Abstract
Induction of Foxp3 gene expression and acquisition of regulatory T cell fate is, understandably, a highly controlled process and one which many investigators want to illuminate. In studying the regulation of Foxp3 gene expression, several conserved non-coding regions have been identified and the role of various transcription factors at these sites has been explored. What emerges, is that many factors, some positive, some negative, interact to collectively drive Foxp3 gene expression and then maintain its expression in Foxp3+ regulatory T cells. TCR signaling is imperative for Foxp3 gene expression and TGF-β is a key cytokine for initiating Foxp3 gene expression in naïve T cells. But other signaling pathways are also known to play a role in properly orchestrating Foxp3 gene expression and regulatory T cell expansion. Here we review the recent progress in understanding the complex molecular events that drive Foxp3 gene expression and allow functional regulatory T cells to develop.
Introduction
The vital role of CD4+CD25+Foxp3+ regulatory T cells (Tregs) in maintenance of a healthy immune system is starkly demonstrated in mice and humans with mutations in the Foxp3 gene. The severe lympho-proliferative diseases, consequential morbidity and reduced life-span seen in scurfy mice and in Immune dysregulation, Polyendocrinopathy, enteropathy, X-linked syndrome (IPEX)-patients are due to Treg deficiencies resulting from Foxp3 mutation [1–3]. Thus Tregs mediate dominant tolerance to self and have also been shown to be equally important in the control of autoimmune diseases, allergy, fetal-maternal tolerance, allograft tolerance and immunopathology. Tregs can develop intrathymically, from CD4+ single positive precursors [4, 5], which commit to the Treg lineage before exiting to the periphery. In addition Tregs can also be generated extrathymically from CD4+CD25− naïve T cells in which Foxp3 expression is induced in a TGF-β-dependent manner [6].
As central as Tregs are to immune homeostasis, Foxp3 is central to Treg development and function. Indeed Foxp3 is considered the “master regulator” of Tregs. The current paradigm suggests that only CD4+CD25+ peripheral T cells and single positive (SP) thymocytes express Foxp3 [7]. Forced expression of Foxp3 in naïve (CD4+CD25−) T cells is sufficient to endow them with Treg properties; transduced cells not only express surface molecules associated with the Treg signature, such as CTLA-4, GITR and CD25, but are also able to suppress other cell types in vitro and in vivo [7, 8]. Generation of mixed bone-marrow chimeras consisting of wild-type and Foxp3−/− cells demonstrated that CD4+CD25+ T cells capable of suppression could not be generated from Foxp3−/− cells [8]. T cell specific deletion of Foxp3 induced severe lympho-proliferative diseases similar to that observed in Foxp3−/− mice [9]. Thus Foxp3 is critical for and defines the CD4+CD25+ regulatory T cell population. Although the importance of Foxp3 is well established the exact pathways by which its expression is induced, maintained and stabilized remain loosely defined, with the stimulus for Foxp3 induction intrathymically being debated. Here we outline the current knowledge of the molecular mechanisms governing Foxp3 expression, with a particular emphasis on the role played by TGF-β.
Foxp3 gene conserved regions
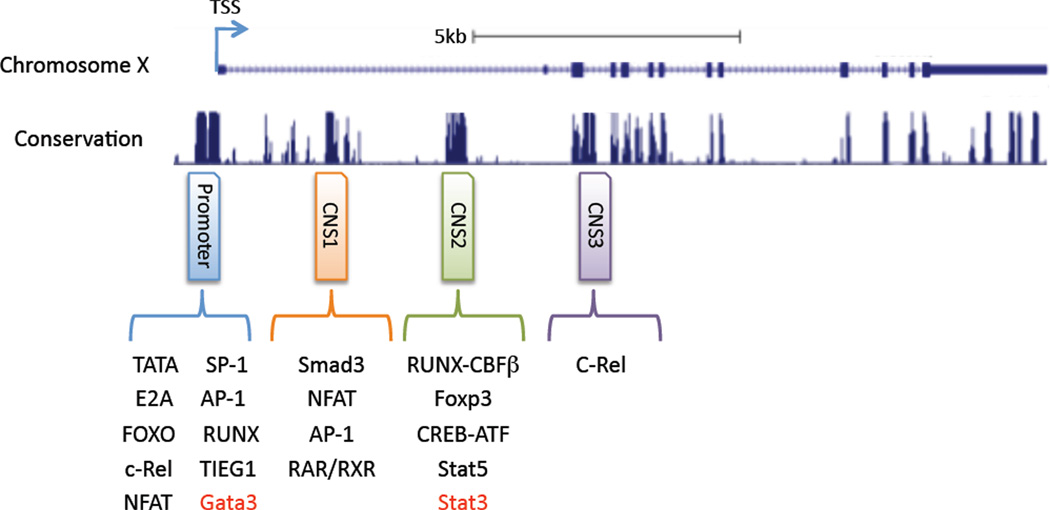
The Foxp3 gene is found on the X chromosome. Critical for the initiation and regulation of Foxp3 transcription is the Foxp3 conserved promoter sequence, located up-stream of the transcriptional starting site (TSS) [10]. Detailed analysis of the Foxp3 gene promoter in humans shows it is located in the region −511/+176 base pairs (bp) relative to the TSS. The promoter contains features common to most eukaryotic promoters; GC (138bp upstream of the TSS), TATA (44bp upstream of the TSS) and CAAT (218bp upstream of the TSS) boxes, mutation of which reduces core promoter activity [11]. Identification of proteins that bind this highly conserved region have shown that mutation of AP-1 and NFAT sites in this region decrease Foxp3 promoter activity, highlighting a role for these transcription factors in the transactivation of the Foxp3 promoter [11]. Other positive regulators that interact at the promoter to drive Foxp3 expression may include Sp1 (an activator of transcription) and Sp3, which can bind the GC box.
