged periods of secretion during eating. Indeed, up-regulation of the ER stress response and acinar cell damage is associated with high levels of secretagogues in the pancreas [38].
A recent initial comparison of the transcriptomes of submandibular glands, stomach, and duodenum between wild type and Car6−/− mice by cDNA microarray analysis indicated that potentially 94, 56, and 127 genes are up- or down-regulated, respectively, at least 1.4-fold in Car6−/− mice [18]. Interestingly, the great majority of the putative differentially expressed genes are not shared among the different tissues. Functional clustering of the transcripts identified by cDNA microarray in each tissue suggests that catabolic processes, stress responses and/or immune system processes may be affected in Car6−/− mice. Although validation of gene expression differences by real-time RT-PCR remains to be completed, we find that the absence of expression of Car6 type A and/or type B transcripts has no apparent affects on salivary or gastrointestinal physiology as assessed by salivary structure/function, weight gain, general appearance and activity level. Any changes in gene expression may be due, in large part, to the loss of type B proteins and compensatory stress-related cellular responses to preserve homeostasis.
In light of no significant differences between wild type and Car6−/− mice in the structure and function of salivary glands other than deletion of CA VI expression, we assessed the contribution of CA VI deletion to caries development in mice when inoculated with S. mutans and administered a highly cariogenic diet. An intriguing finding is the marked protective effect of CA VI deletion against both smooth surface and sulcal caries. These results thus contradict our original hypothesis that CA VI serves to protect enamel surfaces from caries. This hypothesis was based upon a human study in which caries prevalence was associated with lower concentrations of CA VI in the saliva of subjects with poor oral hygiene [16]. Other confounding factors may have contributed to this apparent association. For example, patients deficient in CA II also have markedly reduced levels of salivary CA VI [39]. Because CA II is expressed within the cytoplasm of salivary acinar cells that secrete CA VI [9], lower levels of CA VI in saliva may therefore reflect other metabolic affects that compromise the protective qualities of saliva, possibly leading to the perception of poor oral hygiene. Supporting this explanation are previous results indicating a positive correlation between salivary levels of CA VI and amylase in a similar human subject population as the aforementioned caries study [11]. Alternatively, our results may indicate important differences in CA VI function between different species. Comparison of CA VI proteins between mouse (261 residues) and human (308 residues) reveal 74% similarity and 57% identity, but with the larger human enzyme having 57 residues of unique 5′-end sequence of unknown function compared to the mouse enzyme (NCBI BLASTP 2.2.25).
The lower levels of caries associated with Car6 deletion suggests this enzyme does not contribute significantly to the removal of bacterial derived hydrogen ions at enamel surfaces. Nishimori and co-workers [40] suggested that CA VI at enamel surfaces might function to catalyze the hydration of CO2 (derived from the metabolism of plaque bacteria) to produce bicarbonate and protons, therefore promoting the acid-induced demineralization of enamel. They point out that purified human CA VI has moderately high activity for the CO2 hydration reaction and is inhibited in the reverse reaction by the abundant salivary anion, bicarbonate (KI = 0.80 mM). The concentration of bicarbonate in saliva increases from about 2 mM under non-stimulated conditions to about 40 mM at maximal secretion rates [41]. The reverse dehydration reaction of CA VI should therefore be mostly inhibited during all phases of salivary secretion. Human CA VI was shown to be enzymatically active when bound to enamel, in vitro [15] and to retain its activity after incubation for 30 min at pH 2.2 [42]. At pH 6.4, rat CA VI loses about 50% of its peak activity whereas rat CA I is inactive [34]. CA VI activity at lower pH values has not been reported to our knowledge. CA VI is thus acid resistant and may retain significant enzymatic activity as plaque pH falls to levels that promote the further colonization of acidogenic bacteria and the dissolution of enamel surfaces. The bacteria within the acid plaque may also use bicarbonate produced by the hydration reaction to help buffer cytosolic pH.