Alignment of the Foxp3 locus from different species has highlighted the presence of several highly conserved regions. In addition to the promoter, these include three non-coding regions that could play a role in regulation of the foxp3 gene [12, 13]. Here referred to a conserved non-coding sequence (CNS) 1, 2 and 3, although often referred to as enhancers, detailed analysis of these regions has been undertaken and a plethora of transcription factors have been shown to bind and influence foxp3 gene induction. Highlighting well the mutlifactoral mechanism by which Foxp3 expression is driven is the c-Rel enhanceosome model in which c-Rel promotes the formation of an enhanceosome consisting of p65, NFATc2, Smad3 and pCREB. These factors bind to the foxp3 promoter and CNS1, 2 or 3, collectively promoting Foxp3 expression [12–18]. Specifically, NF-κB binding sites have been identified in the promoter region (at lease four within 1kb upstream from TSS) and in a CpG island in CNS2 regions [14, 15]. NFAT binding sites have been located in the promoter region (three conserved sites within 500bp upstream from TSS) and the CNS1 region (+2145) [11, 16]. c-Rel binding site are in the promoter (377bp upstream from TSS) and CNS3 region [12, 17]. Finally, a CpG island within the CNS2 region has one CREB binding site [18]. So from one model we have many transcription factors coordinately acting from different sites to drive foxp3 expression. Importantly, this is a feat one transcription alone cannot achieve.
Examining the CNS regions in more detail, we will first look at CNS2, which is located 4kb down-stream of the Foxp3 TSS. The chromatin structure of the CNS2 region in natural Tregs (Foxp3+ cells from the thymus; nTregs) has been shown to in an “OPEN” chromatin state (defined as demethylated CpG motifs). In contrast, the CNS2 region of TGF-β induced Tregs (iTregs) has been observed to be in the “CLOSED” configuration [19, 20]. Pair this information with the demonstration that iTregs easily loose their regulatory phenotype when re-stimulated under culture conditions which induce Th1, Th2, or Th17 cell differentiation. nTregs on the other hand are relatively “stable” and don’t easily lose Foxp3 expression when re-stimulated under these same conditions. However, caution should taken in making the conclusion that “nTregs” are more stable than “iTregs”, as it is currently impossible to judge if the in vitro generated “iTregs” are at the same differentiation stage as the freshly isolated “nTregs” from a naïve mouse. Nevertheless, these data begin to suggest CNS2 may play a role in maintaining the regulatory potential of the Foxp3+ cell. CNS2 deficient mice have been generated and these mice exhibit less Foxp3+ Tregs in peripheral lymph nodes. Importantly, these CNS2−/− Foxp3+ Tregs loose Foxp3 expression during proliferation in response to TCR stimulation [12]. Using the CNS2−/− Zheng et al showed that the CNS2 region is required to stabilize foxp3 expression in Foxp3+ Tregs [12].
Within the CNS2 region there are two Est-1 binding sites which are important for proper Treg development [21, 22]. Mutation of both Est-1 binding sites in CNS2 has been shown to significantly decrease Foxp3 promoter activity [21, 22]. Est-1 KO mice were shown to have activated T cells and less Foxp3+ Tregs in the spleen. In addition, Est-1 KO T cells fail to generate Foxp3+ Tregs in response to TGF-β [22]. While Est-1 enrichment can be observed on the CNS2 element while de-methylated (or in an OPEN chromatin state) similar enrichment is not found when the CNS2 adopts the closed chromatin configuration [21, 22]. Some components (Azacytidine and Simvastatin amongst others) have been reported to de-methylate the CNS2 region and induce Foxp3+ Tregs, likely leading to the generation of a more stable Treg population [23, 24]. Binding of Foxp3 to the CNS2 has been observed in nTregs, but not iTregs. De-methylation of a CpG island in the CNS2 region in nTregs allows Foxp3 to bind it along with Cbf-β and RUNX-1 [12]. Cbf-β-deficient Tregs progressively lose Foxp3 expression after transfer into lymphopenic mice [25]. Therefore, it has been suggested that the enrichment of these complexes at the CNS2 region may play an important role in the maintenance of Foxp3 gene expression in nTreg cells [22, 26].
CNS3 is located 7 kb down-stream from the TSS and contains a c-Rel binding element [12]. In their examination of the role of the different CNS regions, Rudensky and colleagues, generated CNS3−/− mice. CNS3−/− mice exhibited reduced frequencies of Foxp3+CD4+ SP thymocytes, and CNS3−/− naïve CD4+ T cells failed to develop into Foxp3+ cells following TCR stimulation in the presence of TGF-β [12]. As CNS3 contains a c-Rel binding sequence, the authors then examined Foxp3 frequencies in chimeras generated from WT and c-Rel-deficient cells or WT and CNS3-deficient cells. In the resultant chimeras, the CNS3−/− and c-Rel−/− frequencies of Foxp3+ thymocytes were similar, leading to the suggestion that c-Rel binds the CNS3, which exists in an open chromatin configuration to facilitate Foxp3 gene transcription and nTreg generation. Thus, unlike CNS2, it is suggested that factors bound at the CNS3 play an important role in the induction of Foxp3 expression.
Finally, an examination of the role of CNS1 in Foxp3 expression was also undertaken, however this will be discussed later. But even from this limited description of the factors interacting at the Foxp3 promoter and CNS regions, one can see that many factors co-operate from different sites to co-ordinatly drive Foxp3 gene transcription. But a sequential series of events by which Foxp3 is induced remains elusive.
TGF-β and Foxp3 gene expression
How TGF-β promotes Foxp3 expression remains an area of intense investigation, with a number of factors shown to bind the Foxp3 locus in response to a TGF-β signal. However an exact, detailed pathway has not been determined. Here we outline the current data involving TGF-β and how it drives Foxp3 expression.
We will begin by firstly discussing the remaining CNS region; CNS1 is located around 2kb downstream of the TSS and has been shown to contain NFAT and SMAD protein binding sites [16]. Smad proteins are prominent members of the canonical TGF-β signaling pathway. TGF-β-signaling induces phosphorylation of Smad2 and 3, this allows these proteins to interact with Smad4, which permits translocation of Smad2 and 3 to the nucleus and induction of Smad-mediated transcription [27, 28]. Despite the critical role of TGF-β in induction of Foxp3, the Smad binding site in the CNS1 is the only Smad binding site located in either conserved and/or promoter regions of the Foxp3 gene. Following TGF-β stimulation of T cells, Smad3, although not Smad2 or Smad4, has been shown to rapidly bind the CNS1 region suggesting a role for this transcription factor in Foxp3 expression. However, a role for Smad2 and/or Smad4 is not precluded as in the absence of either protein induction of Foxp3 in response to TGF-β is reduced (at least in cell culture) [29, 30].