Regardless of CA VI enzymatic activity, its expression is associated with increased caries development and recovered S. mutans from molar teeth. Our results do not support a role for CA VI in promoting S. mutans adherence to hard surfaces. Alternatively, we postulated that loss of salivary CA VI alters the commensal oral microbiota. We identified six distinct colony morphotypes from oral swabs of mice, each yielding nearly identical 16S rRNA gene sequences when compared between mice genotypes. Hence, we did not detect species unique to wild type or Car6 −/− mice. For each colony morphotype, the highest scoring aligned sequence from the RDP database is associated with mice. Actinobacillus muris [43], L. murinus [44] and Staphylococcus saprophyticus [45] are murine commensal oral bacteria. Corynebacteria are commensals of the skin and mucous membranes of humans and animals, with C. mastitidis recently identified as an opportunistic infectious resident of the epidermis of laboratory mice [46]. Sequences from the non-hemolytic white colonies of cocci and the hemolytic-grey colonies most closely align to sequences assigned to the genus Streptococcus and each were isolated from either mouse epidermis [47] and cecum [48], respectively. Streptococci represent the most abundant bacteria recovered from oral swabs, with streptococci from the hemolytic-grey colonies the most highly recovered species from mice of both genotypes. The sequences from these two colonies align best to members of the sanguinis group of streptococci from the annotated human oral microbiota. Our results are thus consistent with previous observations in which streptococci represent predominant commensals of the oral microbiota in humans and in laboratory mice [45, 49]. Moreover, as in prior studies that characterized the oral microbiota of mice from cultivatable bacteria [44] or from pyrosequencing [45] we detected a low number of species compared to humans, likely influenced by modern vivarium procedures to establish disease-free colonies. Interestingly, we find higher recoveries from Car6 −/− versus Car6 +/+ mice of L. murinus and of the non-hemolytic species putatively assigned as streptococci, supporting our hypothesis that CA VI expression influences the commensal oral microbiota. Because S. mutans can be antagonized by indigenous oral lactobacilli as well as by other streptococci [50–52], our results suggest these changes in the oral microbiota in response to the absence of salivary CA VI present a more competitive environment for S. mutans colonization.
Collectively, our results suggest salivary CA VI may promote caries by modulating the oral microbiota to favor S. mutans colonization and/or by the enzymatic production of acid within plaque. Correlations to human CA VI may include in vitro comparisons of human and murine CA VI activities under acidic conditions or the assay of caries susceptibility in a human CA VI knock-in mouse model.
Highlights.
All major salivary glands express Car6 type A transcripts (secretory CA VI)
Car6 type B mRNA (intracellular CA VI) are expressed selectively in major glands
Deletion of both CA VI isoforms has no apparent affects on salivary function
The absence of secretory CA VI in saliva is associated with reduced caries in mice
Car6 KO mice have altered oral microbiota that may antagonize Streptococcus mutans.
Acknowledgments
The authors wish to acknowledge Robert Nguyen for his technical assistance in adherence assays, and Richard L. Cannon for excellent assistance in the design and construction of the alignment and cutting apparatus for mouse molars. We thank Dr. Robert A. Burne for the UA159 strain of S. mutans, for helpful advice and for critiquing the manuscript. We also thank Dr. William H. Bowen for his insights and constant encouragement. This study was funded by National Institute of Dental and Craniofacial Research grant (NIDCR RO1 grant DE16362), European Union DeZnIT project, Academy of Finland, Sigrid Juselius Foundation, and Competitive Research Funding of the Tampere University Hospital (9L071, 9M075). The funders had no role in the study design, data collection and analysis, decisions to publish, or preparation of the manuscript.
Abbreviations
- CA VI
carbonic anhydrase alpha, isozyme 6
- car6
carbonic anhydrase 6 gene
- CHOP
ER stress-related transcription factor C/EBP, homologous protein
- neo
neomycin
- TK
thymidine kinase
- SMG
submandibular gland
- SLG
sublingual gland
Footnotes
Disclosure statement
No potential conflicts of interest are reported for any of the authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Bently Robinson, Email: berobinson@dental.ufl.edu.
Seppo Parkkila, Email: Seppo.Parkkila@uta.fi.
Pei-wen Pan, Email: Peiwen.Pan@uta.fi.
Melanie N. Cash, Email: mcash@dental.ufl.edu.
Helen N. Truong, Email: helenhi@ufl.edu.
Thomas W. Hussey, Email: twh05@ufl.edu.
Sarah L. Gullett, Email: slgullett@dental.ufl.edu.