Despite a role for Smad(s) in mediating TGF-β signaling and the known role of TGF-β signaling in the induction of Foxp3 expression, there is a grey area between activation of Smad proteins and induction of Foxp3 expression. Firstly, a time lag exists between the immediate Smad2/3 phosphorylation (which occurs within minutes) and the detection of Foxp3 mRNA (which takes at least 10–12 hours) after TGF-β treatment. Secondly, as already stated, there is a lack of Smad binding sequences in the control elements of the Foxp3 gene. These data suggest that there must be some intermediate transcription factor(s) linking TGF-β-signaling and Smad activation to Foxp3 promoter activation. In line with this hypothesis, we have recently demonstrated that id3 (inhibitor of DNA binding protein 3) could serve as one such transcription factor in the control of Foxp3 gene expression [31]. Id3 belongs to a family of helix-loop-helix transcription factors [32]. Id3 lacks a DNA binding domain; instead Id3 complexes with basic Helix-loop-Helix proteins (E-protein) and inhibits their transcriptional activity [32, 33]. Id3 deficient mice have been shown to exhibit a Sjogren’s syndrome-like autoimmune disease, with T cells playing an essential role in disease progression [34, 35]. While the mechanisms for disease development in these mice are still unclear, deletion of the Id3 gene in mice results in less Foxp3+ Tregs within the thymus, spleen and peripheral lymph nodes [31], which was primarily due to a defect in Foxp3+ Treg generation. In cell cultures, Id3−/− CD4+ T cells fail to covert Foxp3+ Tregs in response to TGF-β [31]. As Id3 does not directly bind to the DNA, we sought to understand the mechanisms by which Id3 affects Foxp3 promoter activity. We identified two indispensible, yet complementary, processes that are involved in TGF-β-mediated induction of Foxp3 gene transcription. First, the E-protein E2A, was enriched on the Foxp3 promoter in response to TGF-β [31]; the Foxp3 promoter has multiple E-protein binding sites (CANNTG). Enrichment of E-proteins at the Foxp3 promoter encouraged TATA binding protein enrichment and Foxp3 mRNA production, suggesting that E2A proteins serve as positive regulators of Foxp3 expression. Indeed, by a series of deletion and over-expression experiments, we have provided evidence that the enrichment of E2A binding at the Foxp3 promoter promotes Foxp3 gene transcription. Secondly, we showed that binding of GATA-3 is inhibitory to Foxp3 expression, and it must be removed from the Foxp3 promoter in order for Foxp3 expression to occur. These two processes; enrichment of E2A and removal of GATA-3, were defective in Id3−/− CD4+ T cells. Although many exciting questions remain to be answered, these findings emphasize that proper gene transcription requires not only the enrichment of positive “enhancer”(s) elements, but also (and at the same time) removal of negative “silencer” elements.
Other transcription factors have also been suggested to regulate positive Foxp3 gene activation in naïve CD4+ T cells in cross-talk with TGF-β-signaling. Foxo proteins are reported to be involved in Foxp3 gene expression [36–38], and are negatively regulated by the PI3K-Akt pathway, a pathway also inhibitory to the generation of Tregs [36–38]. In the absence of Foxo 1 and/or 3a decreases in thymic Tregs are seen. In addition, in vitro conversion of naïve CD4+ T cells to Foxp3+ cells by TGF-β is inhibited in Foxo deficient T cells. In fact, the response of Foxo-deficient T cells to TGF-β is misdirected and instead of becoming Foxp3+ cells, treated T cells secrete IFN-γ [38]. In non-lymphoid cells a direct interaction between Foxo proteins and TGF-β signaling components in the induction of TGF-β target gene expression was already demonstrated [39, 40], thus Foxo proteins could be a factor in the integration of signals mediated from both the TCR (via PI3-K) and TGF-β receptor. Another factor downstream of TGF-β and involved in Foxp3 induction is TGF-β inducible early gene 1 product (TIEG1), a transcription factor rapidly induced upon TGF-β stimulation. Ubiquitination of TIEG1 by the E3 ubiquitin ligase Itch promotes transcriptional activation of the foxp3 promoter and foxp3 expression [41]. Finally, RUNX3, which was shown to be induced by TGF-β signaling in CD4+ T cells, has been reported to positively regulate Foxp3 expression [42, 43]. RUNX family proteins form complexes with CBF-β which directly binds to the DNA [44]. In humans, RUNX1 and 3 are important for Foxp3 gene expression [42]. It has been reported that the proximal Foxp3 promoter region has three putative RUNX binding site (−53, −287 and −333 from TSS). RUNX-CBFβ enrichment at the Foxp3 promoter activates Foxp3 gene expression. In addition, RUNX1 can complex with Foxp3 and might be responsible for the suppression of IL-2 and IFN-γ production as well as the up-regulation of Treg associated molecules in response to TGF-β signaling [26].
Shown to synergise with TGF-β for the generation of Foxp3+ cells in the gut, Retanoic acid (RA), a metabolic product of Vitamin A, promotes Foxp3 gene expression. RA has been shown to augment phosphorylation of Smad3 allowing translocation of Smad3 to the nucleus [45], although this was debated [46]. It has been reported that RA, functions to maintain homeostasis within the gut through the generation of Foxp3+ Tregs [46, 47]. Moreover, RA controls Foxp3 expression directly through Retinoic acid Receptor (RAR) and Retinoid X Receptor (RXR), which bind the CNS1 region directly and positively regulate Foxp3 expression [48, 49]. Although these reports demonstrate a role for a number of transcription factors in TGF-β-mediated Foxp3 gene transcription, a systematic analysis of their interaction and regulators is needed to identify their precise effect on and at the Foxp3 promoter and work to place these transcription factors within a network which mediates Foxp3 induction.
There are also other cytokines and signaling stimuli that have been reported to affect Foxp3 and, ultimately, Treg development. Downstream of the TCR signal, Activator Protein 1 (AP-1) and Nuclear Factor of Activated T cells (NF-ATc2) are positive regulators of Foxp3 gene expression [11]. Indeed, at least three NF-AT binding sites have been observed in the proximal Foxp3 promoter region, around 500bp upstream from the TSS which positively regulate Foxp3 expression [11]. During Foxp3+ Treg development in the thymus, TGF-β has been shown to be necessary for the initial generation of Tregs but another cytokine, IL-2, is important for their expansion [50]. IL-2 is required for the activation of Foxp3 gene expression through Stat5, a downstream signaling molecule of the IL-2 receptor, which is able to bind onto the Foxp3 promoter and upregulate foxp3 expression [51, 52].
A chemical component has also been found to regulate Foxp3 expression; 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) can activate the Aryl hydrocarbon receptor (AHR) in T cells and induce Foxp3+ Tregs. So far three non-evolutionarily conserved AHR-binding (NCAB) sites in the Foxp3 promoter region and one conserved AHR-binding (CAB) site in exon 11 have been identified. AHR is enriched on the CAB and NCAB sites in the Foxp3 promoter in response to TCDD, and serves to positively regulate foxp3 gene expression [53]. In addition, AHR-null T cells have less Foxp3+ Treg induction in response to TGF-β and, TGF-β induces AHR gene expression [54].
Repressors of Foxp3 gene transcription
Many cytokines and factors negatively regulate Foxp3 gene transcription. Production of IL-4 following TCR stimulation up-regulates GATA-3 expression (the master Th2 transcription factor), which can bind to the Foxp3 promoter region and suppress gene expression [55, 56]. TGF-β signaling prevents IL-4 production, thereby inhibiting Gata3 expression and consequent binding to the Foxp3 promoter. In addition, the downstream IL-4 signaling protein Stat6 can bind to the Foxp3 gene locus (around 2.2 kb downstream from the TSS) and directly prevent Foxp3 gene activation [57]. Combined, these events negatively regulate Foxp3 expression and, as such, must be inhibited to allow for proper Foxp3 gene transcription.
As well as IL-4, the Th1 cytokine IFN-γ activates Stat1 that also prevents Foxp3 gene expression in murine T cells [58]. However, in human T cells, IFN-γ and IL-27 induced Stat1 can actually amplify TGF-β-mediated Foxp3 gene expression. In addition, the human Foxp3 gene has a Stat1 binding element within proximal region of the promoter [59]. Thus, more thorough studies are required to clarify these results and define the effects of Th1-cytokines on Foxp3 gene transcription.
The inflammatory cytokine IL-6 is well known to inhibit Foxp3 expression by promoting acquisition of the Th17 cell fate, in naïve T cells [60]. Downstream of IL-6 signaling, the Stat3 transcription factor can bind directly onto the IL-17 promoter region and induce IL-17 expression. However, Stat3 has the opposite effect on Foxp3 expression. By binding onto the CNS2 region Stat3 inhibits gene transcription [61]. In fact, the balance of Stat3 and, the IL-2 activated, Stat5 has been shown to influence fate decisions of naïve T cells, in determining whether they differentiate into Tregs or Th17 cells [62]. Interestingly however, Stat3 expression in Tregs is reported to play a crucial role in keeping the regulatory function of Tregs in vivo [63], which seems to be attributable largely to Treg-derived IL-10 [64].
RORγt, is the key transcription factor of Th17 cells and is induced by TGF-β and IL-6 signaling [65]. Both Foxp3 and RORγt are induced by TGF-β, but which one dominates following TGF-β signaling depends on the presence of other cytokines (and their activated Stat proteins as indicated above), the strength of TCR engagement and the local concentration of TGF-β. Once Foxp3 is expressed in response to TGF-β, Foxp3 can complex with the RORγt protein and prevent RORγt transcriptional activity [66]. On the other hand, combined TGF-β and IL-6 signaling strongly induces RORγt which can bind to the Foxp3 promoter directly and prevent Foxp3 expression [67]. In addition, low-dose exposure of TGF-β induces maximal expression of IL-23R and, together with inflammatory cytokines IL-6/IL-21/IL-23, inhibits Foxp3 expression and polarizes the naïve cell towards Th17 cell differentiation. High dose treatment of TGF-β inhibits expression of IL-23R, IL-22 and IL-17, thereby promoting Foxp3+ Treg differentiation [66]. Additionally, RA enhances TGF-β signaling and inhibits the expression of IL-6Rα IL-23R and IRF-4, serving to prevent Th17 cell generation [68].