References
- 1.Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakano K, Inaba H, Nomura R, Nemoto H, Takeda M, Yoshioka H, Matsue H, Takahashi T, Taniguchi K, Amano A, Ooshima T. Detection of cariogenic Streptococcus mutans in extirpated heart valve and atheromatous plaque specimens. J Clin Microbiol. 2006;44:3313–3317. doi: 10.1128/JCM.00377-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernley RT, Wright RD, Coghlan JP. A novel carbonic anhydrase from the ovine parotid gland. FEBS Lett. 1979;105:299–302. doi: 10.1016/0014-5793(79)80634-1. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa Y, Matsumoto K, Maeda T, Tamai R, Suzuki T, Sasano H, Fernley RT. Characterization of lacrimal gland carbonic anhydrase VI. J Histochem Cytochem. 2002;50:821–827. doi: 10.1177/002215540205000608. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa Y, Toyosawa S, Inagaki T, Hong SS, Ijuhin N. Carbonic anhydrase isozyme VI in rat lacrimal gland. Histochem Cell Biol. 1995;103:387–394. doi: 10.1007/BF01457814. [DOI] [PubMed] [Google Scholar]
- 6.Leinonen JS, Saari KA, Seppanen JM, Myllyla HM, Rajaniemi HJ. Immunohistochemical demonstration of carbonic anhydrase isoenzyme VI (CA VI) expression in rat lower airways and lung. J Histochem Cytochem. 2004;52:1107–1112. doi: 10.1369/jhc.4A6282.2004. [DOI] [PubMed] [Google Scholar]
- 7.Kimoto M, Iwai S, Maeda T, Yura Y, Fernley RT, Ogawa Y. Carbonic anhydrase VI in the mouse nasal gland. J Histochem Cytochem. 2004;52:1057–1062. doi: 10.1369/jhc.3A6243.2004. [DOI] [PubMed] [Google Scholar]
- 8.Karhumaa P, Leinonen J, Parkkila S, Kaunisto K, Tapanainen J, Rajaniemi H. The identification of secreted carbonic anhydrase VI as a constitutive glycoprotein of human and rat milk. Proc Natl Acad Sci U S A. 2001;98:11604–11608. doi: 10.1073/pnas.121172598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parkkila S, Kaunisto K, Rajaniemi L, Kumpulainen T, Jokinen K, Rajaniemi H. Immunohistochemical localization of carbonic anhydrase isoenzymes VI, II, and I in human parotid and submandibular glands. J Histochem Cytochem. 1990;38:941–947. doi: 10.1177/38.7.2113069. [DOI] [PubMed] [Google Scholar]
- 10.Leinonen J, Parkkila S, Kaunisto K, Koivunen P, Rajaniemi H. Secretion of carbonic anhydrase isoenzyme VI (CA VI) from human and rat lingual serous von Ebner’s glands. J Histochem Cytochem. 2001;49:657–662. doi: 10.1177/002215540104900513. [DOI] [PubMed] [Google Scholar]
- 11.Kivela J, Parkkila S, Metteri J, Parkkila AK, Toivanen A, Rajaniemi H. Salivary carbonic anhydrase VI concentration and its relation to basic characteristics of saliva in young men. Acta Physiol Scand. 1997;161:221–225. doi: 10.1046/j.1365-201X.1997.00217.x. [DOI] [PubMed] [Google Scholar]
- 12.Henkin RI, Martin BM, Agarwal RP. Decreased parotid saliva gustin/carbonic anhydrase VI secretion: an enzyme disorder manifested by gustatory and olfactory dysfunction. Am J Med Sci. 1999;318:380–391. doi: 10.1097/00000441-199912000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Henkin RI, Martin BM, Agarwal RP. Efficacy of exogenous oral zinc in treatment of patients with carbonic anhydrase VI deficiency. Am J Med Sci. 1999;318:392–405. doi: 10.1097/00000441-199912000-00006. [DOI] [PubMed] [Google Scholar]
- 14.Shatzman AR, Henkin RI. Gustin concentration changes relative to salivary zinc and taste in humans. Proc Natl Acad Sci U S A. 1981;78:3867–3871. doi: 10.1073/pnas.78.6.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leinonen J, Kivela J, Parkkila S, Parkkila AK, Rajaniemi H. Salivary carbonic anhydrase isoenzyme VI is located in the human enamel pellicle. Caries Res. 1999;33:185–190. doi: 10.1159/000016515. [DOI] [PubMed] [Google Scholar]
- 16.Kivela J, Parkkila S, Parkkila AK, Rajaniemi H. A low concentration of carbonic anhydrase isoenzyme VI in whole saliva is associated with caries prevalence. Caries Res. 1999;33:178–184. doi: 10.1159/000016514. [DOI] [PubMed] [Google Scholar]