Recently, a new population has been identified which develops in response to TGF-β and IL-4, IL-9 producing T cells (Th9). Th9 express PU.1, but not Foxp3, RORγt, T-bet or GATA-3 [69–71]. PU.1 is a transcription factor of the Est-family, and is able to bind the IL-9 promoter region and positively regulates Il-9 expression in response to TGF-β and IL-4. Importantly, PU.1 inhibits Foxp3+ Treg differentiation despite TGF-β signaling [72]. In addition, IRF-4 is also suggested to be important, not only for Th17, but also Th9 cell differentiation [73–75]. IRF-4 directly binds onto the IL-17A and IL-9 promoters, and upregulates IL-17 and IL-9 production. As such, IRF-4 deficient T cells fail to generate Th17 or Th9 cells in response to TGF-β and IL-6 or IL-4 co-signaling. Furthermore, IRF-4 KO T cells can be induced to Foxp3+ Tregs in response to TGF-β, and the combination of TGF-β and IL-6 signaling does not decrease Foxp3+ Treg induction. Interestingly, however, IRF-4KO Tregs lack suppressive functionality [76]. Although these studies expand the network of transcription factors that regulate the induction of Foxp3 and how this is balanced with the differentiation to other lineages, detailed cross-talk and interactions between the various lineage transcription factors, remains to be elucidated.
Away from cytokine-mediated effects, S1P1-signaling induces mTOR phosphorylation, and prevents the phosphorylation of Smad3. Thus S1P1-mediated phosphorylation of mTOR, serves to prevent TGF-β induced Foxp3 expression [77, 78]. In contrast, Rapamycin has been reported to dephosphorylate mTOR; and treatment with both Rapamycine and TGF-β enhances phosphorylation of Smad3 and, consequently, Foxp3 gene expression. Another inhibitory transcriptional factor for Foxp3 expression, Hes1, can be induced by Notch signaling. Hes1 can bind on the Foxp3 promoter region (3bp up-stream from TSS, and 66bp/128bp down stream from TSS) and negatively regulate Foxp3 gene expression [79]. Interestingly, Hes proteins have been shown to be suppressors of bHLH molecules, inhibiting many bHLH family transcriptional factors including E47 (E47 and E12 are E2A encoded proteins) [80]. It will be interesting to determine whether Hes mediated suppression of Foxp3 gene activation is mediated by down-regulation of E2A binding at the Foxp3 promoter.
So, TGF-β-mediated Foxp3 gene expression requires not only positive regulatory factors working in cooperation, but also requires the repression of negative transcriptional activity to properly coordinate Foxp3 protein production.
Concluding remarks
In this paper, we have discussed the transcription factor network at play in the controlled expression of Foxp3 and the generation of regulatory T cells. We have emphasized a mutual regulation between the “enhancer” and the “silencer”, i.e. the activation of positive transcription factors and the suppression of negative transcription factors which collectively allow for the coordinated expression of Foxp3 and the generation of regulatory T cells. This balance of transcription factors mediates the subtle changes required for TGF-β -signaling to not only mediate the generation of Tregs, but may well also effect differentiation of Th17 and Th9 cells.
Foxp3 gene conserved regions
TGF-γ and Foxp3 gene expression
Repressors of Foxp3 gene transcription
Figure 1. A map of Foxp3 gene regulation.
Foxp3 is located on chromosome X and it has been reported that several conserved regions exist on this gene (Promoter, CNS1, CNS2 or CNS3). So far, epigenetic studies have revealed the role of each of these conserved regions. The promoter region is necessary for directly controlling gene expression. CNS1 serves as a TGF-β sensor, so called enhancer regions. CNS2 is important for stabilizing foxp3 expression. Transcription factors can bind on each of the conserved regions and are either Positive (Black) or Negative (Red) regulators for Foxp3 protein expression.
Acknowledgement
The authors thank all members of the Chen group. We apologize for those whose work could not be cited because of the space limitation. The research in authors’ laboratory is supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research of the National Institutes of Health. T.M. was supported in part by a JSPS Research Fellowship for Japanese Biomedical and Behavioral Researchers at NIH.