- 17.Pan PW, Kayra K, Leinonen J, Nissinen M, Parkkila S, Rajaniemi H. Erratum to: Gene expression profiling in the submandibular gland, stomach, and duodenum of CAVI-deficient mice. Transgenic Res. 2011 doi: 10.1007/s11248-010-9441-2. [DOI] [PubMed] [Google Scholar]
- 18.Pan PW, Kayra K, Leinonen J, Nissinen M, Parkkila S, Rajaniemi H. Gene expression profiling in the submandibular gland, stomach, and duodenum of CAVI-deficient mice. Transgenic Res. 2010 doi: 10.1007/s11248-010-9441-2. [DOI] [PubMed] [Google Scholar]
- 19.Sok J, Wang XZ, Batchvarova N, Kuroda M, Harding H, Ron D. CHOP-Dependent stress-inducible expression of a novel form of carbonic anhydrase VI. Mol Cell Biol. 1999;19:495–504. doi: 10.1128/mcb.19.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culp DJ, Quivey RQ, Bowen WH, Fallon MA, Pearson SK, Faustoferri R. A mouse caries model and evaluation of aqp5−/− knockout mice. Caries Res. 2005;39:448–454. doi: 10.1159/000088179. [DOI] [PubMed] [Google Scholar]
- 21.Gold OG, Jordan HV, Van Houte J. A selective medium for Streptococcus mutans. Arch Oral Biol. 1973;18:1357–1364. doi: 10.1016/0003-9969(73)90109-x. [DOI] [PubMed] [Google Scholar]
- 22.Tanzer JM. Essential dependence of smooth surface caries on, and augmentation of fissure caries by, sucrose and Streptococcus mutans infection. Infect Immun. 1979;25:526–531. doi: 10.1128/iai.25.2.526-531.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keyes PH. Dental caries in the molar teeth of rats. II. A method for diagnosing and scoring several types of lesions simultaneously. J Dent Res. 1958;37:1088–1099. doi: 10.1177/00220345580370060901. [DOI] [PubMed] [Google Scholar]
- 24.Larson RH, Fitzgerald RJ. Caries Development in Rats of Different Ages with Controlled Flora. Arch Oral Biol. 1964;9:705–712. doi: 10.1016/0003-9969(64)90082-2. [DOI] [PubMed] [Google Scholar]
- 25.Das B, Cash MN, Hand AR, Shivazad A, Culp DJ. Expression of Muc19/Smgc gene products during murine sublingual gland development: cytodifferentiation and maturation of salivary mucous cells. J Histochem Cytochem. 2009;57:383–396. doi: 10.1369/jhc.2008.952853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woodman ME. Direct PCR of intact bacteria (colony PCR) Curr Protoc Microbiol, Appendix 3. 2008 doi: 10.1002/9780471729259.mca03ds9. Appendix 3D. [DOI] [PubMed] [Google Scholar]
- 27.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, Sahasrabudhe A, Dewhirst FE. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–3783. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawanami T, Fukuda K, Yatera K, Kido M, Mukae H, Taniguchi H. A higher significance of anaerobes: the clone library analysis of bacterial pleurisy. Chest. 2011;139:600–608. doi: 10.1378/chest.10-0460. [DOI] [PubMed] [Google Scholar]
- 29.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fallon MA, Latchney LR, Hand AR, Johar A, Denny PA, Georgel PT, Denny PC, Culp DJ. The sld mutation is specific for sublingual salivary mucous cells and disrupts apomucin gene expression. Physiol Genomics. 2003;14:95–106. doi: 10.1152/physiolgenomics.00151.2002. [DOI] [PubMed] [Google Scholar]
- 32.Wen ZT, Burne RA. Construction of a new integration vector for use in Streptococcus mutans. Plasmid. 2001;45:31–36. doi: 10.1006/plas.2000.1498. [DOI] [PubMed] [Google Scholar]
- 33.Lemos JA, Abranches J, Koo H, Marquis RE, Burne RA. Protocols to study the physiology of oral biofilms. Methods Mol Biol. 2010;666:87–102. doi: 10.1007/978-1-60761-820-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark WB, Bammann LL, Gibbons RJ. Comparative estimates of bacterial affinities and adsorption sites on hydroxyapatite surfaces. Infection and immunity. 1978;19:846–853. doi: 10.1128/iai.19.3.846-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redman RS, Peagler FD, Johansson I. Immunohistochemical localization of carbonic anhydrases I, II, and VI in the developing rat sublingual and submandibular glands. Anat Rec. 2000;258:269–276. doi: 10.1002/(SICI)1097-0185(20000301)258:3<269::AID-AR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 36.Maquat LE, Gong C. Gene expression networks: competing mRNA decay pathways in mammalian cells. Biochem Soc Trans. 