Abbreviations
- TGF-β
transforming growth factor beta
- Foxp3
forkhead box P3
- TCR
T-cell receptor
- Id3
inhibitor of DNA binding protein 3
- E2A
basic Helix-Loop-Helix protein E2A
- GATA3
Trans-acting T-cell-specific transcription factor GATA-3
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, et al. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 2.Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, et al. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- 3.Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- 4.Liston A, Nutsch KM, Farr AG, Lund JM, Rasmussen JP, Koni PA, et al. Differentiation of regulatory Foxp3+ T cells in the thymic cortex. Proc Natl Acad Sci U S A. 2008;105:11903–11908. doi: 10.1073/pnas.0801506105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HM, Hsieh CS. Rare development of Foxp3+ thymocytes in the CD4+CD8+ subset. J Immunol. 2009;183:2261–2266. doi: 10.4049/jimmunol.0901304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, et al. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 8.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 9.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 11.Mantel PY, Ouaked N, Rückert B, Karagiannidis C, Welz R, Blaser K, et al. Molecular mechanisms underlying FOXP3 induction in human T cells. J Immunol. 2006;176:3593–3602. doi: 10.4049/jimmunol.176.6.3593. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Review Immunol. 2009;9:83–89. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 14.Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 15.Soligo M, Camperio C, Caristi S, Scottà C, Del Porto P, Costanzo A, et al. CD28 costimulation regulates FOXP3 in a RelA/NF-κ B-dependent mechanism. Eur J Immunol. 2011;41:503–513. doi: 10.1002/eji.201040712. [DOI] [PubMed] [Google Scholar]
- 16.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 17.Ruan Q, Kameswaran V, Tone Y, Li L, Liou HC, Greene MI, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baron U, Floess S, Wieczorek G, Baumann K, Grützkau A, Dong J, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37:2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 21.Polansky JK, Schreiber L, Thelemann C, Ludwig L, Krüger M, Baumgrass R, et al. Methylation matters: binding of Ets-1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med. 2010;88:1029–1040. doi: 10.1007/s00109-010-0642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouly E, Chemin K, Nguyen HV, Chopin M, Mesnard L, Leite-de-Moraes M, et al. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lal G, Zhang N, van der Touw W, Ding Y, Ju W, Bottinger EP, et al. Epigenetic regulation of Foxp3 expression in regulatory T cells by DNA methylation. J Immunol. 2009;182:259–273. doi: 10.4049/jimmunol.182.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim YC, Kim KK, Shevach EM. Simvastatin induces Foxp3+ T regulatory cells by modulation of transforming growth factor-beta signal transduction. Immunology. 2010;130:484–493. doi: 10.1111/j.1365-2567.2010.03269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- 27.Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 28.Derynck R, Zhang Y, Feng XH. Smads: transcriptional activators of TGF-beta responses. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 29.Takimoto T, Wakabayashi Y, Sekiya T, Inoue N, Morita R, Ichiyama K, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185:842–845. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 30.Yang XO, Nurieva R, Martinez GJ, Kang HS, Chung Y, Pappu BP, et al. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maruyama T, Li J, Vaque JP, Konkel JE, Wang W, Zhang B, et al. Control of the differentiation of regulatory T cells and T(H)17 cells by the DNA-binding inhibitor Id3. Nat Immunol. 2011;12:86–95. doi: 10.1038/ni.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 33.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 34.Li H, Dai M, Zhuang Y. A T cell intrinsic role of Id3 in a mouse model for primary Sjogren's syndrome. Immunity. 2004;21:551–560. doi: 10.1016/j.immuni.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 35.Guo Z, Li H, Han M, Xu T, Wu X, Zhuang Y. Modeling Sjögren's syndrome with Id3 conditional knockout mice. Immunol Lett. 2011;135:34–42. doi: 10.1016/j.imlet.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harada Y, Harada Y, Elly C, Ying G, Paik JH, DePinho RA, et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207:1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ouyang W, Beckett O, Ma Q, Paik JH, DePinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11:618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 38.Kerdiles YM, Stone EL, Beisner DL, McGargill MA, Ch'en IL, Stockmann C, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seoane J, Le HV, Shen L, Anderson SA, Massagué J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 40.Gomis RR, Alarcón C, He W, Wang Q, Seoane J, Lash A, et al. A FoxOSmad synexpression group in human keratinocytes. Proc. Natl. Acad. Sci. USA. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venuprasad K, Huang H, Harada Y, Elly C, Subramaniam M, Spelsberg T, et al. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nat Immunol. 2008;9:245–253. doi: 10.1038/niXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, et al. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J Exp Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, et al. Runx proteins regulate Foxp3 expression. J Exp Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nolting J, Daniel C, Reuter S, Stuelten C, Li P, Sucov H, et al. Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J Exp Med. 2009;206:2131–2139. doi: 10.1084/jem.20090639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:177–185. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elias KM, Laurence A, Davidson TS, Stephens G, Kanno Y, Shevach EM, et al. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–1020. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu L, Kitani A, Stuelten C, McGrady G, Fuss I, Strober W. Positive and negative transcriptional regulation of the Foxp3 gene is mediated by access and binding of the Smad3 protein to enhancer I. Immunity. 2010;33:313–325. doi: 10.1016/j.immuni.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. A critical function for TGF-beta signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol. 2008;9:632–640. doi: 10.1038/ni.1607. [DOI] [PubMed] [Google Scholar]
- 51.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. doi: 10.4049/jimmunol.178.1.280. [DOI] [PubMed] [Google Scholar]
- 53.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 54.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185:3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mantel PY, Kuipers H, Boyman O, Rhyner C, Ouaked N, Rückert B, et al. GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLos Biol. 2007;5:e329. doi: 10.1371/journal.pbio.0050329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hadjur S, Bruno L, Hertweck A, Cobb BS, Taylor B, Fisher AG, et al. IL4 blockade of inducible regulatory T cell differentiation: the role of Th2 cells, Gata3 and PU.1. Immunol Lett. 2009;122:37–43. doi: 10.1016/j.imlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Takaki H, Ichiyama K, Koga K, Chinen T, Takaesu G, Sugiyama Y, et al. STAT6 Inhibits TGF-beta1-mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem. 2008;283:14955–14962. doi: 10.1074/jbc.M801123200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caretto D, Katzman SD, Villarino AV, Gallo E, Abbas AK. Cutting edge: the Th1 response inhibits the generation of peripheral regulatory T cells. J Immunol. 2010;184:30–34. doi: 10.4049/jimmunol.0903412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ouaked N, Mantel PY, Bassin C, Burgler S, Siegmund K, Akdis CA, et al. Regulation of the foxp3 gene by the Th1 cytokines: the role of IL-27-induced STAT1. J Immunol. 2009;182:1041–1049. doi: 10.4049/jimmunol.182.2.1041. [DOI] [PubMed] [Google Scholar]
- 60.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 61.Zorn E, Nelson EA, Mohseni M, Porcheray F, Kim H, Litsa D, et al. IL-2 regulates FOXP3 expression in human CD4+CD25+ regulatory T cells through a STAT-dependent mechanism and induces the expansion of these cells in vivo. Blood. 2006;108:1571–1579. doi: 10.1182/blood-2006-02-004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich JM, et al. Interleukin-10 signaling in regulatory T cells is required for suppression of th17 cell-mediated inflammation. Immunity. 2011;34:566–578. doi: 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burgler S, Mantel PY, Bassin C, Ouaked N, Akdis CA, Schmidt-Weber CB. RORC2 is involved in T cell polarization through interaction with the FOXP3 promoter. J Immunol. 2010;184:6161–6169. doi: 10.4049/jimmunol.0903243. [DOI] [PubMed] [Google Scholar]
- 68.Xiao S, Jin H, Korn T, Liu SM, Oukka M, Lim B, et al. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-beta-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181:277–284. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, et al. Transforming growth factor-beta 'reprograms' the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 70.Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hadjur S, Bruno L, Hertweck A, Cobb BS, Taylor B, Fisher AG, et al. IL4 blockade of inducible regulatory T cell differentiation: the role of Th2 cells, Gata3 and PU.1. Immunol Lett. 2009;122:37–42. doi: 10.1016/j.imlet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 73.Brüstle A, Heink S, Huber M, Rosenplänter C, Stadelmann C, Yu P, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nature. 2009;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 74.Mudter J, Yu J, Zufferey C, Brüstle A, Wirtz S, Weigmann B, et al. IRF4 regulates IL-17A promoter activity and controls ROR γ t-dependent Th17 colitis in vivo. Inflam Bowel Dis. 2011 doi: 10.1002/ibd.21476. [DOI] [PubMed] [Google Scholar]
- 75.Staudt V, Bothur E, Klein M, Lingnau K, Reuter S, Grebe N, et al. Interferon-regulatory factor 4 is essential for the developmental program of T helper 9 cells. Immunity. 2010;33:192–202. doi: 10.1016/j.immuni.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 76.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu G, Burns S, Huang G, Boyd K, Proia RL, Flavell RA, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10:769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11:1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ou-Yang HF, Zhang HW, Wu CG, Zhang P, Zhang J, Li JC, et al. Notch signaling regulates the FOXP3 promoter through RBP-J- and Hes1-dependent mechanisms. Mol Cell Biochem. 2009;320:109–114. doi: 10.1007/s11010-008-9912-4. [DOI] [PubMed] [Google Scholar]
- 80.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]