2009;37:1287–1292. doi: 10.1042/BST0371287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura T, Sato T, Aoki T, Yamamoto A, Okada T, Hirai R, Harada R, Mori K, Tagaya M, Harada A. p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol Cell Biol. 2009;29:1869–1881. doi: 10.1128/MCB.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubisch CH, Logsdon CD. Secretagogues differentially activate endoplasmic reticulum stress responses in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1804–1812. doi: 10.1152/ajpgi.00078.2007. [DOI] [PubMed] [Google Scholar]
- 39.Murakami H, Sly WS. Purification and characterization of human salivary carbonic anhydrase. J Biol Chem. 1987;262:1382–1388. [PubMed] [Google Scholar]
- 40.Nishimori I, Minakuchi T, Onishi S, Vullo D, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. DNA cloning, characterization, and inhibition studies of the human secretory isoform VI, a new target for sulfonamide and sulfamate inhibitors. J Med Chem. 2007;50:381–388. doi: 10.1021/jm0612057. [DOI] [PubMed] [Google Scholar]
- 41.Bardow A, Madsen J, Nauntofte B. The bicarbonate concentration in human saliva does not exceed the plasma level under normal physiological conditions. Clin Oral Investig. 2000;4:245–253. doi: 10.1007/s007840000077. [DOI] [PubMed] [Google Scholar]
- 42.Parkkila S, Parkkila AK, Lehtola J, Reinila A, Sodervik HJ, Rannisto M, Rajaniemi H. Salivary carbonic anhydrase protects gastroesophageal mucosa from acid injury. Dig Dis Sci. 1997;42:1013–1019. doi: 10.1023/a:1018889120034. [DOI] [PubMed] [Google Scholar]
- 43.Bisgaard M. Actinobacillus muris sp. nov. isolated from mice. Acta Pathol Microbiol Immunol Scand B. 1986;94:1–8. doi: 10.1111/j.1699-0463.1986.tb03013.x. [DOI] [PubMed] [Google Scholar]
- 44.Rodrigue L, Lavoie MC. Comparison of the proportions of oral bacterial species in BALB/c mice from different suppliers. Lab Anim. 1996;30:108–113. doi: 10.1258/002367796780865853. [DOI] [PubMed] [Google Scholar]
- 45.Chun J, Kim KY, Lee JH, Choi Y. The analysis of oral microbial communities of wild-type and toll-like receptor 2-deficient mice using a 454 GS FLX Titanium pyrosequencer. BMC Microbiol. 2010;10:101. doi: 10.1186/1471-2180-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radaelli E, Manarolla G, Pisoni G, Balloi A, Aresu L, Sparaciari P, Maggi A, Caniatti M, Scanziani E. Suppurative adenitis of preputial glands associated with Corynebacterium mastitidis infection in mice. J Am Assoc Lab Anim Sci. 2010;49:69–74. [PMC free article] [PubMed] [Google Scholar]
- 47.Scharschmidt TC, List K, Grice EA, Szabo R, Renaud G, Lee CC, Wolfsberg TG, Bugge TH, Segre JA. Matriptase-deficient mice exhibit ichthyotic skin with a selective shift in skin microbiota. J Invest Dermatol. 2009;129:2435–2442. doi: 10.1038/jid.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, Gordon JI, Chervonsky AV. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Osteras M, Schrenzel J, Francois P. Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Methods. 2009;79:266–271. doi: 10.1016/j.mimet.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haukioja A, Loimaranta V, Tenovuo J. Probiotic bacteria affect the composition of salivary pellicle and streptococcal adhesion in vitro. Oral microbiology and immunology. 2008;23:336–343. doi: 10.1111/j.1399-302X.2008.00435.x. [DOI] [PubMed] [Google Scholar]
- 51.Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. 2009;28:397–403. doi: 10.1089/dna.2009.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikawa H, Tomiyama Y, Hiramatsu M, Yushita K, Takamoto Y, Ishi H, Mimura S, Hiyama A, Sasahara H, Kawahara K, Makihira S, Satoda T, Takemoto T, Murata H, Mine Y, Taji T. Bovine milk fermented with Lactobacillus rhamnosus L8020 decreases the oral carriage of mutans streptococci and the burden of periodontal pathogens. Journal of Investigative and Clinical Dentistry. 2011;2:187–196. doi: 10.1111/j.2041-1626.2011.00056.x. [DOI] [PubMed] [Google Scholar]